Abstract
Each membrane fusion event along the secretory and endocytic pathways requires a specific set of SNAREs to assemble into a 4-helical coiled-coil, the so-called trans-SNARE complex. Although most SNAREs contribute one helix to the trans-SNARE complex, members of the SNAP-25 family contribute two helixes. We report the characterization of the Drosophila homologue of SNAP-29 (dSNAP-29), which is expressed throughout development. Unlike the other SNAP-25 like proteins in fruit fly (i.e., dSNAP-25 and dSNAP-24), which form SDS-resistant SNARE complexes with their cognate SNAREs, dSNAP-29 does not participate in any SDS-resistant complexes, despite its interaction with dsyntaxin1 and dsyntaxin16 in vitro. Immunofluorescence studies indicated that dSNAP-29 is distributed in various tissues, locating in small intracellular puncta and on the plasma membrane, where it associates with EH domain-containing proteins implicated in the endocytic pathway. Overexpression and RNAi studies suggested that dSNAP-29 mediates an essential process in Drosophila development.
Introduction
Vesicular fusion requires SNAREs (SNAP receptors) located on the transport vesicle (e.g., VAMP/synatobrevin) to interact with cognate SNAREs from the target membrane (e.g., syntaxin and SNAP-25) [1], [2]. Such interaction leads to the formation of a stable 4-helical trans-SNARE complex, whose subsequent disassembly after fusion depends on the actions of an ATPase NSF (N-ethyl-maleimide-sensitive factor) and its cofactor SNAP (soluble NSF attachment protein) [3]. The notion that each trafficking event requires a unique set of SNAREs [4] has been supported by the identification of many SNAREs within the syntaxin and VAMP/synaptobrevin families, with rather unique but sometimes overlapping distribution patterns along the secretory and endocytic pathways. SNAP-25 like proteins are unique in that each contributes two helices to the trans-SNARE complex. The virtually completed human, worm and fly genomes all seem to suggest just 3 or at most 4 members in the SNAP-25 family: SNAP-25, SNAP-23, SNAP-29 [5], and SNAP-47 [6].
SNAP-25 is expressed almost exclusively in the brain and required mainly at the plasma membrane of the nerve terminals [7], [8]. Botulinum neurotoxin A and E cleave the C-terminal domain of SNAP-25, causing a partial inhibition of secretion in the neuronal system [9]. Consistent with this, overexpression of a truncated form of SNAP-25, which lacks the last nine residues at the C-terminus, led to a reduction of exocytosis in chromaffin cells [10]. SNAP-23 appears to be the functional isoform of SNAP-25 in non-neuronal tissues [11], [12]. Like SNAP-25, SNAP-23 localizes mainly to the plasma membrane and is involved in exocytosis from both polarized and nonpolarized cell types [13], [14], [15], [16], [17], [18]. Overexpression of SNAP-24, a putative Drosophila orthologue of SNAP-23, rescues the synaptic transmission defect and lethality caused by the loss of SNAP-25 in the fruit fly [19]. Similarly, ubiquitously-distributed SNAP-47 is able to substitute for SNAP-25 in SNARE complex formation with the neuronal SNARE syntaxin 1 and VAMP/synaptobrevin 2 [6]. Unlike SNAP-25, SNAP-47 is enriched in synaptic vesicle fractions of the neuron rather than in the plasma membrane [6] and is required in postsynaptic rather than presynaptic exocytosis [20].
In contrast to SNAP-23, SNAP-25, and SNAP-47, which are mainly to support membrane fusion at the cell surface, SNAP-29 appears to participate in a wide range of fusion events in the cell. Mammalian SNAP-29 is distributed on multiple membranes including Golgi, endosomes, and lysosomes where it can interact with multiple syntaxins [21] [22], [23]. In mast cells, SNAP-29 is recruited to E. coli-containing phagosomes to facilitate phagocytosis and the subsequent killing of the bacteria [24]. In fibroblast cells derived from CEDNIK (Cerebral Dysgenesis, Neuropathy, Ichthyosis and Keratoderma) patients, the loss of functional SNAP-29 impaired endocytic recycling and cell motility without affecting secretion [25]. Depletion of SNAP-29 in C. elegans blocks secretion in both polarized and nonpolarized cells [26], which leads to cytokinesis defects and sterility [27]. In addition, SNAP-29 inhibits synaptic transmission when introduced into cultured presynaptic superior cervical ganglion neurons, by preventing SNARE complex disassembly [28]. More recently, SNAP-29 has been functionally linked to starvation-induced autophagy in both man and fruit fly [29], [30]. These studies suggest that SNAP-29 mediated fusion is required in diverse biological processes in eukaryotes.
To distinguish the unique function of each SNAP-25 related molecule in membrane traffic, binding partners beyond the SNARE super-family are being identified and studied. To-date, mammalian SNAP-25 has been shown to interact with SNIP (SNAP-25-interacting protein), Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate), and intersectin. SNIP is a brain-specific cytoskeleton-associating protein and may serve as a linker to connect SNAP-25 with the submembranous cytoskeleton [31]. Hrs prefers to localize to the endosomal membrane which generates PI(3)P [32] and has recently been assigned a clear role in the maturation of multi-vesicular body (MVB) [33], [34], [35]. However, Hrs also appears to be implicated in calcium-regulated secretion [36], [37]. Intersectin contains two Eps 15 homology (EH) domains and five SH3 domains, which allow the protein to associate with the endocytic machinery [38]. The observation that intersectin binds SNAP-25 and SNAP-23 has led to the postulation that it may function to couple the endocytic membrane traffic to exocytosis. Interestingly, mammalian SNAP-29 has also been found to interact with a EH domain-containing protein EHD1 [39] [25], although the physiological significance of the interaction has yet to be examined.
Here we report the characterization of the SNAP-29 orthologue in Drosophila, which was initially identified via a yeast 2-hybrid screen using dSNAP as bait. dSNAP-29 is ubiquitously expressed throughout the fly life cycle, interacting with multiple proteins including dSNAP, dsyntaxin1, dsyntaxin 16, dEHD1, and DAP160. Overexpression and RNAi studies show that the function of dSNAP-29 is essential in Drosophila.
Materials and Methods
Cloning and subcloning of Drosophila SNAP-29
A partial cDNA of Drosophila SNAP-29 was cloned by a yeast 2-hybrid screen using dSNAP as bait [40]. A blast search of Flybase allowed the identification of an apparent full-length EST clone LD17127, which was obtained from Research Genetics. Full-length dSNAP29 or that lacking the C-terminal 25 amino acids (dSNAP29ΔC)was amplified by PCR with primers: 5′-GAATTCCATGGCCCATAACTACCTGCAGC and 5′-AGATCTCGAGTCACTTCTTCAGAAGCTTGCTC, or 5′-AGATCTCGAGTCAGTTATCCAGCAATTCATTTTGCG, and then subcloned into the Sma I site of pBluescript SK+ vector (Stratagen) for sequencing
dSNAP-29 was then subcloned into pGEX-KG to generate GST-dSNAP-29 for binding assays, into pQE-30 to generate His-dSNAP-29 for antibody production, and into pRmHa-3-myc for transient transfection. dSNAP-29 and dSNAP-29ΔC were each inserted into the polycloning site of pUAST for microinjection.
The sequences of SNAP-25-related proteins were obtained from GenBank™. Multiple sequence alignments were performed using the ClustalW program (using default parameters), which also generates a phylogenetic tree.
Antibody preparation and immunostaining
Peptide CKQNKDMSKLLKK (the N-terminal C was added to faciliate crosslinking) was synthesized (University of Toronto) and coupled to maleimide-activated carrier protein keyhole limpet hemocyanin (Pierce), which was then used to immunize rabbits. To purify the antibody, the peptide was immobilized onto SulfoLink Coupling Gel (Pierce), incubated overnight with the antiserum and then washed with 2 M NaCl in PBS, 0.1 M boric acid (pH 9.1), and PBS (pH 4.5) consecutively. The purified antibody was eluted with 20 mM glycine (pH 2.5) and neutralized immediately with 100 mM Tris (pH 8.5).
To prepare antibody against the full-length protein, dSNAP-29 was subcloned into the pQE-30 vector (Qiagen) and expressed in E. coli strain BSJ72. His-dSNAP-29 was purified on Ni+ beads (Qiagen) for rabbit immunization. Around 150 μg of His-dSNAP-29 were run on an SDS-polyacrylamide gel, transferred onto nitrocellulose filter, and then isolated for antibody purification.
Immunofluorescence studies of salivary glands, imaginal discs, and transfected S2 cells were carried out as described ([41]. On some occasions, S2 cells were fixed with cold methanol for 5 min on ice. Overnight collections of Oregan R embryos were dechorinated with 50% bleach for 2 min, shaked in a scintillation vail with 50% n-heptane and 50% fixation solution (4% paraformaldehyde in 0.1 M Na3PO4, pH 7.2) for 30 min, washed with methanol 3 times, dehydrated with ethanol (for storage at −80 °C), and then rehydrated before staining. Images of salivary glands, S2 cells embryos were captured by a Zeiss LSM510 confocal microscope.
Fractionation, binding assays, immunoprecipitation and western blotting
To determine the membrane association of dSNAP-29, the membrane fraction of an adult fly lysate was separated from the soluble fraction, and treated as described [41]. Subcellular fractionation was described in [42].
Binding assays measuring the direct interaction between recombinant dSNAP-29 and dSNAP were performed with 200 ng of GST-dSNAP-29 (see [41] for details). To identify potential binding partners of dSNAP-29, approximately 200 million Schneider S2 cells were lysed in 0.5 mL of PLC or NP40 lysis buffer. Supernatant supplemented with 0.1% gelatin and 0.1% BSA was pre-cleared with glutatione beads at 4 °C for 1 hr, before incubation with 0.5 mg of GST-dSNAP-29, GST-dSNAP-25, or GST (each coupled with 120 μL of glutathione beads). Following extensive washes with 50 mM HEPES (pH 7.5), 150 mM NaCl, and 2 mM EDTA, agarose beads were boiled with 120 μL of 2×SDS sample buffer which was then subject to SDS-PAGE and western blot analysis. Binding assays measuring the direct interaction between recombinant dSNAP-29 and dSNAP were performed with 200 ng of GST-dSNAP-29 (see [41] for details).
For immunoprecipitation studies, overnight collections of Oregan R embryos (1.5 mL) were homogenized in 1 mL of homogenization buffer (HKA, 1 mM PMSF, 2 mM benzimidine, 2 μg/mL leupeptin, 2 μg/mL pepstatin, 5 mM EDTA). Following centrifugation at 600 g for 10 min, the supernatant was incubated with 4% Triton X-100 for 1 hr at 4 °C and then subject to a second centrifugation at 13,000 g for 10 min. About 0.5 mL of the supernatant was then incubated with 2.5 μg of affinity purified anti-SNAP-29 or pre-immune serum that had been cross-linked to protein A beads. Following a 2 hr incubation at 4 °C, the beads were washed extensively with HKA, 150 mM NaCl, 2% Triton X-100 before extraction with 45 μL of 2× SDS sample buffer containing 10 mM NEM. 10 μL were loaded each time for SDS-PAGE and subsequent western blot analysis.
To examine SDS-resistant SNARE complexes, comtTP7 or wild-type adult flies were incubated at 37 °C for 10 min, after which the heads were collected and homogenized directly in 1% SDS lysis buffer. Half of the amount of each sample was boiled while the other half was not in order to preserve the complexes.
To measure the relative protein levels on the western blot, the chemi-luminescence from each band was captured directly by the FluorChem™ Imaging System (Alpha Innotech).
Double strand RNA interference
For RNAi on cultured S2 cells, dSNAP29 cDNA was amplified from pQE-30-dSNAP29 by PCR using primers carrying the minimal T7 promoter: 5′-TTAATACGACTCACTATAGGGAGAGGATCGCATCACCATCACC and 5′-TTAATACGACTCACTATAGGGAGATCTATCAACAGGAGTCCAAGC. The PCR product was purified from agarose gel (QIAEX II gel extraction kit, Qiagen), and then used to synthesize RNA with an in vitro transcription kit, MEGAscript (Ambion). The dsRNA was annealed by incubation at 65 °C for 30 min followed by gradual cooling to room temperature. For each RNAi experiment, 15 μg of dsRNA were incubated with 1 million S2 cells for 5 to 6 days [43].
To introduce dsRNA into Drosophila, dSNAP-29 cDNA from pGEX-dSNAP-29 was inserted into pUAST [44] via Xho I and Xba I sites, such that the full-length cDNA is inverted. dSNAP-29ΔC was subsequently subcloned into EcoR I and Bgl II sites upstream of the inverted full-length cDNA. This gave rise to a tail-to-tail repeat of dSNAP-29ΔC (775 bp) located downstream of the GAL4-dependent upstream activating-sequence (UAS), which was used for microinjection to create transgenic lines.
Fly stocks and genetic studies
Stocks were maintained at room temperature on standard cornmeal agar medium unless otherwise indicated. Visible markers and balancer chromosomes have been previously described [45]. Transgenic flies UAS-dSNAP-29, UAS- dSNAP-29ΔC, and UAS-dSNAP-29RNAi were created by standard P-element-mediated transformation [46]. Individual UAS lines were crossed with GAL4 lines to activate the ectopic expression of various forms of dSNAP-29.
Results
Temporal and spatial distribution of dSNAP-29/ubisnap
Drosophila SNAP-29 (CG11173) was identified in a yeast 2-hybrid screen for proteins that interact with Drosophila SNAP [40]. Like the other two SNAP-25 like proteins in Drosophila (i.e., dSNAP-24 and dSNAP-25), dSNAP-29 possesses two terminal alpha-helical SNARE domains that are predicted to form coiled-coils (Fig. 1A). However, dSNAP-29 lacks a cysteine cluster and a QPXR(V/I) motif in between the SNARE domains, which are required for anchoring SNAP-25/23 isoforms to the plasma membrane [47], [48], [49]. In contrast, dSNAP-29 has a unique N-terminal NPF motif (Fig. 1A), which is involved in the interaction with Eps15 homology (EH) domains, a feature shared by all known SNAP-29 isoforms (reviewed by [50]).
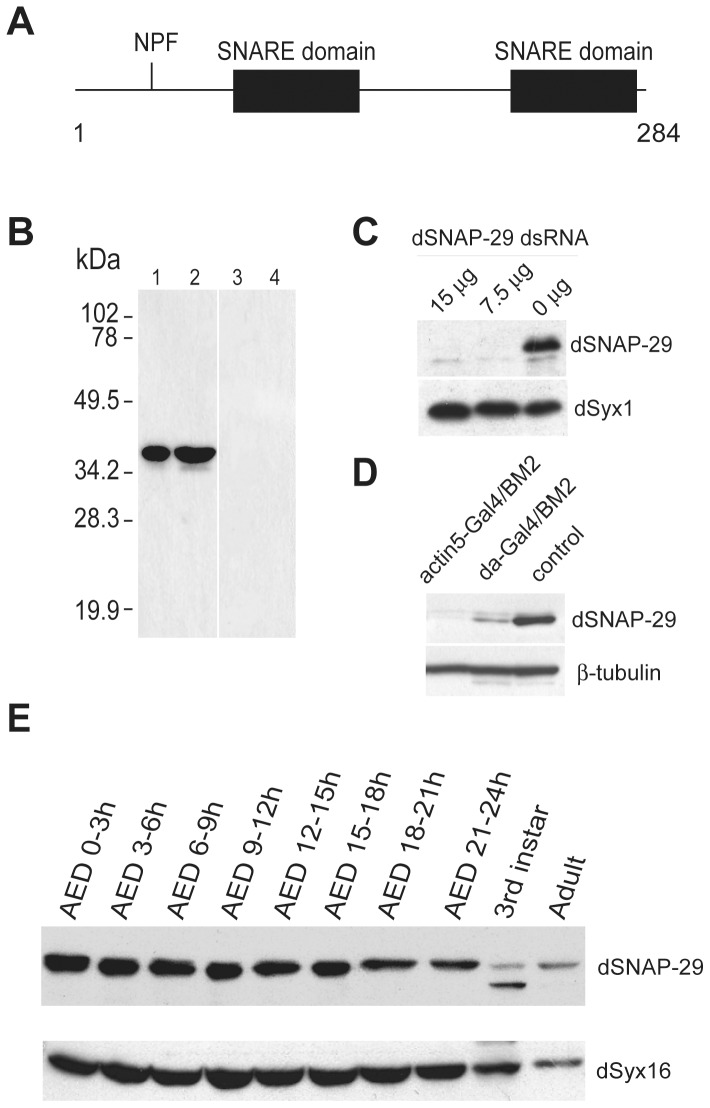
Figure 1. dSNAP-29 is expressed throughout the fly life cycle.
A) Domain structure of dSNAP-29. B). Affinity-purified anti-dSNAP29 preincubated with (lanes 3 and 4) or without (lanes 1 and 2) the peptide was used to blot 10 μg (lanes 1 and 3) and 20 μg (lanes 2 and 4) of total proteins extracted from adult flies. C) S2 cells were treated with dsRNA of dSNAP-29 for 5 days after which the cells were lysed and immunoblotted by anti-dSNAP29 and anti-dSyx1. D) Transgenic line UAS-dSNAP29RNAi-BM2 was crossed with GAL4 lines to allow ectopic expression of dsRNA of dSNAP-29. Inhibition of protein synthesis was demonstrated by western blotting analysis. E) Embryos after deposition (AED) were collected and allowed to develop for indicated period of time. 20 μg of proteins from each of the embryonic collections, larvae, and adults were subject to SDS-PAGE and western blotting analysis.
To distinguish dSNAP-29 from other endogenous fly proteins, antibody against a 12 amino-acid peptide at the C-terminus of the protein was raised in rabbit and then affinity-purified. The peptide antibody recognizes a 35 kDa band from the adult fly lysate (Fig. 1B), slightly above the predicted molecular mass of the protein (32 kDa). The band disappears if the antibody is pre-incubated with the peptide (Fig. 1B, lanes 3 and 4). To further confirm the specificity of the antibody, we applied double-stranded RNA of dSNAP-29 to cultured Schneider 2 (S2) cells, which results in the loss of the immuno-detection of the 35 kDa band from cell lysates (Fig. 1C). The treatment had no effect on the detection of dsyntaxin1 (Fig. 1C). Similarly, in transgenic flies where double-stranded RNA of dSNAP-29 (UAS-dSNAP-29RNAi) was expressed under the control of ubiquitous Gal4 drivers (i.e., actin5-GAL4 or daughterless-Gal4), the level and subsequent detection of dSNAP-29 were selectively reduced (Fig. 1D). We thus conclude that the peptide antibody we generated is specific for dSNAP-29.
To examine the temporal expression of dSNAP-29, embryos from Oregon R were collected every three hours after embryo deposition (AED) and allowed to develop for up to 24 hours. Embryos, 3rd instar larva, and adults were then lysed and equal amounts of total protein were separated by SDS-PAGE and immunoblotted. As shown in Fig. 1E (top lane), dSNAP-29 is expressed in all developmental stages examined, albeit with more abundant levels in early embryos. This is slightly different from dsyntaxin16, which appears to be more evenly expressed throughout the embryonic stages (Fig. 1E, bottom lane). The relative levels of both dSNAP-29 and dsyntaxin16 are reduced in larval and adult stages, probably reflecting the increased production of other proteins in these stages. We went on to determine the spatial localization of dSNAP-29 at various developmental stages. Immunocytochemistry performed on overnight embryonic collections suggests that dSNAP-29 is ubiquitously expressed in embryos (data not shown). Using indirect immunofluorescence, we detected in early embryos, a strong perinuclear staining pattern that appears to be different from that of the Golgi (Fig. 2D to F). However, the narrow cytoplasmic space between the nucleus and the plasma membrane precludes a more defined localization of dSNAP-29 in these cells. In cultured S2 cells, the distribution patten of dSNAP-29 is also different from that of the Golgi marker p120 (Fig. 2 compare G to H), but there appear to be some limited colocalization between the two, suggesting that dSNAP-29 might mediate membrane traffic to and/or from the Golgi. In 3rd instar larva, dSNAP-29 has been found to distribute widely: from imaginal discs to salivary glands (Fig. 2A to C). A close inspection of salivary gland cells with confocal microscopy suggests that dSNAP-29 is not just in the cytoplasm but also on the plasma membrane. This is in contrast to dSNAP-25, which is not detected in salivary glands, and dSNAP-24, which is predominantly localized onto the secretory granules [51]. Because of the ubiquitous distribution of dSNAP-29, we had initially named the gene Ubisnap (usnp).
Figure 2. dSNAP-29 is widely distributed in Drosophila.
Using indirect immuofluorescence, dSNAP29 was detected in wing disc A), salivary gland cells B) and C), early embryos D), and S2 cells G). The samples in D) and G) were co-stained with anti-p120 as shown in E) and H). G) and J) are merged images.
The detection of dSNAP-29 on the plasma membrane of the salivary gland cells persuaded us to investigate the nature of its association with the membrane. Adult fly lysate was separated into soluble and membrane fractions by centrifugation. Subsequent SDS-PAGE and western blotting analysis showed that although dSNAP-29 was predominant in the membrane fraction, a small yet significant portion was also present in the soluble fraction. This is different from dSNAP-25, which was exclusively distributed in the membrane fraction (Fig. 3A). This difference between the two proteins was more evident after the membrane fraction was treated with KCl, Na2CO3 (high pH), urea, Triton X-100 and SDS respectively. While both dSNAP-29 and dSNAP-25 resisted the extraction by KCl and urea, dSNAP-29 appeared to be more sensitive to Na2CO3, suggesting that dSNAP-29 may require hydrophobic interactions to bind its membrane receptors. Similar observations have been reported for mammalian SNAP-29s. Considering the fact that SNAP-29 isoforms lack the cysteine cluster and the QPXR(V/I) motif, it is likely that they use an alternative but less stringent mechanism to associate with the membrane.
Figure 3. Fractionation studies of dSNAP-29.
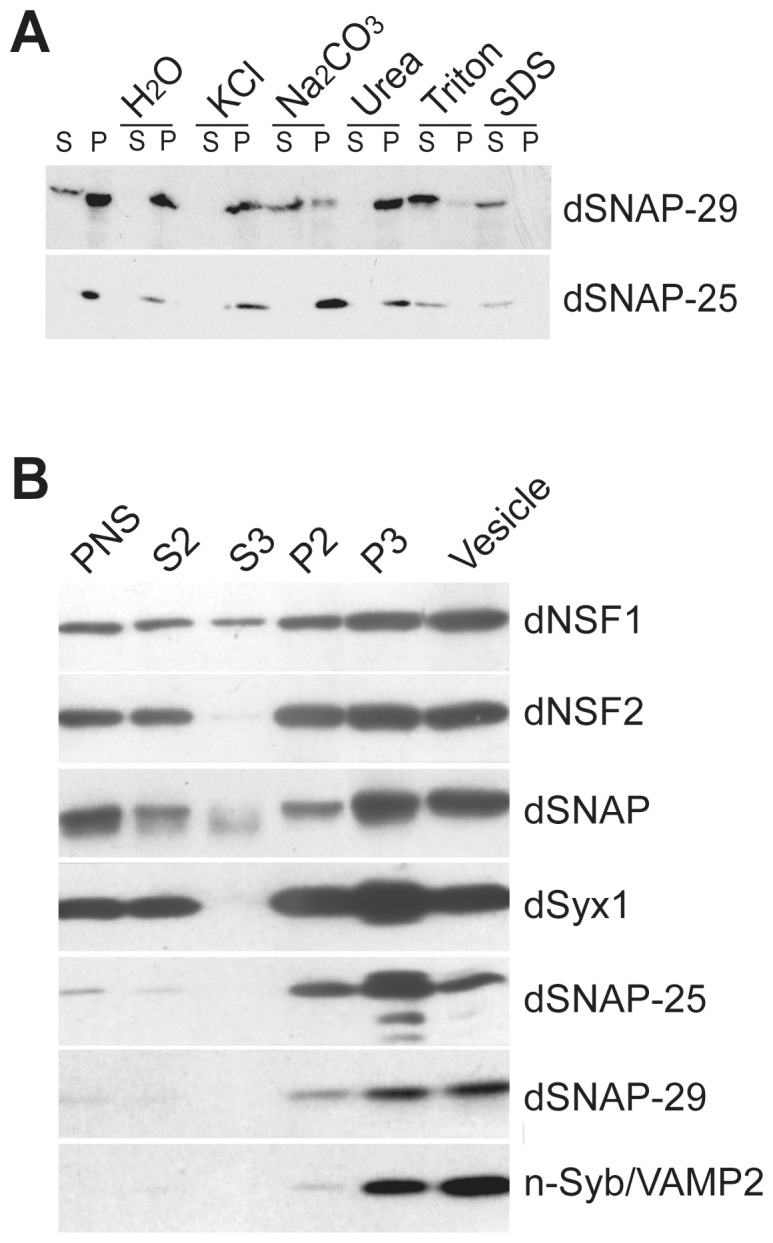
In A), crude membrane fraction from homogenized Oregon R adults was treated with H2O, KCl, Na2CO3, Urea, Triton X-100, or SDS and then centrifuged to separate the soluble from the insoluble. In B), a post nuclear supernatant (PNS) from fly head homogenate was fractionated into the pellet (P2) and the supernatant (S2) fractions by centrifugation. The S2 fraction was then centrifuged on a discontinuous sucrose gradient to separate cytosol (S3), crude vesicles and the plasma membrane (P3). 10 μg of total protein from each fraction were subject to SDS-PAGE and blotted with various antibodies indicated on the right.
To further characterize the subcellular localization of dSNAP-29, the post-nuclear supernatant (PNS) from fly head homogenate was fractionated into the pellet (P2) and the supernatant (S2) fractions. The S2 fraction was then centrifuged on a discontinuous sucrose gradient to separate cytosol (S3), crude vesicles and the plasma membrane (P3). Subsequent western blot analysis suggests that dSNAP-29 is equally enriched in the vesicle and the plasma membrane fractions (Fig. 3B). The fractionation profile of dSNAP-29 is more similar to that of n-Syb, a vesicle SNARE, but in contrast to those of syntaxin1 and SNAP-25, which have demonstrated a preference for plasma membrane. Importantly, both dNSF2 and dSNAP appear to cofractionate with dSNAP-29, implicating the two proteins as potential dSNAP-29 regulators on both the vesicles and the plasma membrane.
dSNAP-29 interacts with dsyntaxin1 and dsyntaxin16
Previous studies suggest that the mammalian SNAP-29 associates with syntaxins on multiple membranes and may thereby participate in multiple trafficking events [21], [23]. To identify the cognate SNARE partners for the Drosophila SNAP-29, we performed a pull-down assay using fresh lysates of cultured Schneider cells (S2). S2 cells are derived from embryonic cells and appear to express dNSF2, dSNAP, and various SNAREs such as dSNAP-29, dSyx1, dSyx 5, and dSyx 16. Using immobilized GST-SNAP-29, we were able to recover about 7% of the SNAP from the lysate (Fig. 4A). Importantly, the same amount of GST did not recover any detectable amount of dSNAP, suggesting that the assay is both specific and efficient. In the meantime, GST-SNAP-29, but not GST, recruited 6% of dsyntaxin1 and 4% of dsyntaxin 16 from the lysate.
Figure 4. dSNAP-29 interacts with dSyx1 and dSyx16 but not Hrs.
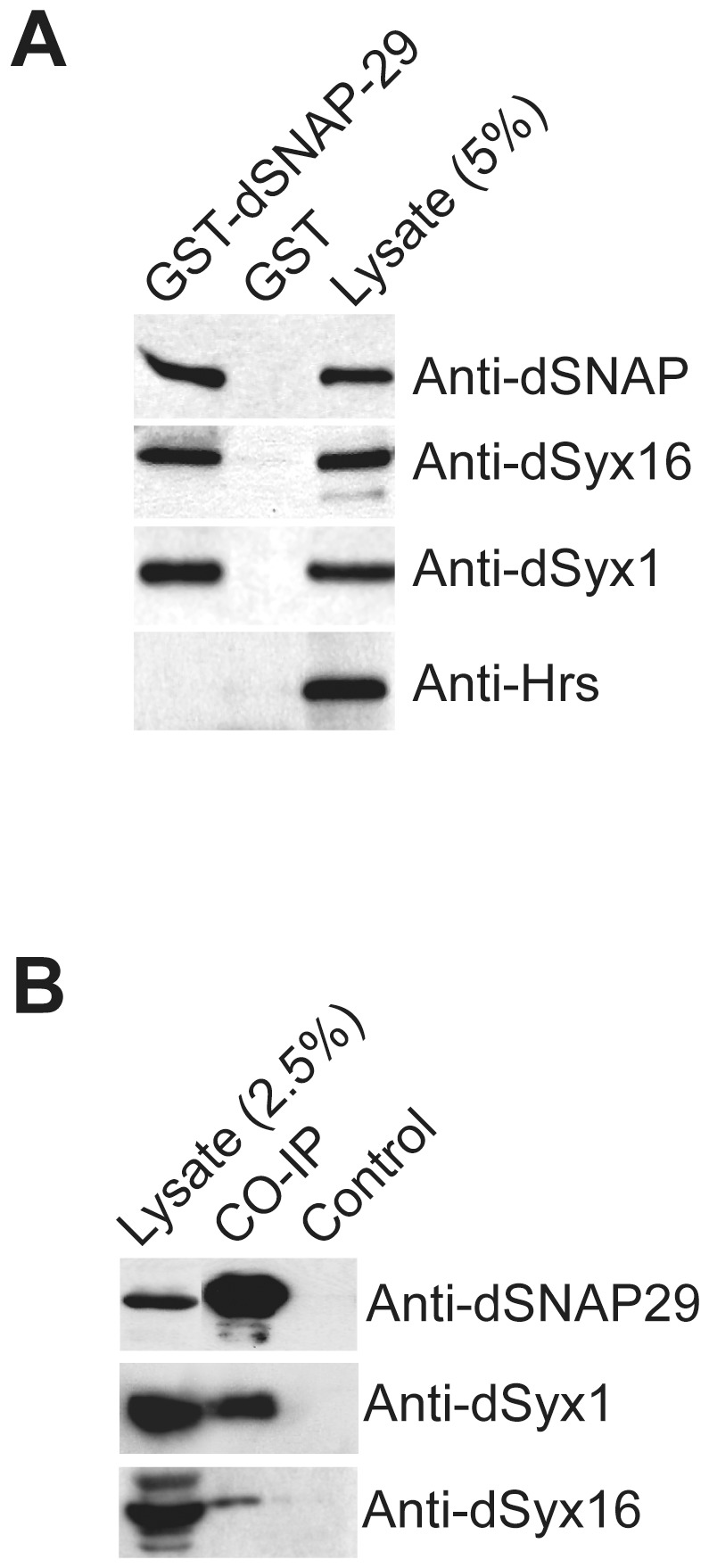
A) Immobilized GST-dSNAP29 and GST were incubated with S2 cell lysate. Following extensive washes, Proteins were eluted and subject to SDS-PAGE and western blotting analysis. B) Drosophila head lysate was incubated with anti-dSNAP29 crosslinked to protein A beads. Pre-immune serum was used as control.
To determine whether the interactions exist in vivo, we performed co-immunoprecipitation experiments using overnight embryonic collections. Instead of the peptide antibody, which may not recognize dSNAP-29 in the SNARE complex, we employed affinity-purified serum raised against the full-length recombinant dSNAP-29. The antibody, but not the pre-immune serum, brought down significant amounts of dSNAP-29 from the embryonic lysate, together with dsyntaxin1 and a small amount of dsyntaxin 16 (Fig. 4B). Thus, our studies suggest that dSNAP-29 may serve on the Golgi or TGN as a partner for dsyntaxin16 [41]. It is intriguing however, to learn that dSNAP-29 may actually interact with syntaxin1, considering their very different distribution patterns in the salivary gland (syntaxin1 is on the apical membrane whereas SNAP-29 is localized to the basolateral membrane and the cytoplasm). Further analysis is warranted to determine the physiological relevance of the biochemical interactions
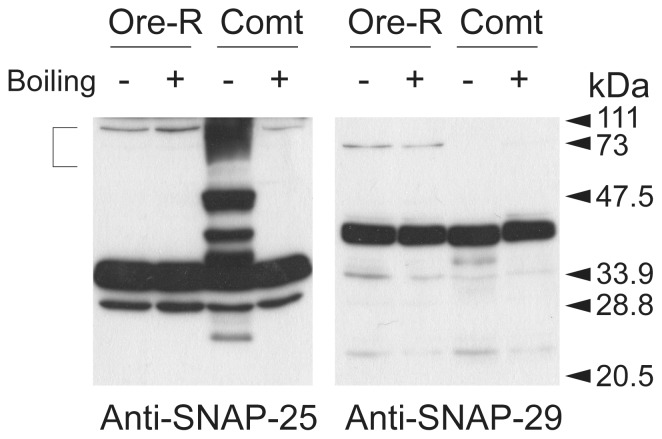
dSNAP-29 does not form SDS-resistant SNARE complexes
Some SNAREs readily form stable SDS-resistant complexes whose dissassembly requires the actions of NSF and SNAP. In Drosophila, blocking the ATPase activity of NSF-1 by incubation of comtTP7 (a temperature-sensitive allele of dNSF-1) adult flies at non-permissive temperatures led to a significant increase of SDS-resistant SNARE complexes detectable by western blot analysis [42] Such complexes appear to contain neuronal SNAREs such as dsyntaxin1 and dSNAP-25, which is in agreement with the notion that dNSF1 is the predominant functional isoform in the adult nervous system [52]. Niemeyer and Schwarz demonstrated that recombinant dSNAP-24 can also form SDS-resistant complexes in vitro [51]. To examine whether dSNAP-29 forms SDS-resistant complex with its cognate SNAREs in vivo, we heat shocked comtTP7 or wild-type adult flies at 37 °C for 10 min, after which the heads were collected and homogenized directly in 1% SDS lysis buffer. Half of the amount of each sample was boiled and the other half was not in order to preserve the complexes. As revealed in Fig. 5, dSNAP-25 from heat-challenged comtTP7 flies accumulated in the SDS-resistant complexes that are disintegrated by boiling. In contrast, dSNAP-29 was not detected in such SDS-resistant complexes when dNSF1 was impaired (Fig. 5). At least three scenarios could account for this. In the first scenario, dSNAP-29 forms SDS-resistant complexes that are sensitive to dNSF-2 rather than to dNSF-1. However, blocking dNSF-2 function in salivary gland cells does not result in any detectable SDS-resistant complex at all (data not shown). In the second scenario, the SNARE complex formed by dSNAP-29 is sensitive to SDS treatment. This is not utterly surprising as most SDS-resistant SNARE complexes reported thus far involve syntaxin1 and are likely responsible for exocytosis. In the third scenario, a very small quantity of dSNAP-29 participates in SDS-resistant complex at a level that is too low to detect. Whether our finding represents a unique case for SNAP-29 in Drosophila or might apply to other SNARE complexes remains to be determined.
Figure 5. dSNAP-29 does not form SDS-resistant complexes.
Heads of heat-challenged Oregon R and ComtTp7 flies were collected in SDS-containing homogenization buffer. Half of each sample was boiled while the other half was not to preserve SNARE complexes. Boiling-sensitive yet SDS-resistant SNARE complexes are indicated by ([).
dSNAP-29 is a partner of dEHD1 but not Hrs
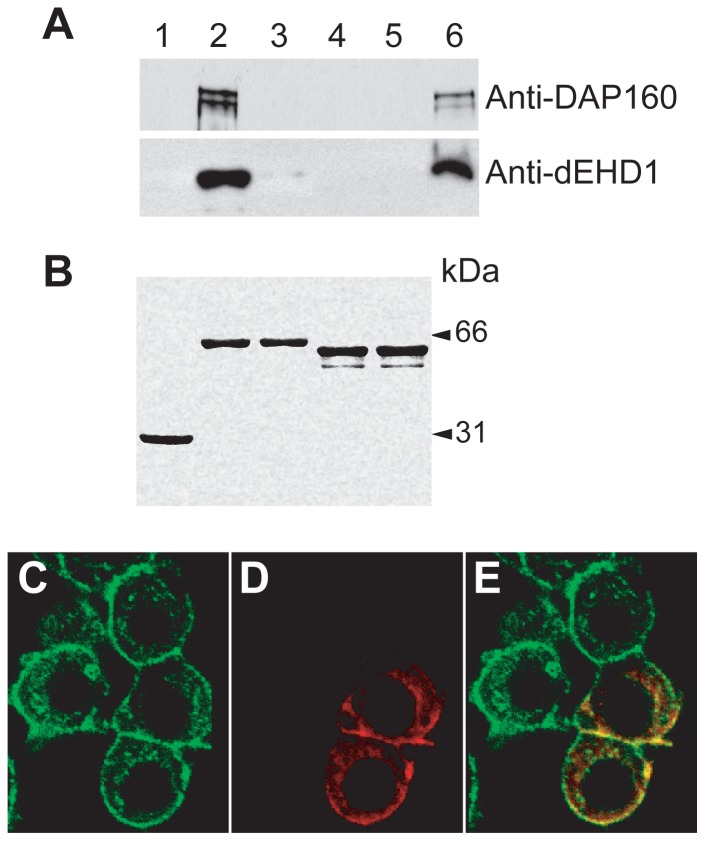
The functions of SNAREs are often regulated through reversible interactions with SNARE modulators. Amino-acid sequence analysis revealed a N-terminal NPF motif in dSNAP-29, suggesting that it may interact with EH domain-containing proteins. In the fly genome, there are at least 4 EH domain-containing proteins: PAST1/EHD1, DAP160/intersectin, CG16932/Eps15 and CG6192 [53]. In this study, we tested whether dSNAP-29 interacts with PAST1/EHD1 and DAP160/intersectin, the two Drosophila EH domain-containing proteins that we had antibodies for. GST-dSNAP-29, GST-dSNAP-25, and GST were immobilized separately onto glutathione-conjugated agarose beads and then incubated either with homogenized S2 sample or homogenization buffer alone. Subsequent western blot analysis (Fig. 6A) revealed a minimal cross-reaction between affinity purified anti-dEHD1 and recombinant GST-dSNAP29 (lane 3), which runs with the same mobility as the endogenous EHD1. Nonetheless, GST-dSNAP-29 pulled down a significant amount of dEHD1 (12%) and DAP160 (15%) from the lysate (compare lanes 2 and 3) whereas GST (lane 1) or GST-dSNAP25 (lane 4) did not at all, suggesting the interaction between dSNAP-29 and dEHD1 is highly selective. To examine their distribution in vivo, S2 cells transfected with myc-dSNAP29 was immuno-stained with mouse anti-myc (Fig. 6C) and rabbit anti-dEHD1 (Fig. 6D). Significant colocalization was observed on cell surface and on punctate intracellular structures (Fig. 6E). Our finding is line with a recent report which suggests the association of mammalian SNAP-29 with both insulin-like growth factor 1 receptor and EHD1 [39].
Figure 6. dSNAP-29 selectively interact and partially co-localize with dEHD1 in S2 cells.
A) S2 cell lysate (lanes 1, 2, 4) or lysate buffer alone (lanes 3 and 5) was incubated with immobilized GST (lane 1), GST-dSNAP29 (lanes 2 and 3), GST-dSNAP25 (lanes 4 and 5). Associated proteins were separated by SDS-PAGE and blotted by anti-dEHD1. 5% of the lysate used in each pull-down experiment were loaded in lane 6. Shown in B) is a Coomassie-blue stained gel to demonstrate that equal amounts of GST chimeras were used for the pull-down assay. To examine the distribution of dSNAP-29 and dEHD1, S2 cells were transfected with myc-dSNAP29, fixed with methanol, and stained with C) mouse anti-myc and D) rabbit anti-dEHD1. Merged image is shown in E.
Modulation of dSNAP-29 levels in cultured cells and transgenic flies
To evaluate the functional involvement of dSNAP-29 in membrane traffic, we took two commonly used approaches. The first is to ectopically overexpress the wild-type or the truncated form of dSNAP-29 in transgenic flies. High levels of wild-type dSNAP-29 may sequester its potential binding partners (e.g., dEHD1), causing an inhibition of the putative trafficking step. As a matter of fact, the ubiquitous overexpression of dSNAP-29 controlled by da-GAL4 or actin5-GAL4 led to lethality at the 1st larval stage (Table 1). However, we did not observe any defect in embryogenesis. It has also been demonstrated that a SNAP-25 lacking the C-terminus was able to form complexes with its cognate SNARE partners but failed to execute membrane fusion [10], [54]. Therefore, we constructed UAS-dSNAP-29ΔC, which carries a partial cDNA that does not encode the C-terminal 25 amino acids. Similar to the wild type, overexpression of dSNAP-29ΔC using actin5-GAL4 or da-GAL4 driver is lethal (Table 1). Ectopic expression of dSNAP-29ΔC using C96-GAL4 (wing margin) causes a notch-wing phenotype whereas expression with elav-GAL4 (the nervous system) causes defects in wing extension and development. However, these non-lethal phenotypes were only observed with one of the two UAS- dSNAP-29ΔC lines generated (Table 1).
Table 1. Phenotypes caused by ectopic expression of various forms of dSNAP-29.
| Gal4 lines | Expression pattern | UAS-dSNAP29 | UAS-dSNAP29ΔC | UAS-dSNAP29RNAi |
| actin5-Gal4 | ubiquitous | 1st instar lethal | lethal | pupal lethal at 29 °C |
| da-Gal4 | ubiquitous | 1st instar lethal | lethal | n.d. |
| C96-Gal4 | wing margin | none | Notched wings* | none |
| 24B-Gal4 | muscle (late) | none | n.d. | pupal lethal at 29 °C |
| mhc-Gal4 | muscle (early) | none | n.d. | none |
| elav-Gal4 | CNS | none | Abnormal wings* | none |
* Not observed in all the independent UAS lines that were tested.
For each UAS construct, several independent lines were tested to rule out positional effect. Visible phenotypes or sterility were scored. The crosses are set at both room temperature and 29 °C unless indicated otherwise. n.d., not determined.
An alternative but more direct approach is to knock down the protein expression by RNA interference. We constructed a UAS-dSNAP-29RNAi transgene to express double-stranded RNA in defined tissues in the fly. When the construct was expressed under the control of the ubiquitous actin5-GAL4 driver at room temperature, we achieved excellent reduction of the protein level (around 95%; Fig. 1D). Although flies appear normal under these conditions, incubation at 29 °C causes lethality at the pupal stage (Table 1). Lethality was also observed when the 24B-GAL4 driver (expressed mainly in the muscle) was used, whereas the elav-GAL4 driver (expressed mainly in the nervous system) does not produce any visible phenotype. Thus, it appears that dSNAP-29 may play an essential role in the post synaptic cells although it may also be actively involved in other cells/tissues.
Discussion
In multicellular systems, SNAP-25/23 functions predominantly on the plasma membrane, whereas SNAP-29 is found mainly in the cytoplasm. An important difference between SNAP-25 and SNAP-29 is that the later possesses an N-terminal NPF motif, which allows for interaction with EH domain-containing proteins. In our study, dSNAP-29 has been shown to interact with dEHD1 and DAP160, two of the four known EH domain-containing proteins in the fly genome [53]. Whereas Drosophila EHD1 has been implicated in endocytosis [55], its worm orthologue RME-1 and its mammalian orthologue EHD1 have been implicated in the recycling of a set of endocytosed molecules back to the plasma membrane [56], [57], [58]. DAP160 on the other hand, may act as a scaffold protein by interacting with the endocytic machinery including dynamin, clathrin and synaptojanin [59]. Intriguingly, SNAP-29, EHD1, and DAP160 do not appear to be present in the yeast genome, suggesting that they are required for a more specialized pathway that has evolved only in multicellular organisms.
In an attempt to address the physiological relevance of the interactions between dSNAP-29 and the EH domain-containing proteins, we transfected S2 cells with the myc-dSNAP-29 construct and performed immunostaining using anti-dEHD1 and anti-myc antibodies. At low expression levels, myc-SNAP-29 appears to partially colocalize with dEHD1 at the plasma membrane as well as the peripheral regions underneath the plasma membrane (Fig. 6 C,D,E). However, the plasma-membrane distribution of dSNAP-29 was not observed when cells were stained with anti-dSNAP-29. Which staining pattern more accurately represents the in vivo distribution of the protein is not clear.
How do we explain the interaction between dSNAP-29 and the syntaxins? It may be counter-intuitive to suggest that dSyx1 and dSNAP-29 can actually function together in the salivary gland. dSyx1 is found on the apical membrane whereas dSNAP-29 is not. Since dSNAP-25 is not expressed in the salivary gland, a likely partner for syntaxin 1 during granule secretion is probably dSNAP-24, which is localized to the secretory granules and which redistributes upon exocytosis. dSNAP-29 is not enriched on those secretory granules, but is found in the remaining cytoplasm, and may very well be on recycling vesicles. In cultured S2 cells, transfected dSNAP-29 appears to partially colocalize with dSyx1 on intracellular punctae, with a pattern differing significantly from that of ER, Golgi, endosomes, or lysosomes (data not shown). It is possible that the two proteins interact only transiently during the recycling pathway. As well, we cannot rule out the possibility that dSNAP-29 may adopt different distribution patterns in different tissues.
The demonstration that dSNAP-29 also interacts with dsyntaxin 16 does not necessarily imply that dSNAP-29 is promiscuous. In a separate study, we showed that dsyntaxin 5 does not interact with dSNAP-29 in vitro (Hsien and Trimble, unpublished), which is in agreement with reports on its mammalian orthologue. However, unlike dsyntaxin 5, which is predominantly on the cis-Golgi, dsyntaxin 16 is localized to a late Golgi compartment, possibly in the trans-Golgi network. More importantly, mammalian syntaxin 16 has recently been implicated in retrograde transport from the endosomes [60], consistent with the notion that SNAP-29 is also involved in the recycling pathway. Recently, dSNAP-29 has been shown to regulate autophagy along with dsynxtin11 and dVAMP7 [30], arguing for a more versatile role for SNAP-29 in intracellular membrane traffic.
Acknowledgments
The authors thank Dr. Jane McGlade for anti-EHD1 antibody, Dr. Jack Roos for anti-DAP160 antibody, Dr. Hugo Bellen for anti-Hrs antibody, Dr. David Deitcher for anti-dSNAP-25 antibody.
Funding Statement
These studies were supported by grants from the Canadian Institutes of Health Research (GLB) and the Natural Sciences and Engineering Research Council of Canada (WST). GLB and WST are Canada Research Chair holders. HX is a USM intramural development grant recipient. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chen YA, Scheller RH (2001) SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol 2: 98–106. [DOI] [PubMed] [Google Scholar]
- 2. Jahn R, Sudhof TC (1999) Membrane fusion and exocytosis. Annu Rev Biochem 68: 863–911. [DOI] [PubMed] [Google Scholar]
- 3. Rothman JE (1994) Intracellular membrane fusion. Adv Second Messenger Phosphoprotein Res 29: 81–96. [DOI] [PubMed] [Google Scholar]
- 4. Pelham HR (2001) SNAREs and the specificity of membrane fusion. Trends Cell Biol 11: 99–101. [DOI] [PubMed] [Google Scholar]
- 5. Bock JB, Matern HT, Peden AA, Scheller RH (2001) A genomic perspective on membrane compartment organization. Nature 409: 839–841. [DOI] [PubMed] [Google Scholar]
- 6. Holt M, Varoqueaux F, Wiederhold K, Takamori S, Urlaub H, et al. (2006) Identification of SNAP-47, a novel Qbc-SNARE with ubiquitous expression. J Biol Chem 281: 17076–17083. [DOI] [PubMed] [Google Scholar]
- 7. Oyler GA, Higgins GA, Hart RA, Battenberg E, Billingsley M, et al. (1989) The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol 109: 3039–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawasaki F, Ordway RW (2009) Molecular mechanisms determining conserved properties of short-term synaptic depression revealed in NSF and SNAP-25 conditional mutants. Proc Natl Acad Sci U S A 106: 14658–14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, et al. (1993) Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature 365: 160–163. [DOI] [PubMed] [Google Scholar]
- 10. Wei S, Xu T, Ashery U, Kollewe A, Matti U, et al. (2000) Exocytotic mechanism studied by truncated and zero layer mutants of the C-terminus of SNAP-25. Embo J 19: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ravichandran V, Chawla A, Roche PA (1996) Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues. J Biol Chem 271: 13300–13303. [DOI] [PubMed] [Google Scholar]
- 12. Wang G, Witkin JW, Hao G, Bankaitis VA, Scherer PE, et al. (1997) Syndet is a novel SNAP-25 related protein expressed in many tissues. J Cell Sci 110 (Pt 4): 505–513. [DOI] [PubMed] [Google Scholar]
- 13. Foster LJ, Yaworsky K, Trimble WS, Klip A (1999) SNAP23 promotes insulin-dependent glucose uptake in 3T3-L1 adipocytes: possible interaction with cytoskeleton. Am J Physiol 276: C1108–1114. [DOI] [PubMed] [Google Scholar]
- 14. Chen D, Lemons PP, Schraw T, Whiteheart SW (2000) Molecular mechanisms of platelet exocytosis: role of SNAP-23 and syntaxin 2 and 4 in lysosome release. Blood 96: 1782–1788. [PubMed] [Google Scholar]
- 15. Galli T, Zahraoui A, Vaidyanathan VV, Raposo G, Tian JM, et al. (1998) A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol Biol Cell 9: 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leung SM, Chen D, DasGupta BR, Whiteheart SW, Apodaca G (1998) SNAP-23 requirement for transferrin recycling in Streptolysin-O-permeabilized Madin-Darby canine kidney cells. J Biol Chem 273: 17732–17741. [DOI] [PubMed] [Google Scholar]
- 17. Low SH, Chapin SJ, Wimmer C, Whiteheart SW, Komuves LG, et al. (1998) The SNARE machinery is involved in apical plasma membrane trafficking in MDCK cells. J Cell Biol 141: 1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lafont F, Verkade P, Galli T, Wimmer C, Louvard D, et al. (1999) Raft association of SNAP receptors acting in apical trafficking in Madin-Darby canine kidney cells. Proc Natl Acad Sci U S A 96: 3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vilinsky I, Stewart BA, Drummond J, Robinson I, Deitcher DL (2002) A Drosophila SNAP-25 null mutant reveals context-dependent redundancy with SNAP-24 in neurotransmission. Genetics 162: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jurado S, Goswami D, Zhang Y, Molina AJ, Sudhof TC, et al. (2013) LTP requires a unique postsynaptic SNARE fusion machinery. Neuron 77: 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steegmaier M, Yang B, Yoo JS, Huang B, Shen M, et al. (1998) Three novel proteins of the syntaxin/SNAP-25 family. J Biol Chem 273: 34171–34179. [DOI] [PubMed] [Google Scholar]
- 22. Wong SH, Xu Y, Zhang T, Griffiths G, Lowe SL, et al. (1999) GS32, a novel Golgi SNARE of 32 kDa, interacts preferentially with syntaxin 6. Mol Biol Cell 10: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hohenstein AC, Roche PA (2001) SNAP-29 is a promiscuous syntaxin-binding SNARE. Biochem Biophys Res Commun 285: 167–171. [DOI] [PubMed] [Google Scholar]
- 24. Wesolowski J, Caldwell V, Paumet F (2012) A novel function for SNAP29 (synaptosomal-associated protein of 29 kDa) in mast cell phagocytosis. PLoS One 7: e49886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rapaport D, Lugassy Y, Sprecher E, Horowitz M (2010) Loss of SNAP29 impairs endocytic recycling and cell motility. PLoS One 5: e9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato M, Saegusa K, Sato K, Hara T, Harada A (2011) Caenorhabditis elegans SNAP-29 is required for organellar integrity of the endomembrane system and general exocytosis in intestinal epithelial cells. Mol Biol Cell 22: 2579–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang J, Bai Z, Zegarek MH, Grant BD, Lee J (2011) Essential roles of snap-29 in C. elegans. Dev Biol 355: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su Q, Mochida S, Tian JH, Mehta R, Sheng ZH (2001) SNAP-29: a general SNARE protein that inhibits SNARE disassembly and is implicated in synaptic transmission. Proc Natl Acad Sci U S A 98: 14038–14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, et al. (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495: 389–393. [DOI] [PubMed] [Google Scholar]
- 30. Takats S, Nagy P, Varga A, Pircs K, Karpati M, et al. (2013) Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol 201: 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chin LS, Nugent RD, Raynor MC, Vavalle JP, Li L (2000) SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J Biol Chem 275: 1191–1200. [DOI] [PubMed] [Google Scholar]
- 32. Komada M, Soriano P (1999) Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev 13: 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, et al. (2002) Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108: 261–269. [DOI] [PubMed] [Google Scholar]
- 34. Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, et al. (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol 4: 389–393. [DOI] [PubMed] [Google Scholar]
- 35. Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, et al. (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394–398. [DOI] [PubMed] [Google Scholar]
- 36. Kwong J, Roundabush FL, Hutton Moore P, Montague M, Oldham W, et al. (2000) Hrs interacts with SNAP-25 and regulates Ca(2+)-dependent exocytosis. J Cell Sci 113 (Pt 12): 2273–2284. [DOI] [PubMed] [Google Scholar]
- 37. Bean AJ, Seifert R, Chen YA, Sacks R, Scheller RH (1997) Hrs-2 is an ATPase implicated in calcium-regulated secretion. Nature 385: 826–829. [DOI] [PubMed] [Google Scholar]
- 38. Okamoto M, Schoch S, Sudhof TC (1999) EHSH1/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. A protein connection between exocytosis and endocytosis? J Biol Chem 274: 18446–18454. [DOI] [PubMed] [Google Scholar]
- 39. Rotem-Yehudar R, Galperin E, Horowitz M (2001) Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J Biol Chem 276: 33054–33060. [DOI] [PubMed] [Google Scholar]
- 40. Xu H, Brill JA, Hsien J, McBride R, Boulianne GL, et al. (2002) Syntaxin 5 is required for cytokinesis and spermatid differentiation in Drosophila. Dev Biol 251: 294–306. [DOI] [PubMed] [Google Scholar]
- 41. Xu H, Boulianne GL, Trimble WS (2002) Drosophila syntaxin 16 is a Q-SNARE implicated in Golgi dynamics. J Cell Sci 115: 4447–4455. [DOI] [PubMed] [Google Scholar]
- 42. Mohtashami M, Stewart BA, Boulianne GL, Trimble WS (2001) Analysis of the mutant Drosophila N-ethylmaleimide sensitive fusion-1 protein in comatose reveals molecular correlates of the behavioural paralysis. J Neurochem 77: 1407–1417. [DOI] [PubMed] [Google Scholar]
- 43. Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, et al. (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A 97: 6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- 45.Lindsley D, Zimm GG (1992) The Genome of Drosophila melanogaster. San Diego: Academic Press.
- 46. Rubin GM, Spradling AC (1982) Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- 47. Gonzalo S, Greentree WK, Linder ME (1999) SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain. J Biol Chem 274: 21313–21318. [DOI] [PubMed] [Google Scholar]
- 48. Gonzalo S, Linder ME (1998) SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol Biol Cell 9: 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Low SH, Roche PA, Anderson HA, van Ijzendoorn SC, Zhang M, et al. (1998) Targeting of SNAP-23 and SNAP-25 in polarized epithelial cells. J Biol Chem 273: 3422–3430. [DOI] [PubMed] [Google Scholar]
- 50. Santolini E, Salcini AE, Kay BK, Yamabhai M, Di Fiore PP (1999) The EH network. Exp Cell Res 253: 186–209. [DOI] [PubMed] [Google Scholar]
- 51. Niemeyer BA, Schwarz TL (2000) SNAP-24, a Drosophila SNAP-25 homologue on granule membranes, is a putative mediator of secretion and granule-granule fusion in salivary glands. J Cell Sci 113 (Pt 22): 4055–4064. [DOI] [PubMed] [Google Scholar]
- 52. Stewart BA, Mohtashami M, Rivlin P, Deitcher DL, Trimble WS, et al. (2002) Dominant-negative NSF2 disrupts the structure and function of Drosophila neuromuscular synapses. J Neurobiol 51: 261–271. [DOI] [PubMed] [Google Scholar]
- 53. Confalonieri S, Di Fiore PP (2002) The Eps15 homology (EH) domain. FEBS Lett 513: 24–29. [DOI] [PubMed] [Google Scholar]
- 54. Chen YA, Scales SJ, Patel SM, Doung YC, Scheller RH (1999) SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell 97: 165–174. [DOI] [PubMed] [Google Scholar]
- 55. Olswang-Kutz Y, Gertel Y, Benjamin S, Sela O, Pekar O, et al. (2009) Drosophila Past1 is involved in endocytosis and is required for germline development and survival of the adult fly. J Cell Sci 122: 471–480. [DOI] [PubMed] [Google Scholar]
- 56. Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, et al. (2002) A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. Embo J 21: 2557–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, et al. (2001) Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol 3: 573–579. [DOI] [PubMed] [Google Scholar]
- 58. Lin SX, Grant B, Hirsh D, Maxfield FR (2001) Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol 3: 567–572. [DOI] [PubMed] [Google Scholar]
- 59. Roos J, Kelly RB (1998) Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J Biol Chem 273: 19108–19119. [DOI] [PubMed] [Google Scholar]
- 60. Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, et al. (2002) Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol 156: 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]