Background: A gene encoding a distant homolog of pterin-4α-carbinolamine dehydratase (PCD) is present in all α-type carboxysome operons.
Results: This conserved PCD-like protein promotes the assembly of native RuBisCO when overexpressed together with GroELS.
Conclusion: The protein, named here acRAF for α-carboxysome RuBisCO assembly factor, is a newly identified molecular chaperone.
Significance: The activity of this chaperone toward RuBisCO has potential agricultural application.
Keywords: Carbon Dioxide, Molecular Chaperone, Protein Assembly, Protein Evolution, Protein Folding, RuBisCO, Carbon Fixation, Carboxysome, Pseudo-enzyme, Pterin Dehydratase
Abstract
Carboxysomes are proteinaceous bacterial microcompartments that increase the efficiency of the rate-limiting step in carbon fixation by sequestering reaction substrates. Typically, α-carboxysomes are genetically encoded as a single operon expressing the structural proteins and the encapsulated enzymes of the microcompartment. In addition, depending on phylogeny, as many as 13 other genes are found to co-occur near or within α-carboxysome operons. One of these genes codes for a protein with distant homology to pterin-4α-carbinolamine dehydratase (PCD) enzymes. It is present in all α-carboxysome containing bacteria and has homologs in algae and higher plants. Canonical PCDs play an important role in amino acid hydroxylation, a reaction not associated with carbon fixation. We determined the crystal structure of an α-carboxysome PCD-like protein from the chemoautotrophic bacterium Thiomonas intermedia K12, at 1.3-Å resolution. The protein retains a three-dimensional fold similar to canonical PCDs, although the prominent active site cleft present in PCD enzymes is disrupted in the α-carboxysome PCD-like protein. Using a cell-based complementation assay, we tested the PCD-like proteins from T. intermedia and two additional bacteria, and found no evidence for PCD enzymatic activity. However, we discovered that heterologous co-expression of the PCD-like protein from Halothiobacillus neapolitanus with RuBisCO and GroELS in Escherichia coli increased the amount of soluble, assembled RuBisCO recovered from cell lysates compared with co-expression of RuBisCO with GroELS alone. We conclude that this conserved PCD-like protein, renamed here α-carboxysome RuBisCO assembly factor (or acRAF), is a novel RuBisCO chaperone integral to α-carboxysome function.
Introduction
Ribulose-bisphosphate carboxylase/oxygenase (RuBisCO)4 catalyzes the rate-limiting reaction of carbon fixation: the covalent attachment of carbon dioxide to the sugar ribulose 1,5-bisphosphate (1). Despite its central role in metabolism, RuBisCO is a notoriously inefficient enzyme. Its turnover rate varies between species but is in the range of seconds (2, 3). The enzyme also shows relatively poor discrimination between its substrate, carbon dioxide, and molecular oxygen (4, 5). Higher plants have evolved various mechanisms to overcome these inherent catalytic inefficiencies, including strategies that rely on compartmentalization of the steps of carbon fixation across multiple cell types. Many bacteria, on the other hand, rely on a giant proteinaceous structure known as the carboxysome to sequester the carbon fixation reactions of the cell. By encapsulating RuBisCO and carbonic anhydrase together, concentrated CO2 can be provided in the vicinity of RuBisCO (6–10). In this way, the carboxysome serves as an essential carbon fixing organelle in many photosynthetic and some chemoautotrophic bacteria.
The major protein constituents of the carboxysome have been well characterized. Bacterial microcompartment (BMC) shell proteins assemble into hexagonal lattices that form the walls of the carboxysome shell (8, 9). Pentameric bacterial microcompartment vertex proteins enable full closure of these icosahedral structures by occupying the spaces at each of the 12 vertices (9, 11–13). A variety of paralogous BMC proteins are typically encoded in carboxysome operons; they are believed to play distinct roles in the shell (9). Carboxysomes are phylogenetically classified as either α- or β-type (14, 15). The two divergent carboxysome lineages are distinguished by differences in RuBisCO phylogeny (RuBisCO type IA and type IB are found in α- and β-carboxysomes, respectively), gene composition, and operon organization. α-Carboxysome genes generally appear within a single operon, whereas β-carboxysome genes typically appear in multiple dispersed gene clusters. Functional differences resulting from this divergence, if any, are not currently understood.
Numerous proteins are implicated in the complex process of RuBisCO folding and activation in diverse organisms. In some instances known RuBisCO chaperones are encoded near α-carboxysome operons or β-carboxysome RuBisCO genes. RbcX, a small (∼15 kDa) dimeric protein encoded between RuBisCO large and small subunits of β-carboxysomes, has been shown to facilitate hexadecameric assembly of RuBisCO in the β-carboxysome system (16–18). RbcX homologs are not present in α-carboxysome containing organisms, suggesting that either the subunit assembly function performed by RbcX is unnecessary for carboxysome type 1A RuBisCO or that a distinct, unknown mechanism promotes hexadecameric assembly of α-carboxysome RuBisCO. CbbX, a RuBisCO activase, modulates RuBisCO activity by dislodging inhibitory ribulose 1,5-bisphosphate molecules from the active site (19). Other ATP-dependent chaperones CbbO and CbbQ regulate and enhance RuBisCO catalysis by altering RuBisCO conformational states in a variety of carbon-fixing prokaryotes (20). CbbO, CbbQ, and CbbX homologs are sometimes present in α-carboxysome operons (Fig. 1). GroELS (also referred to as chaperonin) is a ubiquitous ATP-dependent chaperone complex essential for the proper folding of RuBisCO large subunits (21, 22). RuBisCO assembly factor 1 was recently identified as a novel RuBisCO chaperone in Zea mays (23). RuBisCO assembly factor 1 is conserved in green plant lineages, but is not present in carbon-fixing prokaryotes. To date, plant RuBisCO has not been successfully assembled in vitro or in a heterologous Escherichia coli host, hinting at the probable existence of yet unidentified folding or assembly factors.
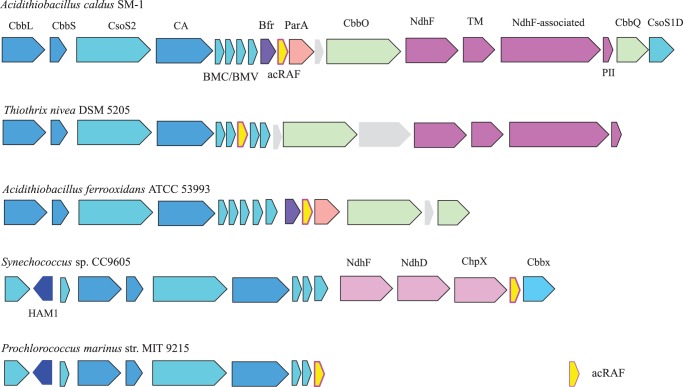
FIGURE 1.
acRAF is a defining member of α-carboxysome operons. Carboxysome operons of A. caldus SM-1, T. nivea DSM 5205, A. ferrooxidans ATCC 53993, Synechococcus sp. CC9605, and P. marinus str. MIT 9215 operons are shown to demonstrate the extent of acRAF association with α-carboxysome operons. acRAFs are present within all α-carboxysome operons examined. CbbL, RuBisCO large subunit; CbbS, RuBisCO small subunit; CsoS2, shell protein of unknown function; CA, carbonic anhydrase; BMV, bacterial microcompartment vertex shell protein; Bfr, bacterioferritin family; acRAF, α-carboxysome RuBisCO assembly factor; ParA, partitioning A family (example YP_003262802); CbbO, CbbQ activase; NdhF (NuoL/Nqo12-like), complex I NADH oxidoreductase chain F family protein (example YP_003262799); TM, transmembrane protein (example YP_003262798); NdhF-associated, ORF commonly encoded adjacent to or near NdhF (example YP_003262797); PII, PII nitrogen regulatory family (example YP_003262796); CbbQ, ATP-dependent RuBisCO activase; CsoS1D, double domain BMC shell protein; HAM1, Ham1 domain containing; NdhF, complex I NADH dehydrogenase oxidoreductase subunit F family; NdhD, complex I NADH dehydrogenase oxidoreductase M family; ChpX, CO2 hydration protein; CbbX, RuBisCO activase.
Recent bioinformatics studies have identified several genes of unknown function that consistently occur near α-carboxysome BMC genes (24, 25). This conserved genetic association strongly suggests that the encoded proteins are involved in the catalytic function, regulation, or structure of the α-carboxysome. One of these proteins is remotely related in sequence (often with BLAST E values ≥ 0.05) to pterin-4α-carbinolamine dehydratase (PCD) enzymes, and is represented in all α-carboxysome operons we examined. It appears to be a defining member of this carboxysome subtype. Fig. 1 illustrates the organization of a number of diverse α-carboxysome operons and highlights the presence of this remote homolog of PCD.
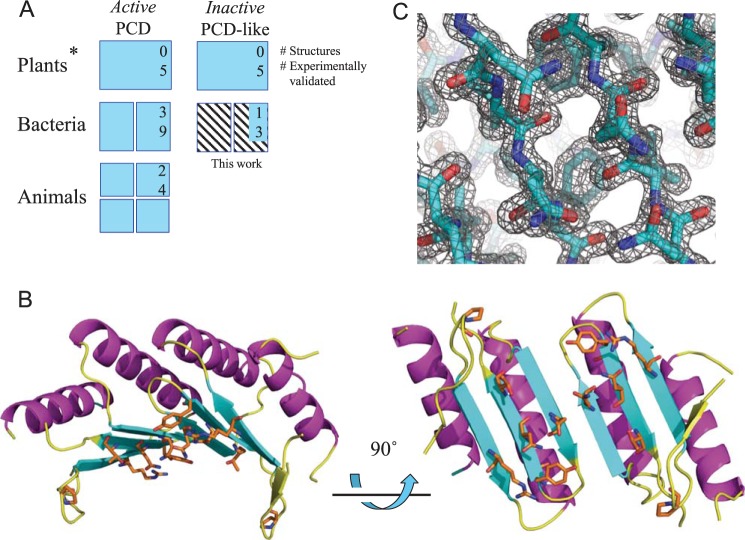
PCD enzymes are ubiquitous throughout the three kingdoms of life, where they act in concert with aromatic amino acid hydroxylases (AAH) in amino acid metabolism pathways (26, 27). AAHs use tetrahydrobiopterin (also known as BH4 and THB) cofactors and molecular oxygen to catalyze the addition of a hydroxyl group onto aromatic amino acids. A specific example of an AAH is bacterial phenylalanine hydroxylase, which adds a hydroxyl group to phenylalanine to produce tyrosine (28). In this reaction, the tetrahydrobiopterin cofactor is simultaneously hydroxylated on the 4α carbon, generating petrin-4α-carbinolamine. Two enzymes are required to regenerate pterin-4α-carbinolamine to tetrahydrobiopterin: dihydropterin reductase and PCD (27). Functional PCDs exist as homodimers in prokaryotes. In animals, DCoH (dimerization cofactor of HNF-1 or hepatocyte nuclear factor-1) is a tetrameric bifunctional enzyme that serves both as a PCD and as a transcriptional activator of HNF-1 (29). Despite its dual role in animals, DCoHs are similar in sequence and tertiary structure to bacterial PCDs (30). In prokaryotes lacking AAHs, “orphan” PCDs are sometimes present and are suspected to participate in other unknown reactions or cellular processes (27). Catalytically inactive PCD-like proteins of unknown function are present both in chloroplasts (27) and, as shown here, in many bacteria, where they occur in close association with α-carboxysome operons. No structures have been reported to date for these PCD-like proteins. Fig. 2A summarizes current information on PCDs and PCD-like proteins.
FIGURE 2.
Crystal structure and conserved surface regions in the acRAF dimer. A, diagram summarizing current knowledge of PCDs and inactive PCD-like proteins. PCDs are capable of pterin-4α-carbinolamine dehydratase activity. PCD-like proteins, whereas structurally similar to PCDs, do not show evidence of pterin-4α-carbinolamine dehydratase activity. Oligomeric states are represented by number of subunits shown, except for plant PCD and PCD-like proteins (denoted by *), whose oligomeric states are unknown. Numbers in upper right hand corners represent the number of representative proteins with crystal structures deposited in the PDB. Numbers below indicate the number of representative proteins examined experimentally, to our knowledge. Information on the bacterial carboxysomal acRAFs (hatched) refers to the present study. B, solvent-exposed, conserved residues of the acRAF dimer are shown as orange sticks. C, σA-weighted 2mFo − DFc electron density map of the dimer interface viewed in the same orientation as the bottom right model in panel B.
Here we report our work investigating the structure and function of the conserved PCD-like family of proteins found in α-carboxysome containing bacteria. We show that the protein is an enzymatically inactive relic of PCD, and that its true cellular function relates to the folding or assembly of hexadecameric RuBisCO. Based on our results, we rename this protein the α-carboxysome RuBisCO assembly factor, or acRAF.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Tillandsia intermedia acRAF
The construct used for crystallization of codon-optimized acRAF (with amino acids “MHHHHHH” replacing the first 5 amino acids of the protein coding sequence) was ordered from Biomatik Corp. Gene Synthesis Co. Codon optimization was performed by DNAworks. The resulting gene sequence was cloned into pET22b(+) vector between the 5′ NdeI and 3′ EcoRI sites. acRAF was overexpressed in E. coli BL21(DE3). One-liter flasks were inoculated with 15 ml of overnight culture, and grown under ampicillin selection in LB media at 37 °C, shaking at 240 rpm, until cells reached an optical density absorbance of 0.8 at 600 nm (A600 = 0.8). Isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 1.0 mm. After 3–5 h, cells were spun down and frozen at −20 °C until further use. Frozen cell pellets collected from 2 liters of culture were resuspended in 50 ml of buffer (50 mm Tris-HCl, pH 7.6, 300 mm NaCl, Roche Complete EDTA-free protease inhibitor mixture) and lysed by sonication. Cell debris was pelleted by centrifugation at 16,500 × g in a SS-34 rotor for 30 min at 4 °C. The resulting supernatant was filtered through a 0.22-μm Whatman filter. Filtered supernatant was then loaded onto a HisTrap 5-ml column by syringe, washed with 30 ml of wash buffer (50 mm Tris-HCl, pH 7.6, 300 mm NaCl, 50 mm imidazole), and eluted in one step with elution buffer (50 mm Tris-HCl, pH 7.6, 300 mm NaCl, 400 mm imidazole). Purified acRAF was dialyzed into 10 mm Tris-HCl, pH 7.6, 50 mm NaCl, 5% glycerol. acRAF was concentrated to 11 mg/ml, as calculated from absorbance at 280 nm using the theoretical extinction coefficient.
acRAF Crystallization
Hanging-drop crystallization screens were performed using the nanoliter liquid handling Mosquito from TTP LabTech at the High Throughput Macromolecular Crystallization Facility at UCLA. Optimization of needle-like crystals obtained in JCSG+ Suite crystallization screen condition A8 were pursued in 24-well (500 μl well) trays. Crystals were collected from 2-μl hanging drops of 1:1 ratio protein:well solution after 16 days of slow but continuous growth at room temperature. The optimized well solution contained 17.5% PEG 3500 and 800 mm ammonium formate. A cryoprotectant was prepared containing 17.5% PEG 3500, 800 mm ammonium formate, and 25% ethylene glycol, with or without 200 mm NH4I2 for subsequent phasing by anomalous scattering. Crystals were soaked in cyroprotectant for 1–2 min before freezing and storage in liquid nitrogen.
Phasing and Refinement
Diffraction data extending to 1.3-Å resolution were collected at the Argonne National Laboratory, Advanced Photon Source (APS), beamline 24-ID-C (Table 1). Phases were obtained from single wavelength data using isomorphous replacement with anomalous scattering from iodide. Five iodide-bound sites were identified by anomalous scattering, and all five sites were used for phasing. Iodide sites were located and phases were calculated using the program HKL2MAP (31). The program Buccaneer (32) was used to automatically trace the main chain. Subsequent rounds of model building were performed with COOT (33) and the structure was refined using BUSTER (34) to a final Rwork and Rfree of 0.203 and 0.204, respectively. 98% of the backbone dihedral angles are within the favored regions of a Ramachandran diagram. Coordinates and structure factors have been deposited with the PDB ID code 4LOW.
TABLE 1.
acRAF X-ray data collection and model refinement statistics
Statistics for the highest resolution shell are shown in parentheses.
| Statistics | Value |
|---|---|
| Wavelength (Å) | 0.9791 |
| Resolution range (Å) | 51.1 − 1.30 (1.34–1.30) |
| Space group | P41 |
| Unit cell (Å,°) | a = b = 51.1, c = 60.9; α = β = γ = 90 |
| Total reflections recorded | 253,115 (169,41) |
| Unique reflections | 37,451 (2610) |
| Multiplicity | 6.7 (6.5) |
| Completeness (%) | 96.9 (92.6) |
| Mean I/σ(I) | 17.2 (2.05) |
| Wilson B-factor (Å2) | 23.1 |
| CC(1/2) | 99.9% (69.4%) |
| Model Rwork | 0.203 (0.211) |
| Model Rfree | 0.204 (0.241) |
| Number of chains | 2 |
| Number of modeled residues (chain A/chain B) | 83/78 |
| Number of non-hydrogen atoms | 1452 |
| Macromolecules | 1305 |
| Formate | 18 |
| Water | 127 |
| Ni2+a | 2 |
| Protein residues | 336 |
| Geometric deviations (rms) | |
| bonds (Å) | 0.001 |
| angles (°) | 1.03 |
| Ramachandran statistics (%) | |
| Favored | 97.8 |
| Additionally allowed | 2.2 |
| Generously allowed | 0 |
| Disallowed | 0 |
| Average B-factor (Å2) | |
| Protein main chain | 29.5 |
| Protein side chains | 33.6 |
| Solvent (water) | 39.1 |
a The divalent metal ion was defined as Ni2+ during the refinement process based on its location between histidine tags, combined with the plausibility of contamination from nickel-bound chromatography columns.
PCD Complementation Assay
All PCD constructs were cloned between the 5′ NdeI and 3′ KpnI restriction enzyme sites in a pBSKII(−) vector. The PCD construct from Pseudomonas aeruginosa (NP_249562) in the vector pBSKII(−) and P. aeruginosa phenylalanine hydroxylase (NP_249563) in vector pACYC177 were gifts from Dr. Andrew Hanson. A cDNA template of the Arabidopsis thaliana chloroplast acRAF gene (NP_199924) was provided by Dr. Sabeeha Merchant. Codon optimized T. intermedia K12 acRAF (YP_003641868) DNA was purchased through Biomatik Corp. The codon-optimized T. intermedia K12 PCD (YP_003642661) gene was synthesized by assembly PCR from oligonucleotides designed by DNAworks and purchased from IDT. Codon-optimized acRAF genes from Halothiobacillus neapolitanus (YP_003262803) and Prochlorococcus marinus (NP_874951) were purchased as IDT gBlocks. All PCDs and acRAFs were transferred into clonal descendants of Hanson's original P. aeruginosa PCD pBSKII(−) construct using Gibson assembly cloning methods, thereby replacing the original insert. Codon optimized T. intermedia K12 phenylalanine hydroxylase (YP_003642727) (purchased from Biomatik) was transferred into that PCR-linearized pACYC177 using Gibson assembly (35). The starting ATG of the pACYC177 ampicillin resistance open reading frame became the beginning ATG of T. intermedia K12 phenylalanine hydroxylase, and largely replaced the ampicillin resistance gene. A pBSKII(−) empty vector control and pACYC177 empty vector controls were selected from failed cloning experiments. All constructs were sequenced by Laragen, Co. Sequences are available upon request.
Cloning of RuBisCO and acRAF
Several experiments were performed to determine the effects of acRAF and/or GroELS co-expression on the solubility and assembly of RuBisCO subunits. Ten different E. coli BL21(DE3) strains were created by triple or double electroporation into Lucigen Crop. “E. cloni EXPRESS BL21(DE3) Electrocompetent cells.” CbbLS, CbbL, and CbbS were PCR amplified from H. neapolitanus genomic DNA and cloned into pRSF-duet vector between 5′ NcoI and 3′ XhoI restriction enzyme sites using Gibson assembly. acRAF, also PCR-amplified from H. neapolitanus genomic DNA, was cloned into pET22b(+) between 5′ NdeI and 3′ EcoRI restriction enzyme sites. Plasmid pBB541, a GroELS expressing vector with a p15a origin of replication, was purchased from Addgene. Empty pRSF and pET22b(+) plasmids were used as negative controls where appropriate. Exact sequences are available upon request.
Heterologous RuBisCO and acRAF Co-expression Assay
Transformed BL21(DE3) cells were grown overnight in LB broth supplemented with the following antibiotics as appropriate: 15 μm kanamycin, 50 μm ampicillin, 12.5 μm streptomycin. 2 ml of overnight cultures were added to 250-ml flasks containing 100 ml of LB. Cells were grown at 37 °C until the A600 reached 0.8. Cells were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, shaking at 240 rpm, for 3 h at 37 °C. Cells were spun down and resuspended in the following buffer: 50 mm Tris-HCl, pH 7.6, 300 mm NaCl, 10 mm MgCl2, 10 mm NaHCO3, 10 mm DTT. Buffer volume was adjusted so that all cell cultures achieved an A600 of 54 (based on serial dilution). Cells were lysed by sonication. Insoluble cell debris was pelleted in 1.6-ml Eppendorf tubes spinning at 20,000 × g in a tabletop microcentrifuge. Supernatant was then loaded onto Bio-Rad MiniProtean TGX gels (catalog number 456-9036) and analyzed by SDS-PAGE (data not shown) and native gels (Fig. 4).
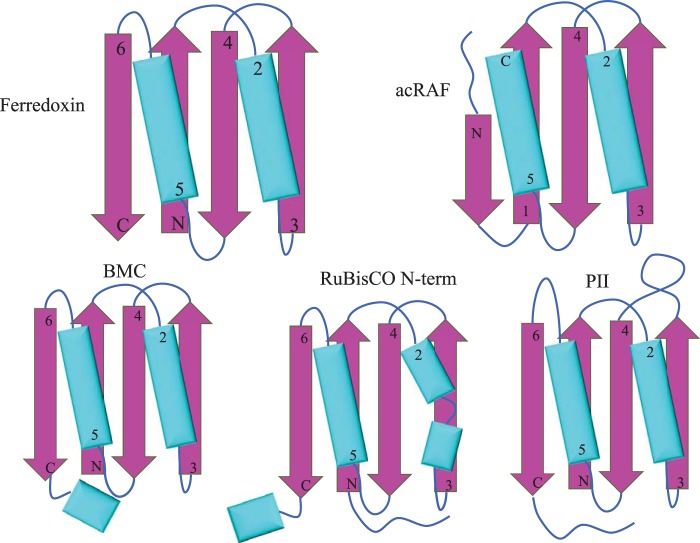
FIGURE 4.
Ferredoxin-like protein folds associated with carboxysomes. Simplified models of secondary and tertiary structural motifs of select carboxysome-associated proteins. α-Helices are colored cyan and β sheets are colored magenta. Top left, ferredoxin. Top right, PCD/acRAF. Bottom left, BMC (bacterial microcompartments shell proteins). Bottom center, N-terminal domain of the RuBisCO large subunit. Bottom right, nitrogen regulatory PII family protein (encoded near several chemoautotrophic α-carboxysome operons).
Associated Gene Analysis
α-Carboxysome operons were detected by searching the non-redundant protein sequence database for sequences homologous to the α-carboxysome protein CsoS2, using the program Blast. Twenty-one α-carboxysome operons were selected with the aim of maximizing diversity of both genomic backgrounds and α-carboxysome components. Operons from the following organisms were analyzed by hand: Nitrobacter winogradskyi NB-255, T. intermedia K12, Nitrosomonas eutropha C91, Thiobacillus denitrificans ATCC 25259, Nitrococcus mobilis NB-231, H. neapolitanus, Thioalkalivibrio sulfidophilus HL-EbGr7, Thiothrix nivea DSM 5205, Acidithiobacillus caldus SM-1, Acidithiobacillus thiooxidans ATCC 19377, Acidithiobacillus ferrooxidans ATCC 53993, Synechococcus sp. CC9311, Synechococcus sp. CC9902, Synechococcus sp. CC9605, Cyanobium sp. PCC 7001, P. marinus MED4, P. marinus str. MIT 9215, P. marinus str. MIT 9202, P. marinus str. NATL2A, and P. marinus str. MIT 9211.
To develop a better picture of the various uncharacterized proteins that appear to be associated with α-carboxysomes, we have used the comparative genomics program SEED to systematize the known α-carboxysome operons and their associated genes. Those results are publicly accessible (SEED curator: nwheatley; subset name: α-carboxysome).
Data Deposition
Atomic coordinates and diffraction data for acRAF have been deposited with PDB code 4LOW.
RESULTS
Structure of T. intermedia acRAF
We determined the structure of T. intermedia acRAF (GI: 296134626, YP_003641868) to 1.3-Å resolution by x-ray crystallography (Fig. 2, B and C). An initial acRAF construct with a C-terminal His6 tag did not crystallize despite many attempts. Secondary structure prediction programs indicated that the first six amino acids of native acRAF were unstructured. We therefore moved the polyhistidine tag to the N terminus. We chose to replace the presumptively disordered N terminus with the polyhistidine tag rather than extend the disordered region, and this construct crystallized. The structure was determined by anomalous scattering methods (Table 1). The asymmetric unit of the P41 crystal contained two protein subunits, or one dimer. A final refined model was obtained in which a total of 148 of the 164 residues of the combined subunits of the acRAF dimer could be reliably modeled (Fig. 2B). The electron density for residues 66–70, corresponding to the region of catalytic activity in PCD enzymes, was not interpretable, indicating either multiple conformations and/or high structural disorder in this region of acRAF. This segment was therefore not included in the final deposited structure, although the loop was included in a plausible conformation for the purposes of structure visualization in some cases. In the crystal structure, the histidine residues that were inserted at the N terminus emerge from an ordered β strand, implying that the deleted N-terminal residues of acRAF might have contributed to a β sheet, contrary to our predictions. Further analysis revealed that these ordered histidine residues are involved in a chelation-dependent crystal contact with a divalent cation, thereby creating a fortuitous interaction important for crystal formation.
Comparisons between acRAF and true PCD enzymes show significant structural similarities despite low sequence identity. Generally, about 75% of a typical acRAF sequence can be aligned to PCD. Within that region of similarity, 25–35% of residues are identically conserved. Structural superposition of acRAF onto Toxoplasma gondii PCD (PDB code 2V6T) resulted in a backbone root mean square deviation of only 1.5 Å (Fig. 3). The main difference between the PCD and acRAF structures is the N-terminal helix. The PCD enzyme from T. gondii contains an 11-residue N-terminal helix that is lacking in acRAF. The tertiary structures of PCD and acRAF are similar to the ferredoxin superfamily, but with a minor circular permutation near the chain termini. We note here the intriguing observation that a number of other carboxysome-related proteins also show a similar fold. This includes the main BMC shell proteins, the N-terminal domain of the RuBisCO large subunit, and a PII nitrogen regulatory-like protein encoded in several proteobacteria α-carboxysome operons (36) (Fig. 4). The oligomeric state of acRAF in the crystal structure is a dimer, which like true PCDs is composed of two subunits arranged as a symmetric dimer. The dimeric interface is formed by a β strand (residues 50–55) and an α helix (residues 28–43). The strand pairs with the cognate strand from the other subunit to form an anti-parallel β sheet.
FIGURE 3.
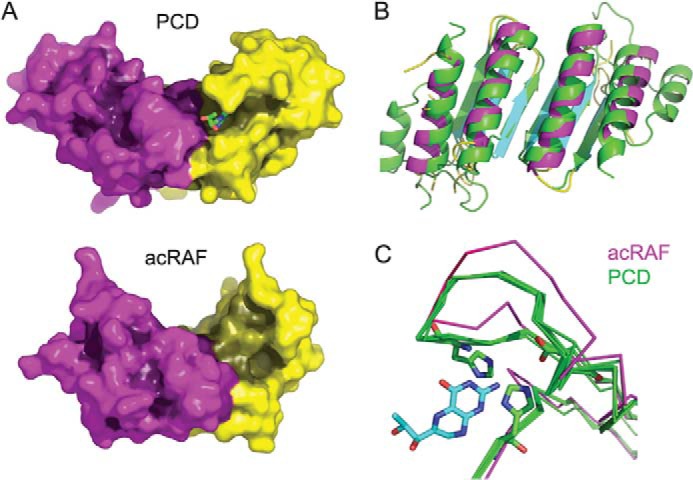
Structural comparison of acRAF and PCD. A, comparison of the surface shapes and binding clefts of a true PCD with acRAF from the α-carboxysome operon. A bound dihydrobiopterin is shown in the active site cleft of PCD (PDB code 2V6T). The two proteins are shown in the same orientation. The active site region of PCD is dramatically altered and much less pronounced in acRAF. Surfaces are shaded according to depth based on a calculation of diffusion accessibility (44). Three partially disordered residues of a loop were included in a plausible configuration to convey a more realistic view of the shape of acRAF. B, structural alignment of acRAF to PCD from T. gondii (PDB code 2V6T). T. gondii PCD is colored green. C, structural alignments of active sites among five PCDs shown in green (PDB codes 1DC0, 2EBB, 2V6T, 3JST, and 4C45). acRAF is shown in magenta. The pterin cofactor (from model 2V6T) is shown in blue. Residues required for PCD catalysis that are present in all PCDs, but not in acRAF, are shown as sticks. The structural alignment is tilted roughly 45° upwards into the plane of the page compared with the orientation of PCD and acRAF in panel A.
A presumptive catalytic motif for PCD enzymatic activity can be inferred by analyzing the arrangement and sequence conservation in representative PCDs known to be active compared with those proven earlier to be inactive. The PCD catalytic motif (27), [EDKH]-X(3)-H-[HN]-[PCS]-X(5,6)-[YWF]-X(9)-[HW]-X(8,15)-D, is not conserved in acRAF; only one histidine residue (residue 48) remains in a potentially catalytic position in the structure determined here. Consistent with this divergence, the structure of acRAF showed that its active site region (vis à vis PCD) is dramatically altered (Fig. 3, A and C). The narrow cleft that binds the pterin cofactor in PCD is substantially opened up in acRAF.
Despite structural divergence in the active site, we deemed it necessary to experimentally test acRAFs for PCD activity for two reasons. First, the catalytic mechanism of PCD is not well understood, making it difficult to rule out enzymatic activity of acRAF from the structure alone. Second, previous studies demonstrating a lack of catalytic activity in PCD-like proteins from plant chloroplasts (also referred to as type 2 PCDs) left open questions about the reason for this inactivity. The plant PCD-like proteins previously studied show a lack of conservation in the active site region, but they also bear a 40–60-amino acid extension at their N termini (not including the predicted chloroplast targeting sequence), which could, in principle, block or regulate the active site. No such extension is present in the acRAF proteins from α-carboxysome operons.
Sequence alignments among 73 carboxysome-associated acRAFs were performed using the program PRALINE (37) to investigate potentially conserved motifs specific to this family. Pro47 and Phe52 are the only fully conserved residues; both likely fulfill structural roles required for overall acRAF stability. Several other solvent accessible residues, however, also show substantial conservation (Fig. 2B). Interestingly, these conserved residues localize to a contiguous surface region of acRAF, distant from the vestigial active site, implicating this surface region as a putative functional interface.
Absence of PCD Enzyme Activity
To test acRAF for PCD enzyme activity, we employed a cell-based complementation assay that requires PCD activity for cell growth (18). The complementation assay was performed in E. coli strain JP2255, a tyrosine and phenylalanine auxotroph that can replicate on media lacking tyrosine if supplemented with both a functional phenylalanine hydroxylase and a functional PCD. E. coli species naturally lack AAHs and PCDs but do have genes encoding dihydropterin reductases, which are required (in combination with dehydration by PCD) to regenerate the pterin cofactor. We used E. coli strain JP2255 to confirm the catalytic inactivity of three different acRAFs: those from T. intermedia (the same organism as our crystal structure), H. neapolitanus C2, and P. marinus CCMP1375. As a negative control, we tested the type 2 PCD from A. thaliana and its N-terminal truncation mutant. Two positive controls were used: a well characterized PCD from P. aeruginosa POA1 and a predicted canonical PCD from T. intermedia, a paralog to the acRAF protein structurally characterized here. T. intermedia also contains a presumptive phenylalanine hydroxylase. We used this phenylalanine hydroxylase in our complementation assay along with another well characterized phenylalanine hydroxylase from P. aeruginosa PAO1.
Fig. 5 shows representative results of the PCD complementation experiments. In combination with either of the phenylalanine hydroxylases, none of the acRAFs we tested restored cell growth on media lacking tyrosine. In contrast, both the PCD from P. aeruginosa and the non-carboxysomal PCD from T. intermedia supported cell growth when co-transformed with plasmids encoding either of the phenylalanine hydroxylases. These results are consistent with the suggestion from structural and sequence analysis that acRAFs are not pterin dehydratases. These results also show that T. intermedia encodes enzymes capable of phenylalanine hydroxylation independent of acRAF, further supporting the hypothesis that acRAF fulfills a novel, non-enzymatic role in the function and/or structure of α-carboxysomes.
FIGURE 5.
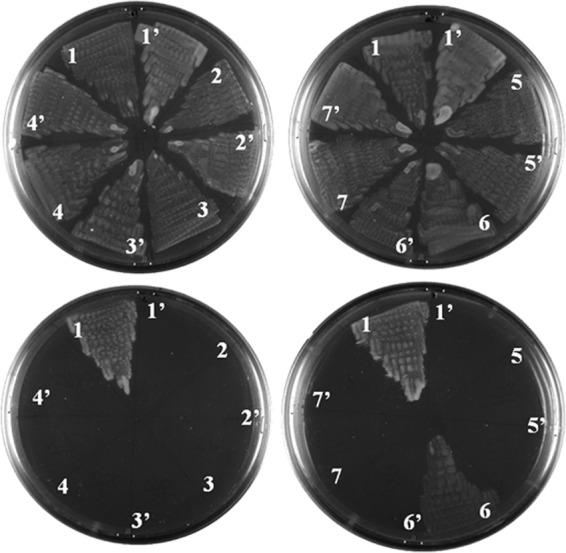
acRAFs do not compliment PCD activity in vivo. An E. coli tyrosine auxotroph, strain JP2255, was doubly transformed with two compatible plasmids, one plasmid containing the phenylalanine hydroxylase from T. intermedia and the second plasmid containing a putative or experimentally verified PCD. Cells were streaked in wedges on minimal media agar plates supplemented with or without tyrosine (top and bottom rows, respectively). Putative PCDs are numbered as follows: 1, P. aeruginosa PCD (NP_249562). 2, type-2 PCD from A. thaliana missing an N-terminal 46 residue chloroplast-targeting sequence (NP_199924). 3, type-2 PCD from A. thaliana missing 101 residues of N terminus. 4, T. intermedia acRAF with a 5-residue N-terminal truncation (YP_003641868). 5, H. neapolitanus acRAF (YP_003262803). 6, T. intermedia PCD (YP_003642661). 7, P. marinus acRAF (NP_874951). The prime symbol, as in 1′, indicates an empty plasmid transformed in place of a putative PCD. None of the PCDs or acRAFs were able to support tyrosine-independent growth without co-transformation with a phenylalanine hydroxylase (data not shown).
The absence of enzyme activity prompted us to search for an alternative function for this apparent enzyme relic. Clues regarding acRAF function arose in comparing gene content between α- and β-carboxysomes. The two types of carboxysomes are functionally similar in their abilities to concentrate and fix CO2. They are morphologically similar, and their shells are assembled mainly from homologous proteins that follow similar architectural principles. Yet those similarities belie key differences in genomic organization and protein composition. The presence of mutually exclusive genes in the two carboxysome systems suggests that some functions might be satisfied by functionally analogous but evolutionarily distinct proteins. The use of highly divergent, or even unrelated, carbonic anhydrases in the two systems is a case in point (38). We wondered if acRAF might play a role in RuBisCO folding and assembly similar to that played by RbcX in the β-carboxysome.
Heterologous RuBisCO Assembly Assay
RbcX is a RuBisCO chaperone understood to function in β-carboxysomes by stabilizing RuBisCO L2 dimers before they assemble into L8S8 hexadecamers (16, 18). Noting that α-carboxysome-containing organisms do not contain RbcX proteins but always encode acRAF, we tested the effects of co-expressing H. neapolitanus acRAF with H. neapolitanus RuBisCO. In a first set of experiments, we found that co-expressing acRAF with RuBisCO in E. coli did not improve the yield of soluble native RuBisCO. However, RuBisCO folding and assembly is a complex process: GroELS is important in subunit folding, but in β-carboxysome-containing cyanobacteria RbcX is required for assembly of the native (hexadecameric) complex. Indeed, in a second set of experiments we found that when acRAF was expressed together with GroELS, the yield of natively assembled RuBisCO, as judged by native PAGE, increased dramatically compared with when GroELS was expressed with RuBisCO but without acRAF (Fig. 6). This result identifies the PCD-like family of proteins as a new RuBisCO chaperone.
FIGURE 6.
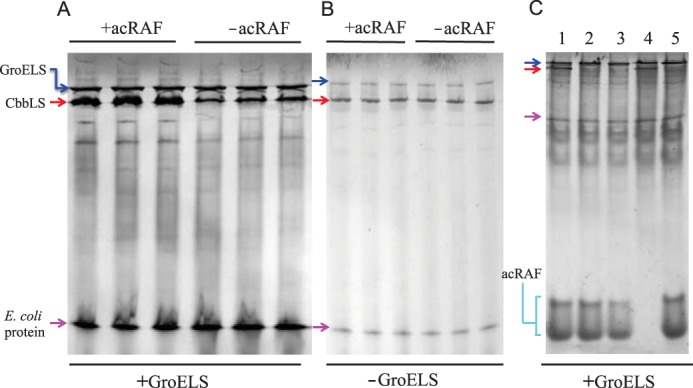
acRAF increases the quantity of natively assembled RuBisCO when heterologously co-expressed with GroELS in E. coli. A, the RuBisCO CbbLS genes were co-overexpressed with GroELS in E. coli BL21(DE3) either with (+) or without (−) acRAF. Soluble, whole cell lysates were analyzed by native non-denaturing PAGE. Independent co-expression experiments were performed in triplicate. The amount of natively assembled RuBisCO is substantially increased by the presence of acRAF (when GroELS is also overexpressed). The band corresponding to assembled CbbLS was deduced by comparisons to control gels run on lysates from cells expressing or lacking specific proteins (panel C). B, CbbLS was overexpressed in E. coli BL21(DE3) either with (+) or without (−) acRAF, but without GroELS overexpression. Soluble, whole cell lysates were analyzed by native non-denaturing PAGE. Independent co-expression experiments were performed in triplicate. No effect of acRAF on RuBisCO production is seen. C, CbbLS and acRAF bands were identified from soluble fractions of lysates of E. coli BL21(DE3) cells, which were transformed with different combinations of compatible plasmids. Strain 1, CbbLS, acRAF and GroELS. Strain 2, CbbL, acRAF, GroELS. Strain 3, CbbS, acRAF, GroELS. Strain 4, CbbLS, empty pET22b, GroELS. Strain 5, empty pRSF, acRAF, GroELS. RuBisCO (hexadecameric L8S8) and acRAF band identities were deduced by comparisons of strain-specific band patterns. The acRAF from H. neapolitanus migrated as two species, in contrast to acRAF from T. intermedia, which migrates as one species (not shown). Note that only strains 1 and 4 are expressing both large and small RuBisCO subunits, consistent with the assignment of native RuBisCO to the band present exclusively in those lanes. Blue, red, and magenta arrows indicated bands that correspond to GroELS, CbbLS, and an unidentified E. coli protein, respectively.
DISCUSSION
Previous studies have noted the occurrence of genes for PCD-like proteins, proteins related to the pterin dehydratase enzyme family, in α-carboxysome operons from both cyanobacteria (25) and chemoautotrophs (24). A systematic examination suggests that these proteins (now renamed acRAFs) are universally conserved in α-carboxysome operons, but absent from genomes that encode β-carboxysomes, making acRAF a defining gene of α-carboxysome operons. By determining the three-dimensional structure of acRAF from T. intermedia we showed that, despite overall structural similarity to bacterial PCDs, the active site cleft of acRAF is drastically altered. Conserved residues believed to be catalytically important in PCDs are not replaced by potentially catalytic residues in the acRAF structure. An in vivo enzyme complementation assay in E. coli confirmed that carboxysome-related acRAFs lack PCD enzymatic activity. After confirming its catalytic inactivity, we were able to establish a role for this protein in RuBisCO assembly.
When heterologously co-expressed in combination with GroELS, acRAF enhances the production of assembled RuBisCO. The mechanism by which acRAF interacts with and supports RuBisCO expression, stability, and/or assembly is not yet understood. All that can be asserted based on these first experiments is that acRAF likely acts at the subunit assembly stage (in a fashion analogous to RbcX), because its effect is only manifested in the presence of GroELS, which acts generally to promote subunit folding. Further mechanistic details await additional studies.
The evolution of enzymes for alternate functions is well known, as are examples of pseudoenzymes or “dead enzymes” wherein the loss of catalytic activity evolves in concert with the acquisition of regulatory or structural functions. Lens crystallins are classic examples of pseudoenzymes (39). Genomic and biochemical studies have focused attention on the adaptation of enzymes for other purposes as a widespread evolutionary phenomenon (40, 41) (reviewed in Refs. 42 and 43).
Our findings here regarding a cellular function for α-carboxysomal acRAFs align with previous studies of Type 2 PCDs from plant chloroplasts. Proteins from that group have been shown to lack catalytic activity (27), and genetic analysis has demonstrated that a homolog of acRAF in plants is required for RuBisCO assembly in vivo.5 Now that a function has been identified, detailed mechanistic studies on acRAF and its homologs in plants can be undertaken.
Acknowledgments
We thank Dr. Andrew Hanson and Mike Ziemak for providing PCD constructs, Dr. Sabeeha Merchant and Dr. Rikard Fristedt for valuable discussions and Arabidopsis constructs, Drs. Alice Barkan, Leila Feiz, and David Stern for discussions on acRAF homologs in plants. We thank M. Capel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov, and F. Murphy at NECAT for synchrotron x-ray assistance. We thank Mike Collazo and Michael Sawaya in the UCLA-DOE x-ray Crystallization and Crystallography Core Facilities. We are indebted to an anonymous reviewer for making suggestions that were critical in establishing the function of acRAF. The UCLA-DOE X-ray Crystallography Core Facility is supported by Department of Energy Grant DE-FC02-02ER63421. APS beamline 24-ID is supported by National Institutes of Health Grants P41RR015301 and P41GM103403. Use of the APS is supported by the Department of Energy under contract DE-AC02–06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI08114 (to T. O. Y.) and the BER program of the Department of Energy Office of Science.
L. Feiz, A. Barkan, and D. Stern, personal communication.
- RuBisCO
- ribulose-bisphosphate carboxylase/oxygenase
- acRAF
- α-carboxysome RuBisCO assembly factor
- PCD
- pterin-4α-carbinolamine dehydratase
- BMC
- bacterial microcompartment shell protein
- AAH
- aromatic amino acid hydroxylases.
REFERENCES
- 1. Andersson I., Backlund A. (2008) Structure and function of RuBisCO. Plant Physiol. Biochem. 46, 275–291 [DOI] [PubMed] [Google Scholar]
- 2. Sage R. F. (2002) Variation in the kcat of Rubisco in C(3) and C(4) plants and some implications for photosynthetic performance at high and low temperature. J. Exp. Bot. 53, 609–620 [DOI] [PubMed] [Google Scholar]
- 3. Marcus Y., Altman-Gueta H., Wolff Y., Gurevitz M. (2011) Rubisco mutagenesis provides new insight into limitations on photosynthesis and growth in Synechocystis PCC6803. J. Exp. Bot. 62, 4173–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson I. (2008) Catalysis and regulation in RuBisCO. J. Exp. Bot. 59, 1555–1568 [DOI] [PubMed] [Google Scholar]
- 5. Satagopan S., Scott S. S., Smith T. G., Tabita F. R. (2009) A RuBisCO mutant that confers growth under a normally “inhibitory” oxygen concentration. Biochemistry 48, 9076–9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Espie G. S., Kimber M. S. (2011) Carboxysomes. Cyanobacterial RuBisCO comes in small packages. Photosynth. Res. 109, 7–20 [DOI] [PubMed] [Google Scholar]
- 7. Iancu C. V., Ding H. J., Morris D. M., Dias D. P., Gonzales A. D., Martino A., Jensen G. J. (2007) The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 372, 764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kerfeld C. A., Sawaya M. R., Tanaka S., Nguyen C. V., Phillips M., Beeby M., Yeates T. O. (2005) Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938 [DOI] [PubMed] [Google Scholar]
- 9. Tanaka S., Kerfeld C. A., Sawaya M. R., Cai F., Heinhorst S., Cannon G. C., Yeates T. O. (2008) Atomic-level models of the bacterial carboxysome shell. Science 319, 1083–1086 [DOI] [PubMed] [Google Scholar]
- 10. Yeates T. O., Kerfeld C. A., Heinhorst S., Cannon G. C., Shively J. M. (2008) Protein-based organelles in bacteria. Carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6, 681–691 [DOI] [PubMed] [Google Scholar]
- 11. Wheatley N. M., Gidaniyan S. D., Liu Y., Cascio D., Yeates T. O. (2013) Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci. 22, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cai F., Menon B. B., Cannon G. C., Curry K. J., Shively J. M., Heinhorst S. (2009) The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS ONE 4, e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sutter M., Wilson S. C., Deutsch S., Kerfeld C. A. (2013) Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species. Photosynth. Res. 118, 9–16 [DOI] [PubMed] [Google Scholar]
- 14. Badger M. R. (2003) CO2 concentrating mechanisms in cyanobacteria. Molecular components, their diversity and evolution. J. Exp. Bot. 54, 609–622 [DOI] [PubMed] [Google Scholar]
- 15. Rae B. D., Long B. M., Badger M. R., Price G. D. (2013) Functions, compositions, and evolution of the two types of carboxysomes. Polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Onizuka T., Endo S., Akiyama H., Kanai S., Hirano M., Yokota A., Tanaka S., Miyasaka H. (2004) The rbcX gene product promotes the production and assembly of ribulose-1,5-bisphosphate carboxylase/oxygenase of Synechococcus sp. PCC7002 in Escherichia coli. Plant Cell Physiol. 45, 1390–1395 [DOI] [PubMed] [Google Scholar]
- 17. Liu C., Young A. L., Starling-Windhof A., Bracher A., Saschenbrecker S., Rao B. V., Rao K. V., Berninghausen O., Mielke T., Hartl F. U., Beckmann R., Hayer-Hartl M. (2010) Coupled chaperone action in folding and assembly of hexadecameric RuBisCO. Nature 463, 197–202 [DOI] [PubMed] [Google Scholar]
- 18. Bracher A., Starling-Windhof A., Hartl F. U., Hayer-Hartl M. (2011) Crystal structure of a chaperone-bound assembly intermediate of form I RuBisCO. Nat. Struct. Mol. Biol. 18, 875–880 [DOI] [PubMed] [Google Scholar]
- 19. Mueller-Cajar O., Stotz M., Wendler P., Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199 [DOI] [PubMed] [Google Scholar]
- 20. Hayashi N. R., Arai H., Kodama T., Igarashi Y. (1997) The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem. Biophys. Res. Commun. 241, 565–569 [DOI] [PubMed] [Google Scholar]
- 21. Chen D.-H., Madan D., Weaver J., Lin Z., Schröder G. F., Chiu W., Rye H. S. (2013) Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell 153, 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ellis R. J. (2001) Molecular chaperones: inside and outside the Anfinsen cage. Curr. Biol. 11, R1038–1040 [DOI] [PubMed] [Google Scholar]
- 23. Feiz L., Williams-Carrier R., Wostrikoff K., Belcher S., Barkan A., Stern D. B. (2012) Ribulose-1,5-bisphosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell 24, 3435–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jorda J., Lopez D., Wheatley N. M., Yeates T. O. (2013) Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 22, 179–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roberts E. W., Cai F., Kerfeld C. A., Cannon G. C., Heinhorst S. (2012) Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J. Bacteriol. 194, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thöny B., Auerbach G., Blau N. (2000) Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347, 1–16 [PMC free article] [PubMed] [Google Scholar]
- 27. Naponelli V., Noiriel A., Ziemak M. J., Beverley S. M., Lye L.-F., Plume A. M., Botella J. R., Loizeau K., Ravanel S., Rébeillé F., de Crécy-Lagard V., Hanson A. D. (2008) Phylogenomic and functional analysis of pterin-4α-carbinolamine dehydratase family (COG2154) proteins in plants and microorganisms. Plant Physiol. 146, 1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Volner A., Zoidakis J., Abu-Omar M. M. (2003) Order of substrate binding in bacterial phenylalanine hydroxylase and its mechanistic implication for pterin-dependent oxygenases. J. Biol. Inorg. Chem. 8, 121–128 [DOI] [PubMed] [Google Scholar]
- 29. Cronk J. D., Endrizzi J. A., Alber T. (1996) High-resolution structures of the bifunctional enzyme and transcriptional coactivator DCoH and its complex with a product analogue. Protein Sci. 5, 1963–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sourdive D. J., Transy C., Garbay S., Yaniv M. (1997) The bifunctional DCOH protein binds to HNF1 independently of its 4-α-carbinolamine dehydratase activity. Nucleic Acids Res. 25, 1476–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pape T., Schneider T. R. (2004) HKL2MAP. A graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 32. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER version X.Y.Z. Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 35. Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., 3rd, Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 36. Starkenburg S. R., Chain P. S., Sayavedra-Soto L. A., Hauser L., Land M. L., Larimer F. W., Malfatti S. A., Klotz M. G., Bottomley P. J., Arp D. J., Hickey W. J. (2006) Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 72, 2050–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Simossis V. A., Heringa J. (2005) PRALINE. A multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 33, W289–W294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. So A. K., Espie G. S., Williams E. B., Shively J. M., Heinhorst S., Cannon G. C. (2004) A novel evolutionary lineage of carbonic anhydrase (ϵ class) is a component of the carboxysome shell. J. Bacteriol. 186, 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Piatigorsky J. (2003) Crystallin genes. Specialization by changes in gene regulation may precede gene duplication. J. Struct. Funct. Genomics 3, 131–137 [PubMed] [Google Scholar]
- 40. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 41. Zeqiraj E., Filippi B. M., Deak M., Alessi D. R., van Aalten D. M. (2009) Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326, 1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leslie M. (2013) “Dead” enzymes show signs of life. Science 340, 25–27 [DOI] [PubMed] [Google Scholar]
- 43. Jeffery C. J. (2004) Molecular mechanisms for multitasking. Recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 14, 663–668 [DOI] [PubMed] [Google Scholar]
- 44. Yeates T. O. (1995) Algorithms for evaluating the long-range accessibility of protein surfaces. J. Mol. Biol. 249, 804–815 [DOI] [PubMed] [Google Scholar]