Abstract
Ca2+ signals control cell migration by regulating forward movement and cell adhesion. However, it is not well understood how Ca2+-regulatory proteins and second messengers are spatially organized in migrating cells. Here we show that receptor tyrosine kinase and phospholipase C signaling are restricted to the front of migrating endothelial leader cells, triggering local Ca2+ pulses, local depletion of Ca2+ in the endoplasmic reticulum, and local activation of STIM1, supporting pulsatile front retraction and adhesion. At the same time, the mediator of store-operated Ca2+ influx STIM1 is transported by microtubule plus ends to the front. Furthermore, higher Ca2+ pump rates in the front relative to the back of the plasma membrane enable effective local Ca2+ signaling by locally decreasing basal Ca2+. Finally, polarized phospholipase C signaling generates a diacylglycerol gradient towards the front that promotes persistent forward migration. Thus, cells employ an integrated Ca2+ control system with polarized Ca2+ signaling proteins and second messengers to synergistically promote directed cell migration.
Introduction
Migration is a fundamental property of many metazoan cells that allows organisms to develop, repair tissues, and defend against pathogens. Cells can move in a directed fashion in response to soluble chemicals or ligands (chemotaxis), mechanical cues (mechanotaxis), and substrate-bound chemo-attractants (haptotaxis)1,2. Directed migration is often studied in single cells but is also critical for groups of cells that migrate collectively towards an open space or chemoattractant3. Leader cells at the front of the group respond to environmental stimuli similarly to migrating single cells, while follower cells located behind the leader cells migrate based on cues from their neighboring cells4.
To move forward and turn, cells require spatial and temporal coordination of force-generating components such as actin and myosin5–7, as well as regulatory proteins such as Rac, RhoA and Cdc428,9. Nevertheless, how these molecular processes are coordinated for successful cell migration is still incompletely understood.
Ca2+ signals are one such coordinator of cell migration10,11 partly through local Ca2+ pulses near the leading edge that activate myosin light chain kinase (MLCK) and modulate nascent focal adhesions6,12,13. Nevertheless, it remains unclear why Ca2+ levels are often lower in the front than in the back of migrating cells11,14,15, whether receptor tyrosine kinase (RTK), phospholipase C (PLC) or stromal interaction molecule 1 (STIM1) signaling is polarized, whether the co-generated second messenger diacylglycerol (DAG) regulate cell migration in parallel, and whether Ca2+ signaling differs between leader cells and follower cells during collective sheet migration.
Many receptor stimuli induce PLC to generate inositol-1,4,5-trisphosphate (IP3), which activates IP3 receptor (IP3R) in the endoplasmic reticulum (ER), and locally or globally release Ca2+ stored in the ER. Ca2+ signals are terminated by removal of released Ca2+ through plasma membrane (PM) Ca2+ ATPase (PMCA) to the outside, and through ER Ca2+ ATPase (SERCA) back into the ER16. PLC also produces the lipid second messenger DAG which often acts synergistically with Ca2+ in activating cellular processes17,18. In addition, STIM1 proteins sense low luminal ER Ca2+ and signal across the ER membrane to activate PM Ca2+ influx channels (SOC) at junctions where the ER contacts the PM.
Here we use live-cell imaging of migrating sheets of endothelial cells to determine if and how this Ca2+ signaling system is spatially organized during migration. We identified gradients in cytosolic and ER Ca2+ levels as well as polarized distributions of growth factor receptor signaling, Ca2+ pulses, DAG, Ca2+ pumps and STIM1, together generating an integrated Ca2+ control system that is uniquely suited to regulate directionality, speed and turning of endothelial leader cells as they move into open space.
Results
Receptor tyrosine kinase signaling is polarized in migrating leader cells
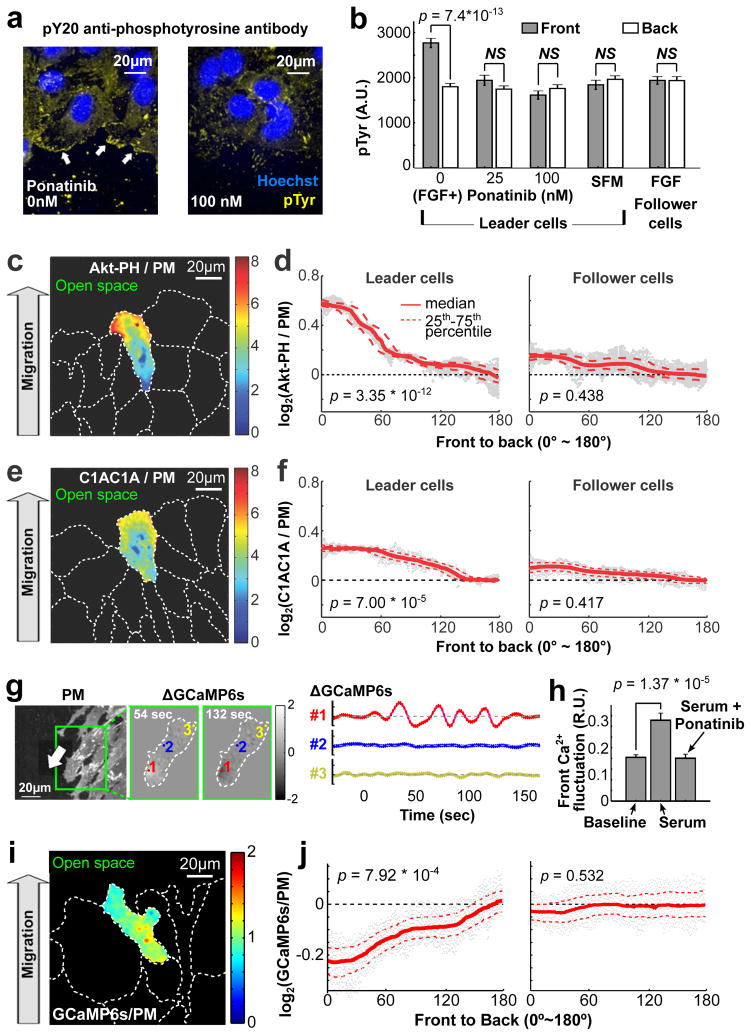
We investigated the collective migration of human umbilical vein endothelial cells (HUVEC) plated as confluent monolayers. Growth factors promote the migration of HUVECs into a band of open space that can be generated by removing cells using a scratch tool4,19. In the presence of uniform fibroblast growth factor (bFGF), phospho-tyrosine signals were higher in the front than in the back of leader cells. In contrast, cells in serum-free medium or cells stimulated with bFGF, but inhibited by the pan-tyrosine kinase inhibitor Ponatinib20, lost this phospho-tyrosine gradient (Fig. 1a,b and Supplementary Fig. 1a), arguing that receptor tyrosine kinase (RTK) signaling is polarized. The phospho-tyrosine gradient was restricted to leader cells, as it was not observed in follower cells inside the monolayer (Fig. 1b).
Figure 1.
Receptor tyrosine kinase (RTK) signaling is restricted to the front of migrating leader cells. (a,b) bFGF-induced tyrosine phosphorylation was higher in the front of migrating cells (white arrows). Addition of the pan-RTK inhibitor Ponatinib blocked tyrosine kinase signaling in the front, but not in the back of leader cells. Follower cells did not respond to bFGF. HUVECs were fixed and stained with pY20 anti-phospho-tyrosine antibody (n = 107, 105, 115, 110 and 107 cells for SFM, follower cells, and Ponatinib 0 nM, 25 nM and 100 nM, respectively). SFM: serum-free medium. (c,e) Fluorescence ratio images of leader cells co-expressing YFP-Akt-PH (PIP3 sensor) or YFP-C1AC1A (DAG sensor) and a plasma membrane marker (CFP-mCD4). PIP3 (c) and DAG (e) were enriched in the front of migrating cells. (d,f) Front-to-back gradients of PIP3 and DAG were present in leader, but not follower cells (24 leader and 42 follower cells in (d), 28 and 62 cells in (f)). (g) Ca2+ pulses in migrating HUVECs were measured as relative increases in local PM targeted GCaMP6s fluorescence intensity. Higher activities were observed in the front (#1) compared to the middle (#2) or back (#3) of migrating cells. (h) Relative mean amplitudes of local Ca2+ fluctuations measured over 3 minutes in the front of migrating cells in response to serum or serum plus Ponatinib (see also Supplementary Fig. 1e,f). Amplitudes of Ca2+ fluctuations were normalized to basal cytosolic levels (0.3 R.U. means the fluctuation is 30% of the average cytosolic [Ca2+] level; n = 24 cells). (i,j) Migrating HUVECs expressing GCaMP6s-CAAX and the reference membrane marker mCD4 were used to measure Ca2+ gradients in leader and follower cells (n = 83 leader and n = 86 follower cells). Bars denote mean ± SEM in Fig. 1b,h. Student t test was used for Fig. 1b,d,f,h,j. In Fig. 1d,f,j, p values were calculated by comparing the ratio of the sensor / PM intensity ratios in the front and back (both regions were 10% of cell length).
We next tested whether PLC, a downstream target of RTK signaling, was also activated in a polarized fashion, using an improved DAG sensor to monitor whether PLC-generated DAG was polarized in migrating cells (PKCγ-C1AC1A; See Methods). A PM marker was used as a reference (Supplementary Fig. 1b,d). Strikingly, the ratio of membrane-localized DAG sensor over the PM marker was higher in the front versus the back of the cell, arguing that a gradient in PLC activation is converted into a gradient in DAG signaling (Fig. 1e,f and Supplementary Fig. 1d). As has been shown in other cells, we used an Akt-PH translocation sensor for phosphatidylinositol (3,4,5)-trisphosphate (PIP3) to confirm that this co-produced lipid second messenger is polarized21 (Fig. 1c,d and Supplementary Fig. 1d).
PLC activation also generates IP3, which releases Ca2+ from the ER. Indeed, use of a plasma membrane targeted Ca2+ indicator GCaMP6s-CAAX22 revealed a higher frequency of local Ca2+ release pulses in the front than in the back of cells (Fig. 1g and Supplementary Movie 1; see Methods). Furthermore, these local Ca2+ pulses in the front were dependent on the presence of either bFGF or serum, and were suppressed by Ponatinib (Fig. 1h; Supplementary Fig. 1e–h). Together, these data support the interpretation that cyclic local Ca2+ pulses in the front of the cell as well as a gradient of DAG are both induced by local RTK-mediated PLC activation (Fig. 1g). These signals and gradients were exclusively observed in leader but not follower cells. Nevertheless, despite the polarized Ca2+ pulses in the front, the average cytosolic Ca2+ levels distributed in an inverted manner with markedly lower levels in the front than in the back of the cell (Fig. 1i,j), similar to observations in other cell types11,14,15.
Identification of Ca2+ signaling proteins that regulate collective cell migration
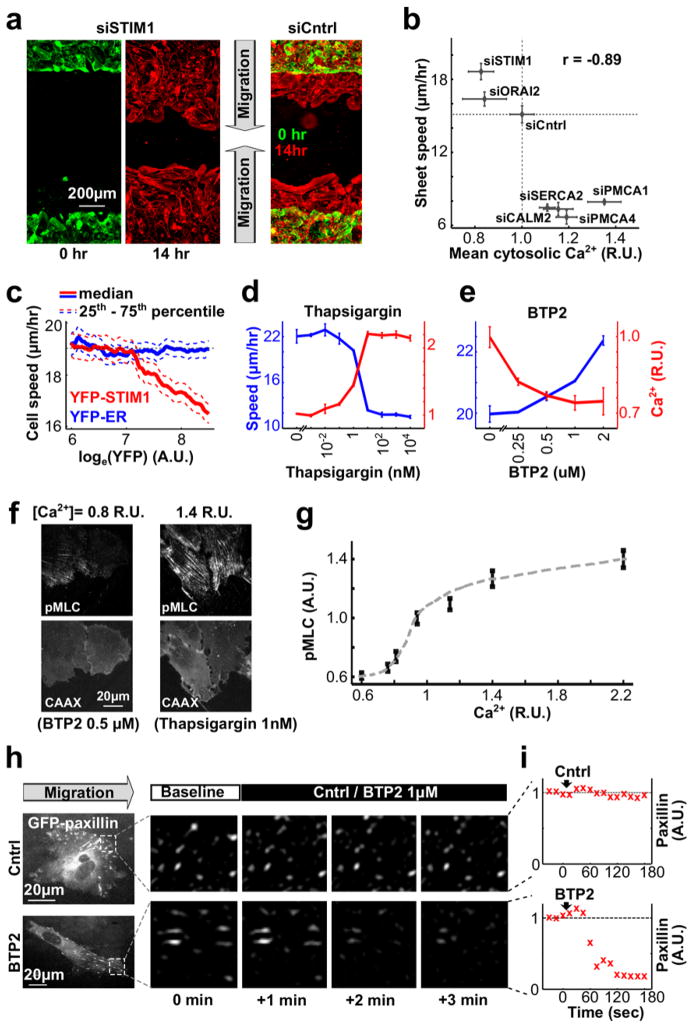
We next conducted siRNA knockdown experiments targeting Ca2+ regulators and used automated microscopy to compare changes in cytosolic Ca2+ level and cell migration speed of leader cells12,19 (Fig. 2a; see Methods). Knocking down the components of the store-operated Ca2+ (SOC) influx pathway, STIM1 and Orai, decreased cytosolic Ca2+ levels, while knocking down the Ca2+ pumps PMCA (which pumps Ca2+ out of cells through the plasma membrane) and SERCA (which pumps Ca2+ from cytosol into ER) or calmodulin (CALM, which regulates PMCA and other Ca2+ regulatory proteins), increased intracellular Ca2+ levels (Fig 2a,b and Supplementary Fig. 2b). Unexpectedly, these changes in basal Ca2+ were paralleled by opposing changes in migration speed (Fig 2b). This inverse correlation was confirmed by the over-expression of YFP-conjugated STIM1, which increased basal Ca2+ and at the same time reduced speed (Fig. 2c and Supplementary Fig. 2c). The same opposing effect on migration speed was observed when we increased or decreased basal Ca2+ levels using the SERCA Ca2+ pump inhibitor thapsigargin and the SOC Ca2+ influx inhibitor BTP223 (Fig. 2d,e). An inhibitory role of local Ca2+ signal pulses12 was confirmed by observing that loading migrating HUVECs with increasing amounts of Fura-2 dampened local Ca2+ pulses and increased the speed of migration (Supplementary Fig. 2e; See Methods, Supplementary Fig. 1g and Supplementary Table 1 for estimation of basal free cytosolic Ca2+ levels).
Figure 2.
Store-operated Ca2+ (SOC) influx controls cell migration by regulating cell-matrix adhesion in the front of migrating cells. (a) HUVEC migration into open space was monitored by staining cells with CellMask (see Methods). Accelerated sheet migration was observed in STIM1-depleted compared to control cells. (b) Comparing changes in the rate of sheet migration and cytosolic Ca2+ levels in HUVECs treated with siRNAs targeting different Ca2+ signaling regulators. Average cytosolic [Ca2+] was normalized to the level of cells treated with siCntrl (n = 4 experiments for each siRNA). (c) Reduced single cell migration speed in cells over-expressing YFP-STIM1. Cells expressing YFP-ER were used as control (n ~ 10,000 cells per condition). (d,e) Effects of the ER Ca2+ pump blocker thapsigargin (d) and the SOC inhibitor BTP2 (e) on cytosolic Ca2+ levels and on sheet migration speed. Notice that increasing cytosolic Ca2+ levels by thapsigargin decreased migration speed, and lowering Ca2+ levels by BTP2 increased migration speed (n = 4 experiments per condition). (f,g) Migrating HUVECs were treated with different concentrations of BTP2 or thapsigargin to reduce or elevate cytosolic Ca2+ levels. Cells were then fixed and stained with anti-phospho-myosin light chain (pMLC) antibody. (f) pMLC signals were lower when SOC was blocked by BTP2 but higher when ER Ca2+ pumps were blocked by thapsigargin. CAAX: plasma membrane marker. (g) pMLC levels increased with increasing cytosolic [Ca2+] (n = 123, 134, 127, 126, 123, 117 and 142 cells per condition from left to right). (h,i) Effect of BTP2 treatment on cell-matrix adhesion. Focal adhesion formation was monitored by expressing GFP-paxillin. BTP2 treatment rapidly decreased the intensities of GFP-paxillin puncta in the front of migrating cells, consistent with SOC influx promoting cell-matrix adhesion. Bars are mean ± SEM in Fig. 2b,d,e,g.
In order for pulses of Ca2+ to have functional consequences, they must be of sufficiently high amplitude to activate targets, i.e. by Ca2+ forming a complex with calmodulin and MLCK. We generated small decreases or increases in the cytosolic Ca2+ levels by titrating BTP2 or thapsigargin. HUVECs were then fixed and stained with anti-phospho-myosin light chain (pMLC) antibody to monitor the activity of MLCK. Indeed, we noticed an increase of pMLC staining upon small increases in cytosolic [Ca2+] (Fig. 2f,g). This argues that changes in Ca2+ in the front of less than two-fold can activate MLCK and myosin II. The tight binding of Ca2+/calmodulin to MLCK (Kd of 1.1 nM) lowers the required level of Ca2+ needed to activate MLCK24, providing a plausible explanation why small Ca2+ signals are sufficient for activation. In support of this interpretation, treatment with the SOC inhibitor BTP2 decreased local Ca2+ pulses (Supplementary Fig. 2f,g) and nascent focal adhesions (Fig. 2h,i and Supplementary Fig. 2h,i) with a rate similar to the one caused by the myosin II inhibitor Blebbistatin. Thus, small Ca2+ pulses have a critical physiological role in the cyclic regulation of local MLCK activity in the front of migrating HUVEC cells.
The relative strength of cell-matrix adhesion determines whether STIM1 enhances or suppresses the speed of migration
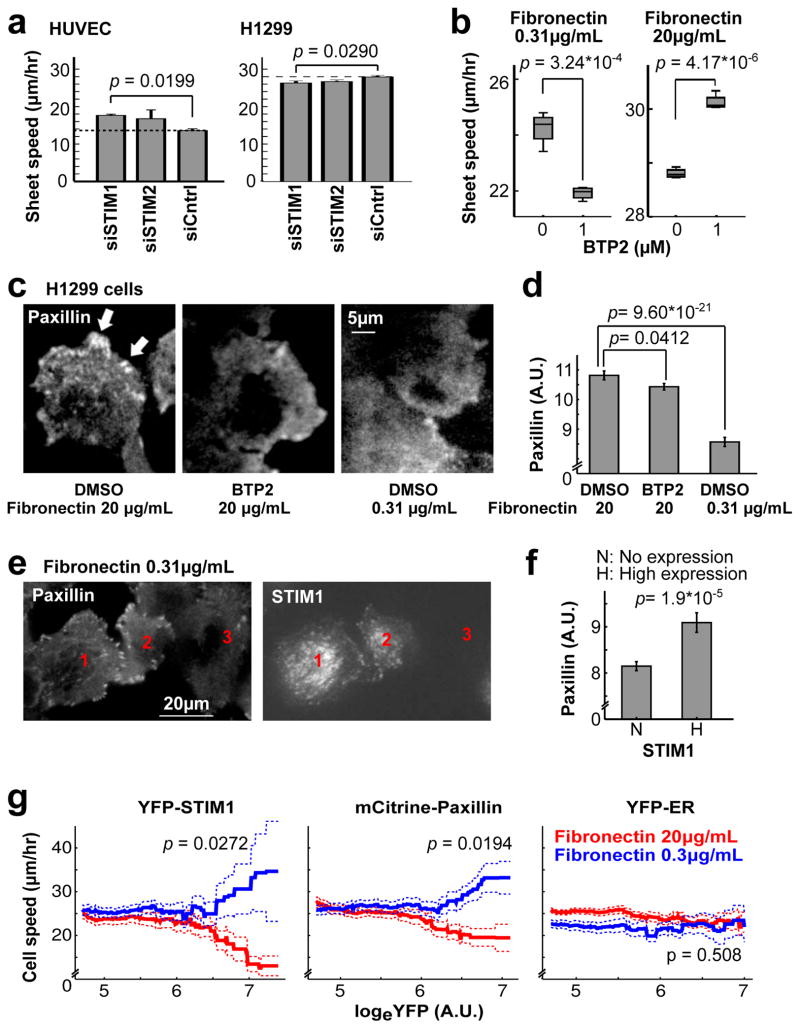
Our result for STIM1 knockdown on HUVEC migration speed differed from previous studies performed in cancer25,26 and smooth muscle27,28 cells which showed reduced migration following STIM1 knockdown. We verified STIM1 knockdown by Western blotting (Supplementary Fig. 2a and Supplementary Fig. 3g) and further validated our findings by performing rescue experiments, where the effect of STIM1 depletion was reversed by overexpression of exogenous STIM1, and where BTP2 treatment abrogates rescue by exogenous STIM1 (Supplementary Fig. 2c,d). STIM1 knockdown using the same siRNAs also had a small but opposing effect on migration of H1299 metastatic lung cancer cells compared to HUVEC cells (Fig. 3a).
Figure 3.
SOC increases migration speed when cell-matrix adhesion is weak, but slows down migration when adhesion is strong. (a) Knocking down STIM isoforms increased sheet migration speed in HUVEC (left) but caused a small and significant reduction in speed in H1299 cells (right) (n = 4 experiments per condition). (b) H1299 cells migrated faster on high fibronectin. BTP2 treatment decreased migration speed of H1299 cells on low, but increased migration speed on high fibronectin. Each box shows the median (horizontal line) and the 25th and 75th percentiles. Lower and upper whiskers are minima and maxima, respectively (n = 3 experiments per condition). (c–f) Fibronectin and SOC jointly regulate focal adhesions. H1299 cells were plated on low or high fibronectin and treated with BTP2 or DMSO before fixation and staining with anti-paxillin antibody to label endogenous focal adhesions. (c) Paxillin puncta (white arrows) were prominent in cells on high fibronectin but were decreased in low fibronectin or when treated with BTP2. (d) Integrated punctate paxillin signals for different treatments in (c) (n = 115, 121 and 104 cells from left to right per condition). (e) Overexpression of YFP-STIM1 in H1299 cells (#1, 2) on low fibronectin increased cell substrate adhesion (paxillin staining) compared to control cells (#3). (f) Average paxillin puncta intensity for control cells and for cells expressing high levels of YFP-STIM1 (n = 42 and 23 cells from left to right per condition). (g) Single cell speed in sheet migration as a function of YFP-STIM1, mCitrine-paxillin or YFP-ER overexpression in H1299 cells. Overexpression of YFP-STIM1 and mCitrine-paxillin accelerated sheet migration on low fibronectin but decreased migration speed on high fibronectin. Solid lines and dashed lines are mean ± SEM; n = 4714 (red) & 4538 (blue) cells with STIM1, 3873 (red) & 4327 (blue) cells with paxillin, and 5698 (red) & 5368 (blue) with ER marker. p value compares the blue and red group for cells with logeYFP > 6.5. Student’s t test was used in Fig. 3a,b,d,f,g. Bars are mean ± SEM in Fig. 3a,d,f.
A possible reconciliation between the opposite results in the two cell types is the role of SOC and local Ca2+ pulses in enhancing the assembly rate of focal adhesion complexes as shown here (Fig. 2h,i) and in previous studies12,29. Although cell-matrix adhesion is critical for cell migration, strong cell-matrix adhesion can impede cell motility30. Therefore, increased cell-matrix adhesion might have contrasting effects on cell migration in weakly (metastatic cancer cells) versus strongly (HUVEC) adherent cell types. Consistent with this hypothesis, cell migration speed for H1299 cells was higher when plated on high (20 μg/ml), comparted to low (0.3 μg/ml) density fibronectin (Fig. 3b). Interestingly, when SOC was blocked using the inhibitor BTP2, cells on low fibronectin slowed down (Fig. 3b, left) while cells on high fibronectin accelerated migration (Fig. 3b, right). Furthermore, using the focal adhesion component paxillin as a marker, H1299 cells formed more focal adhesion complexes on high fibronectin than on low fibronectin, while BTP2 decreased focal adhesions in both conditions (Fig. 3c,d and Supplementary Fig. 3a,b). Cells over-expressing STIM1 protein also formed more focal adhesions (Fig. 3e,f and Supplementary Fig. 3c–f), indicating that SOC is sufficient to enhance focal adhesion. Finally, both STIM1 and paxillin over-expression enhanced migration speed for H1299 cells on low fibronectin but decreased speed for cells on high fibronectin (Fig. 3g). Thus, STIM1 and SOC-mediated Ca2+ signals enhance matrix adhesion, which in turn either accelerates or slows cell migration depending on whether adhesion is weak or strong, respectively.
STIM1 is polarized to the front of migrating cells by microtubule plus end-based transport
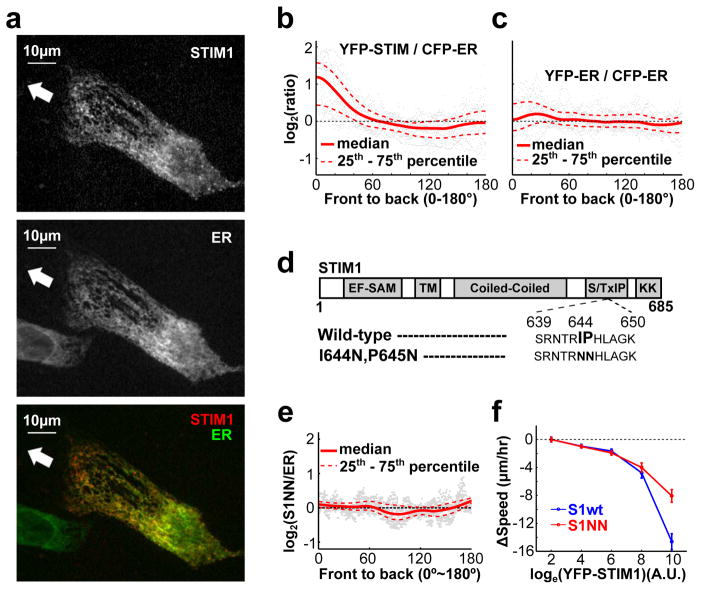
We discovered that expressed YFP-STIM1 was significantly enriched in the front of migrating leader cells (HUVEC) when compared to a CFP conjugated ER marker protein (Fig. 4a–c and Supplementary Fig. 4a). STIM1 can be transported within the ER membrane by binding to the microtubule plus end protein EB131. To test whether microtubule-mediated transport was responsible for the polarization of STIM1, we expressed a YFP-conjugated EB1-binding deficient mutant STIM132 (S1NN) (Fig. 4d). YFP-S1NN increased overall SOC influx to a similar degree as wild-type YFP-STIM1 (YFP-S1wt), indicating that the mutant maintained its full ability to control Ca2+ influx (Supplementary Fig. 4b,c). However, YFP-S1NN failed to polarized in migrating cells (Fig. 4e), arguing that wild-type STIM1 is actively transported by microtubule plus ends to the front and that microtubule-based targeting is important for STIM1 localization but not its activity. Nonetheless, HUVEC cells expressing high levels of wild-type STIM1 dramatically decreased their motility, whereas cells expressing similar levels of YFP-STIM1-S1NN showed a smaller reduction in speed (Fig. 4f). This suggests that the localization of STIM1 towards the front is important for its role in cell migration.
Figure 4.
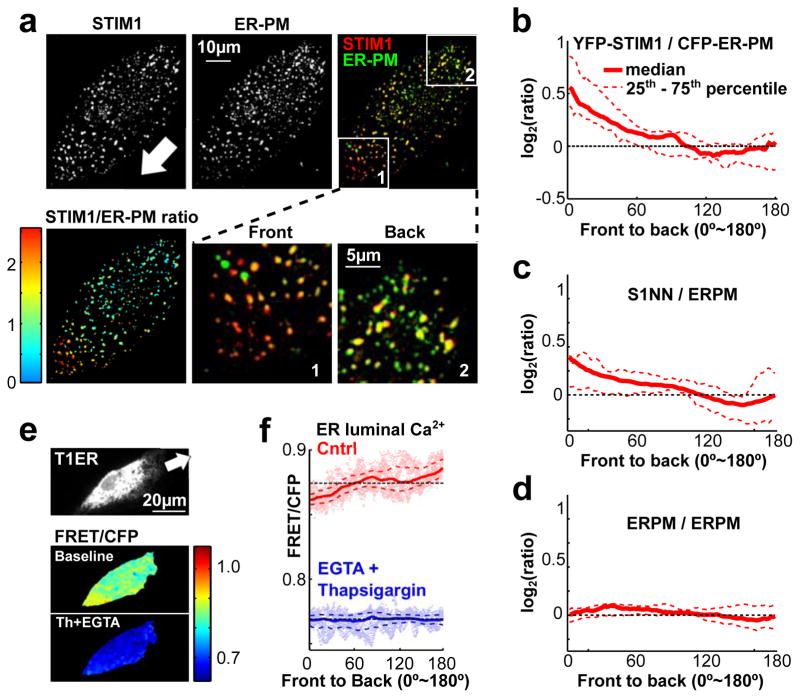
STIM1 is enriched in the front of migrating cells. (a,b) Migrating HUVEC expressed YFP-STIM1 (STIM1) and a CFP-tagged ER marker (ER). Merged and ratio images are shown here and in Supplementary Fig. 4a to show the relative enrichment of STIM1 compared to an ER marker towards the front. White arrow indicates the direction of cell migration. (b) Quantification of the ratio of YFP-STIM1 / CFP-ER marker from front to back (see Methods). YFP-STIM1 was enriched in the front, whereas (c) the control YFP-ER was not (n = 36 cells per condition). (d–f) Enrichment of STIM1 in the front of migrating cells is mediated by binding to the microtubule plus-end binding protein EB1. (d) Domain structure of STIM1 and mutations preventing binding to EB1. (e) Unlike wild-type STIM1 (S1wt) protein (Fig. 4a,b and Supplementary Fig. 4a), the S1NN mutant was not enriched in the front of migrating cells. (n = 27 cells) (f) Over-expression of the S1NN mutant suppressed cell migration less than overexpression of S1wt. Bars are mean ± SEM (n ~ 10,000 cells per condition).
Since STIM1 only regulates SOC after its localization to ER-PM junctions, localization of STIM1 at ER-PM junctions can be used as an indicator for SOC Ca2+ influx. We expressed YFP-tagged STIM1 together with a CFP-tagged marker for ER-PM junctions in migrating HUVECs. The ER-PM marker includes a single ER membrane-spanning region, a cytoplasmic linker that is several nm long, and a polybasic PM interaction domain (See Methods). Confocal images focused at the bottom membrane of the cells showed enhanced accumulation of YFP-STIM1 at ER-PM junction sites in the front compared to the back, arguing that not only is STIM1 polarized towards the front but, in addition, there is enhanced STIM1 activity to activate SOC in the front (Fig. 5a–d and Supplementary Fig. 4d). In support of this interpretation, the S1NN STIM1 mutant, even though it was evenly distributed within the ER (Fig. 4e), was also enriched at ER-PM junctions in the front (Fig. 5c).
Figure 5.
STIM1 is locally activated in the front of migrating cells. (a) HUVEC cells were co-transfected with YFP-STIM1 and the ER-PM junction marker CFP-ER-PM. Confocal images show focal planes at the bottom of the cell. The white arrow marks the direction of migration.YFP-STIM1 was enriched at front ER-PM junctions in migrating cells. (b) Quantification of the ratio of YFP-STIM1 / CFP-ER-PM from front to back in migrating cells. (n =14 cells) (c,d) Similar analysis as in (b) but for cells coexpressing YFP-S1NN and CFP-ER-PM (c) or coexpressing YFP-ER-PM and CFP-ER-PM. A smaller increase in relative S1NN activity was observed in the front (n = 10 cells for S1NN and n = 12 cells for the control group). (e,f) Decreasing luminal ER Ca2+ levels towards the front of migrating leader cells. (e) Ratio-imaging of a modified luminal ER Ca2+ FRET probe T1ER (see Methods) in migrating HUVECs. Adding the SERCA inhibitor thapsigargin (2 μM) and EGTA (3 mM) decreased ER Ca2+ levels (lower panel). (f) Gradient in luminal ER Ca2+ measured using the T1ER probe. Note that the lower Ca2+ levels in the front are still sensitive to EGTA+thapsigargin treatment (n = 79 cells for the control group; n = 49 cells for the thapsigargin + EGTA group).
A gradient in the level of Ca2+ in the lumen of the ER of polarized HUVEC cells
Since STIM1 localization to ER-PM junctions is directly regulated by ER luminal Ca2+ levels, we tested if the ER luminal Ca2+ level is lower in the front. An ER luminal Ca2+ FRET probe33,34 (Fig. 5e and Supplementary Fig. 4e,f) revealed that intra-ER Ca2+ was lower towards the front of migrating cells compared to the back (Fig. 5e,f). Because local Ca2+ pulses are more frequent in the front than the back of migrating cells (Fig. 1g and Supplementary Movie 1), the lowerlevel of luminal ER Ca2+ in the front was likely a result of local IP3-mediated Ca2+ release16. In support of this hypothesis, the pan-RTK inhibitor Ponatinib reduced the translocation of STIM1to ER-PM junctions in the front of migrating cells (Supplementary Fig. 4g–i).
Low basal Ca2+ in the front of migrating cells is maintained by enhanced pump activity of the plasma membrane Ca2+ ATPase (PMCA)
Our previous12 and current studies showed that MLCK is dynamically regulated by local Ca2+ pulses near the front of migrating cells. Therefore, cells have to maintain a basal Ca2+ level in the front below a critical threshold to prevent persistent MLCK activation (Fig. 2f,g)12,24. Experiments with the Ca2+ indicator Fura-2 (Fig 6a), confirmed (Fig. 1j) that HUVEC cells exhibit a Ca2+ gradient with lower basal Ca2+ levels in the front. This gradient is reduced by increased buffering of Ca2+ when we increased Fura-2 in the cytosol (Fig 6b). When we elevated cytosolic Ca2+ levels using the SERCA inhibitor thapsigargin35, the previously reported correlation between local Ca2+ pulses and lamellipodia retraction12 was lost (Supplementary Fig. 5a,b). This suggests that low basal Ca2+ levels in the front are critical for cyclic regulation of lamellopodia retraction and adhesion by Ca2+ pulses.
Figure 6.
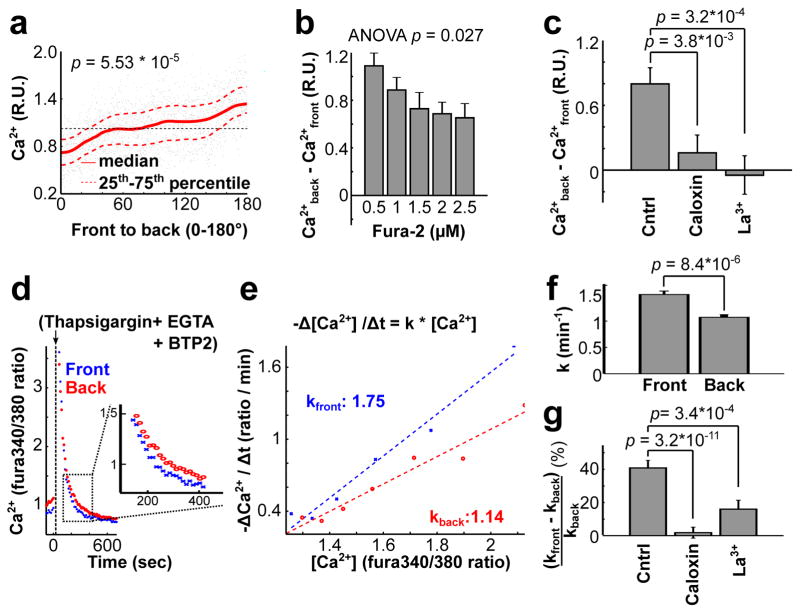
Polarized plasma membrane Ca2+ pump activity keeps cytosolic Ca2+ low in the front. (a) Basal cytosolic Ca2+ levels (measured using Fura-2) in the front were about 50% of the levels in the back of migrating cells (n = 14 cells). Local [Ca2+] was normalized to the average cytosolic level. Cytosolic measurements using GCaMP6s-CAAX are shown in Fig. 1 i,j. (b) High Fura-2 increased the diffusion speed of Ca2+, so as to decrease the Ca2+ gradient between the front and back of migrating cells. Bars are mean ± SEM (n = 160, 160, 80, 160 and 80 cells from left to right for each group). (c) Addition of the PMCA inhibitors Caloxin (200 μM) and La3+ (200 mM) significantly decreased cytosolic Ca2+ gradients in migrating cells. Bars are mean ± SEM (n = 47 cells per condition). (d–g) Cytosolic Ca2+ is transported out of the cell faster in the front than in the back. (d) Addition of thapsigargin, SOC inhibitor BTP2, and Ca2+ chelator EGTA to migrating HUVECs caused a transient increase in cytosolic Ca2+. Local extrusion pump rates were measured as a function of the Ca2+ level in the front or back over time. (e) Graph showing the relative pump rates (derivative of Ca2+ change) in the front and the back as a function of local [Ca2+]. The slopes reflect relative pump activity differences in the front and the back for each cell. (f) Statistical analysis of the relative Ca2+ pump activities in the front versus the back of migrating cells. Bars are mean ± SEM (n = 25 cells). (g) Cells pretreated with inhibitors of PMCA lost their differential Ca2+ pump activities. Bars are mean ± SEM (n = 47 cells for each group). Student t test was used for Fig. 6a,c,f,g. One-way ANOVA was used for Fig. 6b.
To attain low cytosolic Ca2+ in the front, cells may pump Ca2+ either more strongly into the ER through SERCA pumps, or, alternatively, out of the cell through PM-localized Ca2+ pumps (PMCA). Polarized SERCA pump activity was likely not involved in setting up the gradient since inhibition of SERCA using thapsigargin increased rather than an decreased the Ca2+ gradient (Supplementary Fig. 5c,d). However, inhibition of PMCA in migrating cells using either the inhibitor Caloxin 2A1 or La3+, reduced the Ca2+ gradient (Fig. 6c) while overall cytosolic Ca2+ levels were elevated (Supplementary Fig. 5e) and sheet migration was decreased (Supplementary Fig. 6c).
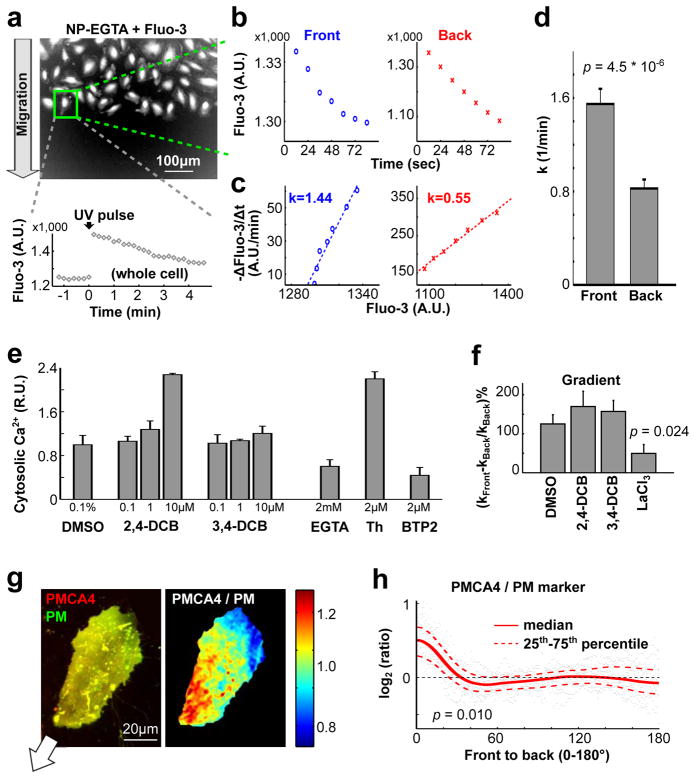
In a second protocol, we simultaneously treated migrating cells with the SERCA inhibitor thapsigargin, the SOC inhibitor BTP2 and also chelated external Ca2+ using EGTA. This protocol immediately released Ca2+ from the ER into the cytosol, followed by a slower removal of Ca2+ from the cytosol to the extracellular space. The time-course of the change in local Ca2+ level (the slope in Fig. 6d) during the removal phase can be used to derive the local Ca2+ pump rate of PMCA (pump rate over local cell volume). A comparison of the local Ca2+ level (x-axis) and the local Ca2+ pump rate (y-axis) for each time point showed that the pump activity (slope in Fig. 6e) in the front of migrating cells is significantly higher than in the back (Fig. 6e,f). The same differential pump activity was confirmed by uniformly releasing Ca2+ using a UV pulse from an intracellular NP-EGTA Ca2+ buffer, which induced a cell-wide transient Ca2+ spike (Fig. 7a–d). Furthermore, the differential pump activity could be eliminated by inhibitors of PMCA (Fig. 6g and Supplementary Fig. 5f), but not by inhibitors of Na+-Ca2+ exchangers, known to contribute to basal Ca2+ homeostasis (Fig. 7e,f and Supplementary Fig. 6a,b). This confirmed a specific role of PMCA in generating Ca2+ gradients in HUVEC.
Figure 7.
Higher Ca2+ pump activity in the front compared to the back generates a gradient of basal [Ca2+] in migrating cells. (a–d) UV flash photolysis experiments confirmed differential Ca2+ pump activities in migrating HUVECs as shown in Fig. 6c–e. (a) Migrating HUVECs were pre-loaded with Fluo-3/AM and NP-EGTA. Thapsigargin and EGTA were added 10 minutes before imaging to block the activity of SERCA and influx Ca2+ channels. A UV pulse was used to induce a Ca2+ spike 1 minute after recording began. (b,c) Ca2+ pump activities (k) in the front and in the back were calculated based on Fluo-3 measurements following UV photolysis, similar to Fig. 6d,e. (d) Quantification of relative Ca2+ pump activities in the front and in the back of migrating cells. (n = 41 cells.) (e) Inhibitors of Na+-Ca2+ exchangers 2,4-DCB or 3,4-DCB both caused a small increase cytosolic Ca2+ levels as measured by Fura-2. EGTA, Thapsigargin (Th) and BTP2 were used as positive controls. [Ca2+] was normalized using average cytosolic levels in the DMSO group (n = 4 wells for each group). (f) Addition of 10 μM 2,4-DCB or 3,4-DCB did not affect the differential Ca2+ pump activities. LaCl3 was used as a positive control (n = 41, 40, 35 & 42 cells in DMSO, 2,4-DCB, 3,4-DCB and LaCl3). (g) PMCA4 is enriched in the front of migrating cells. HUVEC co-expressing GFP-PMCA4 and the plasma membrane marker tdimer2-lyn were imaged by confocal microscopy. The merged (left) and the ratio-image (right) indicated enrichment of GFP-PMCA4 in the front (see also Supplementary Fig. 6d). The white arrow depicts the direction of cell migration. (h) Statistical analysis of the relative spatial distribution of GFP-PMCA4/tdimer2-lyn from the front to the back (n = 13 cells). A Student t test was used for Fig. 7d,f,h. In Fig. 7h, p values were calculated based on the ratio of the sensor / PM ratio in the front 10% region to that in the back 10% region of migrating cells. In Fig. 7d,e,f, Bars are mean ± SEM.
A gradient in PMCA pump rate could either be mediated by differential regulation of local PMCA activity36–38, by differences in the surface-to-volume ratio between the front and the back or by having more relative PMCA in the front compared to the back. We observed a polarized distribution of GFP-tagged PMCA4, an isoform that is prominently expressed in HUVEC39, compared to a tdimer2-tagged PM marker (Fig. 7g,h and Supplementary Fig. 6d,e). Thus, cells lower basal Ca2+ in their front during migration by having locally higher PM Ca2+ pump rates in the front and this higher rate is at least in part the result of a polarized distribution of PMCA.
Gradient in DAG is necessary for effective directed migration
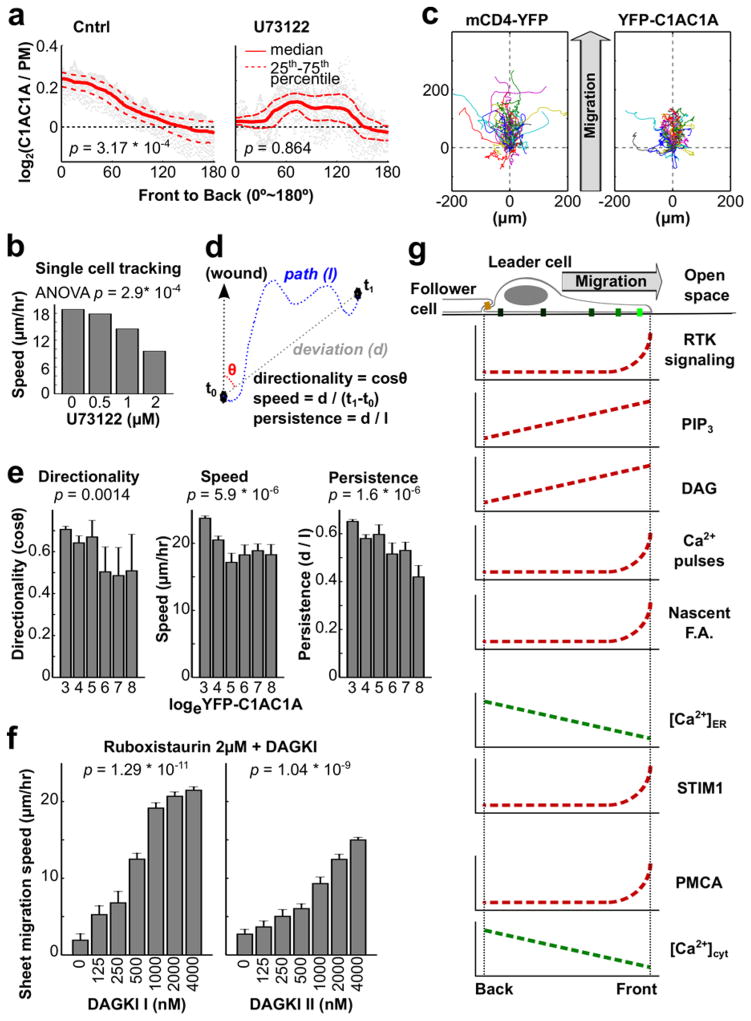
When we treated migrating cells with the PLC inhibitor U73122, the DAG gradient observed in Fig. 1 disappeared (Fig. 8a). To clarify if the DAG gradient was the result of a gradient of its precursor PI(4,5)P2, we monitored the distribution of PI(4,5)P2 using the PI(4,5)P2 sensor PH-PLCδ. Ratio imaging of PH-PLCδ and a PM marker showed no significant difference in PI(4,5)P2 distribution (Supplementary Fig. 1c,d), further supporting the interpretation from Fig. 1 that migrating leader cells establish a gradient in DAG from front to back by inducing locally higher PLC activity in the front.
Figure 8.
A phospholipase C (PLC)-induced gradient of diacylgycerol (DAG) controls cell motility and directionality. (a) Addition of the PLC inhibitor U73122 (1μM) suppressed the DAG sensor accumulation observed in the front of control cells (n = 64 cells for control; n = 22 cells for U73122). (b) The PLC inhibitor U73122 slowed down single cell speed of HUVEC as well as sheet migration speed in a dose-dependent manner (Supplementary Fig. 7a). U73122 was added to the cell sheets prior to time-lapse imaging (n = 2 experiments for each group). (c–e) Overexpression of YFP-C1AC1A reduced the measured migration parameters directionality, single cell speed and directional persistence. (c) Cell migration traces of 50 randomly chosen cells not expressing C1AC1A (left) and 50 cells overexpressing C1AC1A (right) are shown. The traces were aligned to start at the origin (0 μm, 0 μm) with the direction of the wound at the top. Cells expressing C1AC1A had relatively shorter traces and partially lost their orientation towards the open space. (d) Schematic of how the migration parameters speed, persistence and directionality shown in (e) were determined. (e) Statistical analysis of the change in directionality, speed, and persistence in response to YFP-C1AC1A overexpression were calculated for each migrating cell and correlated with binned levels of YFP-C1AC1A expression. Increasing YFP-C1AC1A expression resulted in decreasing directionality, speed, and persistence. Bars are mean ± SEM. (n = 1200 cells) (f) Partial inhibition of PKC by Ruboxistaurin decreased the rate of sheet migration. The reduction could be rescued by inhibiting DAG kinase using two types of inhibitors. Bars are mean ± SEM (n = 3 experiments per condition). (g) Schematic representation of the identified gradients in the Ca2+ and diacylglycerol signaling system. In Fig. 8a, p values were calculated by Student t test based on the ratio of the sensor / PM ratio in the front 10% region to that in the back 10% region of migrating cells. One-way ANOVA was used for Fig. 8b,e,f to determine the significance of difference between multiple groups.
However, treatment of HUVEC cells with the PLC inhibitor U73122 slowed the speed of migration (Fig. 8b and Supplementary Fig. 7a), which was opposite from the expected result since PLC inhibition suppresses Ca2+ signaling which we showed accelerates migration (Fig. 2a–e). This raised the possibility that the co-generated gradient in DAG has an important parallel role in enhancing the speed of cell migration. In support of such a role of DAG, we observed that high expression of the DAG binding translocation sensor YFP-PKC-C1AC1A caused a dose-dependent decrease in the speed and directionality of cell migration (Fig. 8c–e), likely caused by a dominant negative effect whereby the DAG biosensor partially sequesters PM DAG. Furthermore, application of two different DAG kinase inhibitors that increase the intracellular concentration of DAG40 both increased the speed and persistence of leader cells in a concentration-dependent manner (Supplementary Fig. 7b).
Since protein kinase C (PKC) is a main intracellular target of DAG and since PKCs have a role in regulating actin polymerization by phosphorylating actin regulators such as MARCKS, adducin, fascin, and ERM proteins41, we tested whether DAG enhances directed cell migration through PKC. Indeed, addition of the PKCβ inhibitor Ruboxistaurin decreased the sheet migration speed in a concentration-dependent manner (Supplementary Fig. 7c, the left most bars). Interestingly, at intermediate concentrations of ruboxistaurin, inhibition of DAG kinase restored migration (Fig. 8f and Supplementary Fig. 7c), consistent with the interpretation that DAG signals through PKCβ to promote directed cell migration. Together, this suggests that local RTK signaling in the front causes local PLC activation and DAG production, which in turn selectively activates PKCβ in the front.
Discussion
Our study shows that migrating endothelial leader cells establish a gradient in basal free Ca2+ levels with Ca2+ levels being lowest in the front. This is at least in part a result of higher localized PM Ca2+ pump activity in the front (Supplementary Fig. 8c and Fig. 7g,h) and is facilitated by a slow diffusion coefficient of Ca2+ (D=10 μm2/s, reduced by Ca2+ buffers)42 and a relatively extended length of HUVECs of x0 ~ 55 ± 16 μm (Supplementary Fig. 7d). Given these parameters, the diffusion-mediated equilibration time of x02/(2*D) ~ 150 sec is longer than the time required for Ca2+ pump-mediated Ca2+ extrusion, enabling a Ca2+ gradient to form (Fig. 1i,h and Fig. 6a). We further identified a parallel gradient of Ca2+ levels in the lumen of the ER (Fig. 5e,f) that we could explain by bFGF4,19,43 signaling being restricted towards the front of migrating leader cells and generating local PLC activation (Fig. 1a–f). We also showed that RTK and PLC signaling as well as Ca2+ gradients are largely absent in follower cells inside the sheet (Fig. 1b). The absence of significant RTK signaling both in the back of leader cells and in the front and back of follower cells suggests that cell-cell contacts may locally suppress receptor signaling.
Localized receptor signaling towards the front has two major consequences; it explains the localized cyclic IP3-triggered small local Ca2+ release pulses (Supplementary Fig. 1e,f) and the establishment of a DAG gradient (Fig. 1e). While little was previously known about the existence of gradients in DAG during migration, local DAG signals have been observed in pollen tube germination in plants44,45, after activation of T-cell receptors46–48, during phagocytosis49–52, and in neuronal synapses to regulate secretion of neuro-transmitters53,54. Finally, we show that STIM1 is activated locally in the front of migrating cells (Fig. 4) as a result of (i) directed STIM1 transport to the front mediated by microtubule plus end transport and (ii) lower ER Ca2+ levels in the front mediated by local RTK signaling. The resulting polarized SOC signaling provides a key mechanism to maintain the spatial and temporal dynamics of the Ca2+ signaling system.
The functional relevance of our study builds on previous findings on the roles of MLCK in membrane retraction12 and the role of STIM125–29 in regulating cell migration and adhesion. Here we investigated whether Ca2+, diacylglycerol, and STIM1 act in a polarized fashion and have synergistic roles in regulating directed migration. We show that microtubule plus-end-mediated transport of STIM1 to the front and that local STIM1 activation in the front is critical for its role in regulating migration. Our study argues that STIM1 likely acts indirectly on adhesion by enhancing local Ca2+ influx and by reloading ER Ca2+ stores in the front to permit local Ca2+ pulses to be cyclically triggered and MLCK to be locally activated. It is interesting that our study identified an opposite effect of SOC influx on cell migration compared to previous studies in cancer cells25,26. We were able to explain this discrepancy by showing that the relative strength of cell-matrix adhesion can decide whether SOC influx and additional adhesion decelerates or accelerates migration.
Our study further argues that the polarization of DAG that we discovered selectively activates the actin machinery in the front to enhance forward movement and to promote a more persistent polarized migration state. While several actin regulatory proteins, including classic and novel protein kinase C (PKC), protein kinase D, and RasGRP proteins45,55, are activated by DAG, our kinase inhibitor data suggests that the classical PKC pathway, which relies on combined Ca2+ and DAG signals, is significantly involved. Previous studies showed that PKCs can regulate migration56, likely in a synergistic fashion, by phosphorylating myosin57, by regulating the actin cytoskeleton41 or by the turn-over of integrin complexes58. Together with our finding of DAG polarization, this suggests that selective DAG signaling and PKC activation of actin regulators in the front promotes polarization and persistence of migration.
In summary, our study provides an integrated model of the spatial organization of the PLC-Ca2+-DAG-STIM1 signaling system. We show that core Ca2+ systems components are polarized in migrating endothelial leader cells including upstream receptor signaling, local Ca2+ pulses, DAG, Ca2+ pumps, basal cytosolic and ER Ca2+ levels as well as STIM1 distribution and STIM1-mediated store-operated Ca2+ influx (Fig. 8g and Supplementary Fig. 8). Together with the actin regulators Rac, Cdc42, RhoA and PIP3, this Ca2+ and diacylglycerol regulatory system dynamically controls polarization and persistence of migration as well as local adhesion and turning.
Methods
Cell culture
HUVEC (Lonza, C2519A) were cultured in EGM2 (Lonza, CC-3162) kit, as described previously12. HUVEC was tested to be free from mycoplasma by H.W.Y and A.H. H1299 cells (provided by Dr. Chang, Stanford University) were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin / streptomycin. For migration experiments, glass bottom 96-well plates (Greiner) were coated with fibronection (Life Technologies) at concentrations ranging from 0.31 μg to 20 μg/mL in 1X PBS (GIBCO) for 2 h. H1299 cells were plated at 50,000 /cm2 (for single cell tracking) or 100,000 /cm2 (for sheet migration assay) 12 hours prior to experiments.
Reagents and dyes
Fura-2/AM, Fluo-3/AM, thapsigargin, basic fibroblast growth factor (bFGF), and CellMask membrane dye were purchased from Invitrogen. BTP2 was purchased from Calbiochem. Ponatinib from Selleck, U73122 from Tocris, Diacylglycerol kinase inhibitor I, Diacylglycerol kinase inhibitor II, 3,4-DCB and lanthanum chloride (LaCl3) from Sigma Aldrich, Ruboxistaurin from AG Scientific, Caloxin 2A1 from Ana Spec. 2,4-DCB from Santa Cruz. mAb anti-phospho-tyrosine antibody was from Santa Cruz (PY20, sc-508) and was used at 1:500 dilution for immunofluorescence. Paxillin:FITC antibody was purchased from BD Biosciences (P13524-050 / 610053, Mouse IgG1 clone 349) and used as 1:100 for immunofluorescence. Anti-Phospho-myosin light chain 2 (Ser19, #3671, Rabbit) was purchased from Cell Signaling and used at 1:500 for immunofluorescence.
siRNA and DNA constructs
In vitro diced siRNA pools were generated as described previously12,17. In brief, a ~600 bp DNA fragment selected from the coding region of each gene was amplified by two nested polymerase chain reactions (PCR) using human cDNA as template and primers with T7 promoters. Sequences of primers for nested PCR are available in Supplementary Table 2. dsRNA was then generated by in-vitro transcription using T7 polymerase and treated with Giardia Dicer to produce fragments of 24–27 bp. A synthetic pool of siRNAs targeting STIM1 was purchased from Dharmacon (MU-011785-00-0002). mCD4-YFP and mCD4-CFP were generated by replacing GFP by YFP or CFP in mCD4-GFP using AgeI and AauI sites (New England BioLabs). The YFP-S1NN construct was derived form YFP-STIM1 by site-directed mutagenesis using primers S1NN-F: 5′-AGC CGA AAC ACA CGC AAT AAC CAC CTG GCT GGC AAG -3′ and S1NN-R: 5′-CTT GCC AGC CAG GTG GTT ATT GCG TGT GTT TCG GCT -3′. mCD4-GFP63, GFP-PMCA4b39, YFP-STIM17, PLCδ-PH64, tdimer2-lyn65, T1ER (an enhanced version of D1ER, Addgene plasmid #47928)34, GFP-Paxillin and mCitrine-Paxillin12 have been described. C1A domain of C1A-C1A-EYFP (labeled as YFP-C1AC1A) and CFP were from rat PKCγ 26–89 aa. We generated tandem C1A constructs to increase affinity of the sensor to diacylglycerol. The insert was synthesized by Epoch Life Science Inc. (Texas, USA) with EcoR1/ BamH1 restriction enzyme sites and ligated into pEYFP or pECFP N1 vector (Clontech). Two Gly-Ala linkers were inserted into the constructs. First Gly-Ala x6 is in between two C1A domains and second Gly-Ala repeats plus polylinker from the N1 vector bridges C1A and the fluorescent tag. The ER membrane marker CFP-ER was made by ligating the transmembrane domain of STIM1 to the N-terminus of CFP in pECFP-N1 (Clontech). The CFP-luminal ER marker was from Clontech. The ER-PM junction marker was also generated by ligating the transmembrane and polybasic domains17,66 of STIM1 into the pEYFP-N1 or pECFP-N1 (Clontech). GCaMP6s-CAAX was based on pGP-CMV-GCaMP6s22 (Addgene plasmid #40753). A CAAX motif (amino acids: KEKMSKDGKKKKKKSKTKCVIM) was added to GCaMP6s by PCR, cut with BamHI and NotI, and ligated into pEGFP-N1 (Clontech). Primers for GCaMP6s-CAAX: 5′-ATGGATCCGCCACCATGGGTTCTCATC-3′ (forward) and 5′-ATGCGGCCGCTTACATAATTACACACTTTGTCTTTGACTTCTTTTTCTTCTTTTTACCA TCTTTGCTCATCTTTTCTTTTGCTCCTGCTCCCTTCGCTGTCATCATTTGTA-3′ (reverse).
Transfection
HUVEC or H1299 cells were plated on collagen (30 μg/mL) or fibronectin (concentration described above)-coated 96-well plates (Costar) or 8-well LabTek chamber slides 12 hours prior to transfections. For siRNA transfection, 40 nM of in-vitro diced or synthetic siRNA was transfected using Lipofectamine RNAiMAX (Invitrogen) in OPTI-MEMI for 6–8 h, following the manufacturer’s protocol. Migration assays or live-cell imaging were performed 48 h after transfection. For DNA transfection, 20 ng/μl of maxi-prepped DNA was transfected using Lipofectamine 2000 (Invitrogen) in OPTI-MEMI for 2–4 hours according to the manufacturer’s protocol. Cells were analyzed 6–12 h after transfection.
Cell migration
Wound healing assays were done as previously described4,12,19. In brief, HUVECs were plated as monolayers of 31,250 / cm2 in collagen (30 μg/mL)-coated 96-well plates. A uniform cell-free band was generated at the center of each well using a custom-built scratch tool12 for 96-well plates or a pipet tip and a ruler for LabTek chambers. Wounded monolayers were washed 3 times using 1X PBS before the addition of imaging medium. Sheet migration speed was measured by following the advancement of the cell sheet boundaries using cells labeled with CellMask plasma membrane dye, or by tracking individual cells stained with Hoechst 33342. As imaging medium for HUVEC, either EGM2, SFM, or SFM supplemented with bFGF (20ng/ml)(all buffered with 20 mM HEPES) was used as noted. H1299 cells were imaged in full growth medium (buffered with 20 mM HEPES).
Ca2+ Measurements
Fura-2/AM or GCaMP6s were used to measure cytosolic Ca2+ levels. Fluo-3/AM was used in UV photolysis experiments when Fura-2/AM could not be used. T1ER33,34, an enhanced version of D1ER construct, was used to measure luminal ER Ca2+ (Addgene plasmid #47928).
GCaMP6s-CAAX or T1ER were transiently transfected into HUVEC one day before the experiments. Dye loading was done as follows. Fura-2/AM and Fluo-3/AM were loaded at 0.5–2 μM with 0.1% Pluronic F127 (Invitrogen) and 1 μM probenecid (Invitrogen) in endothelial serum-free medium (GIBCO) at 37°C for 30 minutes. The cytosolic Ca2+ was then measured and calibrated as described previously12. In brief, images were taken using 20X Plan Fluar (ImageXpress microscope) or 40X NeoFluar (Nipkow microscope) objectives and epifluorescence illumination, with excitation wavelengths of 340 nm and 380 nm for Ca2+-bound and Ca2+-free dyes, respectively, and the emission wavelength of 510 nm. Relative Ca2+ levels were determined based on the ratio between the images acquired using 340 nm 380 nm excitation. For calibration, a set of solutions with Fura-2 and various concentrations of Ca2+ was prepared (10–1000 nM) by appropriately mixing Ca2+EGTA and K2EGTA, as previously described12. The calibration curve is shown in Supplementary Fig. 1g; the estimated Kd of Fura-2 was 101 nM. Because the Kd of Fura-2 is affected by ionic strength, sub-cellular localization, protein binding and temperature67,68, the actual intracellular Kd may be higher. Therefore, we included estimates for absolute intracellular [Ca2+] using also the Kd of 145 nM from the manufacturer59, and 224 nM from reference60. (Supplementary Table 1) Depending on the Kd values, the estimated basal cytosolic Ca2+ level falls between 45–105 nM in HUVEC, compatible with the measurement from previous reports61,62.
As indicated above, there are challenges for measuring absolute intracellular [Ca2+] in HUVEC. Therefore, instead of using absolute [Ca2+] values, we included relative units (R.U.) by normalizing [Ca2+] relative to the measured average basal cytosolic level. 1 R.U. means the Ca2+ level is close to the average cytosolic [Ca2+], while 2 R.U. means the Ca2+ level is approximately twice as high as the basal cytosolic [Ca2+]. In some cases we used the Fura-2 340/380 ratio where appropriate (Fig. 6d,e and Supplementary Fig. 1e–g, Supplementary Fig. 4c) or arbitrary signal intensity values for the control experiments using Fluo-3 (Fig. 7a,b).
Live-cell imaging
For cell tracking, cells were plated in 96-well plates and automated microscopy was performed using a ImageXpress 5000A (Molecular Devices), equipped with a temperature control unit, 4X S Fluor & 20X Plan Fluar objectives (Nikon) and a 300W xenon arc lamp. Cell nuclei were labeled using 100 ng/ml Hoechst 33342 for 1h at 37°C. As imaging medium, EGM2 supplemented with 20 mM HEPES and 1 mM of L-ascorbic acid was used. Plates were sealed and images captured every 15–30 minutes for 4–12 hours. For measuring local Ca2+ signals and DAG signals, cells were imaged using a custom-assembled spinning disc confocal/epifluorescence microscope system built around a Zeiss Axiovert 200M microscope. The system was outfitted with an automated x–y stage (ASI) and a custom-built environmental chamber (Haison). The confocal light path was equipped with three lasers, 442 nm (He-Cd, 300 mW, Kimmon), 514 nm (Ar-Kr, 400 mW, Melles Griot), and 593.5 nm (DPSS, 100 mW, CNI), a CCD camera (CoolSNAP HQ, Photometrics), and appropriate excitation and emission filters. The epifluorescence light path was equipped with a 100W HBO lamp, a CCD camera (CoolSNAP HQ, Photometrics) and appropriate filtersets. 40X 1.3 NA or a 63X 1.4 NA Plan Apochromat objectives (Zeiss) were used and the system was controlled using μManager69. The cells were plated on the 8-well LabTek chamber slide with 40 mM Hepes, 1 mM L-ascorbic acid and 1 mM probenecid added to the EGM2 medium. Images were taken by 2 × 2 binning in 37°C every 20 seconds for 6–15 minutes depending on the specific experiments. One pixel equals to 2.5 μm, 0.5 μm or 0.31 μm when 4X, 20X or 40X objectives were used in the ImageXpress or Nipkow microscope.
Image processing
All images were processed using Matlab 2010b (Mathworks) as described previously12. In brief, local background subtraction was applied to every image as (new value of each pixel) = (old value of each pixel) – (the median value (for punctate analysis) or the 5th percentile value (for other analysis) of the pixels within 40 μm of that pixel). Cell borders and the nuclei were determined by the modified Otsu’s method70 for thresholds using the signals from the membrane markers and Hoechst, respectively. For STIM1 & ER images as shown in Fig. 4a and Supplementary Fig. S4a, and for FRET images using T1ER as shown in Fig. 5e and Supplementary Fig. 4f, the mask of ER from a single image was determined by summation of the YFP / FRET and CFP image. The CAAX image as shown in Supplementary Fig. 4e was not used to generate the mask because ER signals were not present within 5μm of the cell boundary. Cell tracking was conducted by searching the nearest neighbor surrounding the specific nucleus. Specifically, the speed of sheet migration was determined by the advancement of the boundaries or the nuclei of leader cells in the front of the sheets. The parameters of individual migrating cells (speed, persistence, directionality, etc.) were then calculated based on the migration track of each cell. To determine the temporal changes of GFP-paxillin signal intensities after drug treatments (Fig. 2h and Supplementary Fig. 2h), a 10 μm × 10 μm square box was chosen at the leading edge of migrating cells. The average signal intensity of each time point was recorded accordingly as described previously12. To quantify the gradients of STIM1 (Fig. 4b,c,e, Fig. 5b–d and Supplementary Fig. 4d), cytosolic or ER luminal Ca2+ (Fig. 1j, 4h and Fig. 6a), DAG (Fig. 1f), PIP3 (Fig. 1d) and PI(4,5)P2 (Supplementary Fig. 1b–d), a ring covering the outer 20% area of the specific mask (ER or plasma membrane where applicable) was used to calculate the ratio in the front (0°), the back (180°), and the sides (between 0° and 180°). The local Ca2+ fluctuations shown in Fig. 1h and Supplementary Fig. 1h were calculated by averaging the absolute difference between local Ca2+ peaks and the median cytosolic level of each cell, followed by averaging the results from all cells. To compare the signal differences of phospho-tyrosine (Fig. 1a,b) or Ca2+ (Fig. 6,7 and Supplementary Fig. 5, Supplementary Fig. 6) in an individual cell, the front and back signal were calculated by averaging the signal intensities at the front 10% and the back 10% of the specific mask (ER or plasma membrane where applicable) along the direction of sheet migration.
Annotated data analysis codes were attached as the Supplementary Note, which includes Supplementary Code 1 (basic processing), Supplementary Code 2 (gradient measurement) and Supplementary Code 3 (nuclear tracking).
Statistical tests
Multi-site imaging was done by either an automated microscope for images using 4X or 20X objectives, or a Nipkow spinning disk confocal microscope for images using 40X or 63X objectives. For sheet migration assays, 3–4 duplicates were performed for each condition depending on available wells and time-lapse routines in the 96-well plates. For imaging using the Nipkow microscope, 8 independent areas were taken over each of the 2–4 wells per condition. Therefore, although the sample sizes were not estimated before the experiments, they were generally larger than needed for adequate statistical results. For images from the ImageXpress or Nipkow microscope, every live cell at the border of the wound was used for quantitative analysis of fluorescent signals or migration parameters throughout the experiments. (To avoid too many traces in Fig. 8c, 50 cells were randomly selected from each group for demonstration, using the rand(.) function in Matlab. All cells were used for quantitative analysis as shown in Fig. 8e.) All Bars in the figures are ± standard errors of the mean (SEM). All statistical tests were performed using Matlab (Mathworks), as described preciously12. In brief, Student’s t test was used to compare the difference between two groups while one-way ANOVA was used for three or more groups. The statistical tests were chosen based on the assumption that the values were normally distributed, which was validated as shown in Supplementary Fig. 7d. In addition, based on the Central Limit Theorem, the mean values of samples will approximate normally distribution with the increase of sample size, justifying our choice of statistical tests. Specific p values were provided on the figure panels and p < 0.05 was considered statistically significant.
Repeatability of experiments
In main and supplementary figures, representative images were presented as Fig 1a,c,e,g,i, Fig 2a,f,h, Fig 3c,e, Fig 4a, Fig. 5a,e, Fig 7a,g, Supplementary Fig 1e,f, Supplementary Fig 2a,f,h, Supplementary Fig 4a,e,f,g, Supplementary Fig 5a,c and Supplementary Fig 6d,e. Each representative image is complemented by data quantification with biological repeats, the number of which is mentioned in the figure legends. Throughout the paper, experiment were generally repeated at least three times, except those repeated twice including Fig. 2b,g, Fig. 3d,f,g, Fig. 5f,i, Fig. 6a, Fig. 7d,e,f,h, Fig. 8b,e,f, Supplementary Fig. 3, Supplementary Fig. 5b,d, Supplementary Fig. 6a–c and Supplementary Fig. 7a.
Supplementary Material
Acknowledgments
We thank Drs. S.R. Collins, M. Galic, and R. Wollman for technical support and discussions, Dr. S. Bandara for the modified T1ER construct, Dr. N. Borghi for the paxillin construct, Dr. X. Ge for the CD4 construct, Dr. C.J. Lin for H1299 cells, Dr. E.E. Strehler for the PMCA constructs, and A. Winans for critical reading of the manuscript. The research was supported by a Stanford Graduate Fellowship (F.T.) and the NIGMS (T.M.)
Footnotes
Author Contributions
F.T. conceived, designed and performed experiments, analyzed the data and wrote a draft of the manuscript. A.S. developed the DAG sensor and helped with the diacylglycerol-related experiments and Western blotting. H.W.Y. repeated and validated experiments with the lipid sensors, and compared gradients in leader and follower cells. A.H. helped generating the paxillin constructs, prepared cells stably expressing reference membrane markers, and helped writing the manuscript. S.C. developed the ER-PM and ER-membrane markers. S.M. developed the membrane-targeted version of GCaMP6s and helped with Ruboxistaurin experiments. T.M. conceived the project together with F.T., helped interpret the data, and write the manuscript.
References
- 1.Vorotnikov AV. Chemotaxis: movement, direction, control. Biochemistry Mosc. 2011;76:1528–1555. doi: 10.1134/S0006297911130104. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Huang NF, Hsu S. Mechanotransduction in endothelial cell migration. J Cell Biochem. 2005;96:1110–1126. doi: 10.1002/jcb.20614. [DOI] [PubMed] [Google Scholar]
- 3.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 4.Vitorino P, Hammer M, Kim J, Meyer T. A steering model of endothelial sheet migration recapitulates monolayer integrity and directed collective migration. Mol Cell Biol. 2011;31:342–350. doi: 10.1128/MCB.00800-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giannone G, et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 6.Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesionsite formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnette DT, et al. A role for actin arcs in the leading-edge advance of migrating cells. Nat Cell Biol. 2011;13:371–381. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tkachenko E, et al. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat Cell Biol. 2011 doi: 10.1038/ncb2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans JH, Falke JJ. Ca2+ influx is an essential component of the positive-feedback loop that maintains leading-edge structure and activity in macrophages. Proc Natl Acad Sci USA. 2007;104:16176–16181. doi: 10.1073/pnas.0707719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei C, et al. Calcium flickers steer cell migration. Nature. 2009;457:901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai FC, Meyer T. Ca2+ pulses control local cycles of lamellipodia retraction and adhesion along the front of migrating cells. Curr Biol. 2012;22:837–842. doi: 10.1016/j.cub.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franco SJ, et al. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- 14.Brundage RA, Fogarty KE, Tuft RA, Fay FS. Calcium gradients underlying polarization and chemotaxis of eosinophils. Science. 1991;254:703–706. doi: 10.1126/science.1948048. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert SH, Perry K, Fay FS. Mediation of chemoattractant-induced changes in [Ca2+]i and cell shape, polarity, and locomotion by InsP3, DAG, and protein kinase C in newt eosinophils. J Cell Biol. 1994;127:489–503. doi: 10.1083/jcb.127.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 17.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitorino P, Meyer T. Modular control of endothelial sheet migration. Genes Dev. 2008;22:3268–3281. doi: 10.1101/gad.1725808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gozgit JM, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–699. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 21.Lamalice L, Boeuf FL, Huot J. Endothelial Cell Migration During Angiogenesis. Circulation Research. 2007;100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 22.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zitt C, et al. Potent inhibition of Ca2+ release-activated Ca2+ channels and Tlymphocyte activation by the pyrazole derivative BTP2. J Biol Chem. 2004;279:12427–12437. doi: 10.1074/jbc.M309297200. [DOI] [PubMed] [Google Scholar]
- 24.Kasturi R, Vasulka C, Johnson JD. Ca2+, caldesmon, and myosin light chain kinase exchange with calmodulin. J Biol Chem. 1993;268:7958–7964. [PubMed] [Google Scholar]
- 25.Chen Y-F, et al. Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1103315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S, Zhang JJ, Huang XY. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell. 2009;15:124–134. doi: 10.1016/j.ccr.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Potier M, et al. Evidence for STIM1- and Orai1-dependent store-operated calcium influx through ICRAC in vascular smooth muscle cells: role in proliferation and migration. FASEB J. 2009 doi: 10.1096/fj.09-131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bisaillon JM, et al. Essential role for STIM1/Orai1-mediated calcium influx in PDGFinduced smooth muscle migration. Am J Physiol, Cell Physiol. 2010;298:C993–1005. doi: 10.1152/ajpcell.00325.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schafer C, Rymarczyk G, Ding L, Kirber MT, Bolotina VM. Role of molecular determinants of store-operated Ca2+ entry (Orai1, phospholipase A2 group 6 and STIM1) in focal adhesion formation and cell migration. J Biol Chem. 2012 doi: 10.1074/jbc.M112.407155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 31.Grigoriev I, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honnappa S, et al. An EB1-Binding Motif Acts as a Microtubule Tip Localization Signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 33.Abell E, Ahrends R, Bandara S, Park BO, Teruel MN. Parallel adaptive feedback enhances reliability of the Ca2+ signaling system. PNAS. 2011;108:14485–14490. doi: 10.1073/pnas.1018266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandara S, Malmersjö S, Meyer T. Regulators of calcium homeostasis identified by inference of kinetic model parameters from live single cells perturbed by siRNA. Sci Signal. 2013;6:ra56. doi: 10.1126/scisignal.2003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, Dawson AP. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci USA. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smallwood JI, Gügi B, Rasmussen H. Regulation of erythrocyte Ca2+ pump activity by protein kinase C. J Biol Chem. 1988;263:2195–2202. [PubMed] [Google Scholar]
- 37.Pérez-Gordones MC, Lugo MR, Winkler M, Cervino V, Benaim G. Diacylglycerol regulates the plasma membrane calcium pump from human erythrocytes by direct interaction. Arch Biochem Biophys. 2009;489:55–61. doi: 10.1016/j.abb.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 38.James P, et al. Modulation of erythrocyte Ca2+-ATPase by selective calpain cleavage of the calmodulin-binding domain. J Biol Chem. 1989;264:8289–8296. [PubMed] [Google Scholar]
- 39.Chicka MC, Strehler EE. Alternative splicing of the first intracellular loop of plasma membrane Ca2+-ATPase isoform 2 alters its membrane targeting. J Biol Chem. 2003;278:18464–18470. doi: 10.1074/jbc.M301482200. [DOI] [PubMed] [Google Scholar]
- 40.De Chaffoy de Courcelles DC, Roevens P, Van Belle H. R 59 022, a diacylglycerol kinase inhibitor. Its effect on diacylglycerol and thrombin-induced C kinase activation in the intact platelet. J Biol Chem. 1985;260:15762–15770. [PubMed] [Google Scholar]
- 41.Larsson C. Protein kinase C and the regulation of the actin cytoskeleton. Cell Signal. 2006;18:276–284. doi: 10.1016/j.cellsig.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Smith G, MacQuaide N. Cytoplasmic versus Intra-SR: the Battle of the Ca2+ Diffusion Coefficients in Cardiac Muscle. Biophys J. 2008;95:1005–1006. doi: 10.1529/biophysj.108.133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nature Reviews Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 44.Arisz SA, Testerink C, Munnik T. Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta. 2009;1791:869–875. doi: 10.1016/j.bbalip.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Almena M, Mérida I. Shaping up the membrane: diacylglycerol coordinates spatial orientation of signaling. Trends Biochem Sci. 2011;36:593–603. doi: 10.1016/j.tibs.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Carrasco S, Merida I. Diacylglycerol-dependent binding recruits PKCtheta and RasGRP1 C1 domains to specific subcellular localizations in living T lymphocytes. Mol Biol Cell. 2004;15:2932–2942. doi: 10.1091/mbc.E03-11-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spitaler M, Emslie E, Wood CD, Cantrell D. Diacylglycerol and protein kinase D localization during T lymphocyte activation. Immunity. 2006;24:535–546. doi: 10.1016/j.immuni.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Quann EJ, Merino E, Furuta T, Huse M. Localized diacylglycerol drives the polarization of the microtubule-organizing center in T cells. Nat Immunol. 2009;10:627–635. doi: 10.1038/ni.1734. [DOI] [PubMed] [Google Scholar]
- 49.May RC, Machesky LM. Phagocytosis and the actin cytoskeleton. J Cell Sci. 2001;114:1061–1077. doi: 10.1242/jcs.114.6.1061. [DOI] [PubMed] [Google Scholar]
- 50.Stephens L, Ellson C, Hawkins P. Roles of PI3Ks in leukocyte chemotaxis and phagocytosis. Curr Opin Cell Biol. 2002;14:203–213. doi: 10.1016/s0955-0674(02)00311-3. [DOI] [PubMed] [Google Scholar]
- 51.Scott CC, et al. Phosphatidylinositol-4,5-bisphosphate hydrolysis directs actin remodeling during phagocytosis. J Cell Biol. 2005;169:139–149. doi: 10.1083/jcb.200412162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Botelho RJ, et al. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol. 2000;151:1353–1368. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Jong APH, Verhage M. Presynaptic signal transduction pathways that modulate synaptic transmission. Curr Opin Neurobiol. 2009;19:245–253. doi: 10.1016/j.conb.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Kim K, Yang J, Kim E. Diacylglycerol kinases in the regulation of dendritic spines. J Neurochem. 2010;112:577–587. doi: 10.1111/j.1471-4159.2009.06499.x. [DOI] [PubMed] [Google Scholar]
- 55.Carrasco S, Mérida I. Diacylglycerol, when simplicity becomes complex. Trends Biochem Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 56.Rosse C, et al. PKC and the control of localized signal dynamics. Nature Reviews Molecular Cell Biology. 2010;11:103–112. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- 57.Ludowyke RI, et al. Phosphorylation of nonmuscle myosin heavy chain IIA on Ser1917 is mediated by protein kinase C beta II and coincides with the onset of stimulated degranulation of RBL-2H3 mast cells. J Immunol. 2006;177:1492–1499. doi: 10.4049/jimmunol.177.3.1492. [DOI] [PubMed] [Google Scholar]
- 58.Woods A, Couchman JR. Protein kinase C involvement in focal adhesion formation. J Cell Sci. 1992;101 (Pt 2):277–290. doi: 10.1242/jcs.101.2.277. [DOI] [PubMed] [Google Scholar]
- 59.Fluorescent Calcium Indicators - 154pdf. at < http://probes.invitrogen.com/media/publications/154.pdf>.
- 60.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 61.Huang AJ, et al. Endothelial cell cytosolic free calcium regulates neutrophil migration across monolayers of endothelial cells. J Cell Biol. 1993;120:1371–1380. doi: 10.1083/jcb.120.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ikeda M, et al. Separate analysis of nuclear and cytosolic Ca2+ concentrations in human umbilical vein endothelial cells. J Cell Biochem. 1996;63:23–36. doi: 10.1002/(sici)1097-4644(199610)63:1<23::aid-jcb2>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 63.Hillman RT, et al. Neuropilins are positive regulators of Hedgehog signal transduction. Genes Dev. 2011;25:2333–2346. doi: 10.1101/gad.173054.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stauffer TP, Ahn S, Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- 65.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Groden DL, Guan Z, Stokes BT. Determination of Fura-2 dissociation constants following adjustment of the apparent Ca-EGTA association constant for temperature and ionic strength. Cell Calcium. 1991;12:279–287. doi: 10.1016/0143-4160(91)90002-v. [DOI] [PubMed] [Google Scholar]
- 68.Petr MJ, Wurster RD. Determination of in situ dissociation constant for Fura-2 and quantitation of background fluorescence in astrocyte cell line U373-MG. Cell Calcium. 1997;21:233–240. doi: 10.1016/s0143-4160(97)90047-6. [DOI] [PubMed] [Google Scholar]
- 69.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010;Chapter 14(Unit14.20) doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Transactions on Systems, Man and Cybernetics. 1979;9:62–66. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.