Abstract
Regulation of DNA repair can be achieved through ubiquitin-mediated degradation of transiently induced proteins. In Saccharomyces cerevisiae, Rad4 is involved in damage recognition during nucleotide excision repair (NER) and, in conjunction with Rad23, recruits other proteins to the site of damage. We identified a synthetic interaction upon UV exposure between Rad4 and Cdc20, a protein that modulates the activity of the anaphase promoting complex (APC/C), a multisubunit E3 ubiquitin ligase complex. The moderately UV sensitive Δrad4 strain became highly sensitive when cdc20-1 was present, and was rescued by overexpression of CDC20. The double mutant is also deficient in elicting RNR3-lacZ transcription upon exposure to UV irradiation or 4-NQO compared with the Δrad4 single mutant. We demonstrate that the Δrad4/cdc20-1 double mutant is defective in double strand break repair by way of a plasmid end-joining assay, indicating that Rad4 acts to ensure that damaged DNA is repaired via a Cdc20-mediated mechanism. This study is the first to present evidence that Cdc20 may play a role in the degradation of proteins involved in nucleotide excision repair.
1. Introduction
Increased expression of genes necessary for detecting and repairing DNA damage can result when cells are exposed to certain genotoxic compounds [1–3]. These same treatments can also induce differential expression of genes in the ubiquitin- (Ub-) mediated proteolytic pathway [4, 5], suggesting interplay between DNA repair and protein degradation [6–9]. Mechanisms to transiently stabilize or reduce the abundance of repair proteins following detection and/or removal of DNA damage could provide a means for regulating these processes. Defects in Ub-mediated protein degradation have been linked to breast cancer [10], Angelman syndrome [11], von Hippel-Lindau disease [12], and altered responses to clinical anesthetics [13], while defects in DNA damage repair are associated with human disorders such as xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy [14]. This faithful transmission of genetic material is critical to cell survival, while proper functioning of DNA repair processes ensures that genomic integrity is maintained.
Ubiquitination results in the modification of protein function, thereby controlling cellular processes such as cell cycle progression, differentiation, and stress responses [15, 16]. This 8.5 kDa protein is first activated by ubiquitin-activating enzymes (E1) before being transferred to ubiquitin-conjugating enzymes (E2) and ubiquitin protein ligases (E3) [17]. It is ultimately attached to lysine residues in target proteins, and multiubiquitin chains form through the activity of ubiquitin chain assembly factors (E4). These ubiquitinated proteins are then targeted to the 26S proteasome for degradation. The anaphase promoting complex/cyclosome (APC/C) is one of several multisubunit E3 ubiquitin ligases that control progression through the cell cycle [18, 19]. In addition to E1 and E2 enzymes, APC/C-mediated ubiquitination depends on the activator proteins, Cdc20 or Cdh1 [20–22], with the former regulating the metaphase to anaphase transition through the degradation of securin. Not surprisingly, APC/CCdc20 is a target of the spindle checkpoint, not allowing the degradation of securin until proper attachment and alignment of all kinetochores to the spindle are completed [23]. Clarke et al. [24] have reported that in budding yeast Cdc20 is capable of acting independently of the APC/C, suggesting an alternative mechanism to its ability to regulate mitotic exit. In metazoans, the APC/C is also active in postmitotic differentiated cells, implying that it has assumed functions in nonproliferating cells as well [25].
Nucleotide excision repair (NER) is a highly conserved mechanism that detects and removes bulky lesions from DNA following chemical treatment or UV irradiation [26–28]. Compromising NER activity has pleiotropic effects, leading to increased mutation frequency and risk of carcinogenesis [29, 30]. Bulky adducts repaired by the NER include N-acetyl-2-aminofluorene (AAF) adducts, cyclobutane pyrimidine dimers (CPDs), (6-4) photoproducts, and cisplatin intrastrand crosslinks [31–33]. Global genome NER (GGR), responsible for the repair of untranscribed regions of the genome, can be divided into five distinct steps: damage recognition, incision, excision, repair synthesis, and DNA ligation [26]. The expression of a number of NER proteins is upregulated as a result of DNA damage; however, once the DNA damage is repaired and the proteins are no longer needed, they are degraded to restore basal levels [1, 34]. Multiple interactions between members of the NER, ubiquitin-associated enzymes, and proteasomal subunits have been revealed by both genetic and biochemical approaches [5–7, 9, 35, 36]. This would indicate that one means of regulating the repair process is through Ub-mediated degradation of NER proteins once they are no longer needed.
Many of the more than 30 NER proteins are essential to the repair processes; however, a subset is considered accessory, and deletion of these results in a less severe sensitivity to DNA damaging agents in comparison to the essential components, which have near 0% survival with UV treatment [37–39]. Rad4 and Rad23 are two such accessory members, interacting strongly with one another to control the damage recognition step of NER [40, 41]. Rad4, which transiently accumulates following UV irradiation [42], is stabilized by Rad23 and this interaction stimulates binding of the Rad4/Rad23 complex to the damaged DNA [5, 43]. Ng et al. [8] have concluded that the primary function of Rad23 in NER is the stabilization of Rad4, although diminished Rad4 stability is not the primary cause of deficient NER in rad23 mutant cells [9, 44].
To examine the interplay between Cdc20 and NER, we utilized yeast strains deleted for RAD4 and harboring the conditional cdc20-1 mutation. We speculate that Cdc20 functions to modulate components of the NER. We report here that the diminished capacity of Cdc20 results in extreme UV sensitivity in NER-compromised mutants of S. cerevisiae, specifically those harboring Δrad4.
2. Materials and Methods
2.1. Strain Construction
All deletion strains used in this study carry a gene deletion linked to a kanamycin-resistance marker kanMX that confers resistance to the antibiotic Geneticin (G418) and were obtained from the Mississippi Functional Genomic Network (MFGN) core facility (University of Southern Mississippi, Hattiesburg, MS) or Open Biosystems (now Thermo Fisher Scientific). A cdc20-1 mutant strain containing the CloNAT resistance marker was generated using a one-step gene replacement. Double mutants harboring various gene deletions and cdc20-1 were also generated by one-step gene replacement, making the single and double mutants isogenic with respect to one another. All other double mutants were generated by traditional crosses and verified by temperature sensitivity and dual resistance to G418 and CloNAT.
2.2. Plasmid Constructions
A plasmid containing cdc20-1 was constructed by amplifying the mutant allele from strain 405-1-1 (gift from D. Burke), which included 400 bp upstream of the start codon and 500 bp downstream of the stop codon. The amplicon was then ligated into pGEM-T (Promega). The CloNAT resistance gene was excised from p4339 (obtained from MFGN) and blunted. The fragment was ligated at a StuI site 150 bp downstream of the stop codon of cdc20-1. DNA sequencing was performed to verify the presence of the cdc20-1 mutation and the resistance marker. The resulting recombinant plasmid was linearized with SpeI to be used for one-step gene replacement.
Plasmids designed to overexpress CDC20 were generated using the pYES.2 vector (Invitrogen), which carries a URA3 marker, 2 μ ori, and multiple cloning site downstream of the gal1 inducible promoter. Plasmids harboring CDC20 or cdc20-1 were constructed by ligating a 2.0 kb HindIII fragment into pYES.2 at the HindIII site and orientation was verified through restriction enzyme analysis. All other overexpression plasmids were obtained from Open Biosystems, in which the genes of interest were subcloned behind the gal1 inducible promoter in the BG1805 plasmid vector. All yeast transformations were performed with the high efficiency LiOAc method [45].
2.3. Drug Cytotoxicity Assays
Yeast strains were incubated at 22°C for 12–16 hours in yeast-peptone-dextrose (YPD) or the appropriate selective medium. Cultures were normalized to an A 660 of 1.0, and a 1 : 10 serial dilution was performed in sterile water. From each dilution, 5 μL was spotted onto YPD agar containing methyl methanesulfonate (MMS) (0.05%), hydroxyurea (100 mM), phleomycin (5 μg/mL), benomyl (12 μg/mL), and 4-nitroquinoline (4-NQO) (0.2, 0.5, and 1.0 μg/mL) or exposed to 10 or 25 J/m2 of UV radiation in a Stratalinker model 1800 (Stratagene). For those strains harboring genes to be overexpressed, transformants were assayed on synthetic complete dextrose media lacking uracil and containing either 2% glucose or 3% galactose/0.5% glucose.
Quantification of survival was determined by plating a known number of cells on selective media, followed by UV irradiation (10 and 25 J/m2) and incubation at 22°C for 3-4 days. UV survival was calculated by dividing the number of surviving colonies following treatment by the number of colonies on untreated plates. Three independent trials were conducted for each treatment.
2.4. β-Galactosidase Assays
The β-gal activity assay was performed as described previously [46]. Briefly, 0.5 mL of an overnight culture was used to inoculate 4.5 mL of fresh selective media. After 8–10 hours at 22°C, cells were treated with 0.2, 0.5, or 1.0 μg/mL of 4-NQO or UV irradiated at 10, 25, or 50 J/m2. Cells were returned to 22°C for another 4 hours prior to performing the β-gal assay. Four hours was found to be optimal for induction of the RNR3:lacZ fusion protein in studies by Jia and Xiao [46]. Activities were determined, and fold induction was calculated as a ratio of β-galactosidase activity in the cultures with and without treatment.
2.5. Plasmid End Joining Assay
The pYES.2 vector (Invitrogen) was digested with PvuII, cutting the plasmid outside of the URA3 marker. Linearized and uncut plasmids of a known concentration were transformed in parallel into multiple strains and plated on SCD lacking uracil. Following 3-4 days of incubation at 22°C to allow for sufficient colony growth, colonies were counted. Transformation efficiencies were calculated using the Transformation Efficiency Calculator (http://www.sciencegateway.org/tools/transform.htm). The results are the average of at least three independent experiments, and standard error of the mean was determined.
2.6. Microscopic Analyses
A strain carrying RAD4 with a C-terminal GFP fusion was obtained from the MFGN core facility. The cdc20-1 allele was introduced into this strain via a traditional cross, and gene replacement was verified by PCR, resistance to CloNAT, and temperature sensitivity at 37°C. Cultures were grown at 22°C in SCD lacking histidine with or without CloNAT for 12–16 hours and subsequently UV irradiated at 25 J/m2 and 100 J/m2. An outgrowth of 4 hours was allowed before cells were briefly centrifuged and washed once with water. This outgrowth time was chosen as Lommel et al. [5] have shown the half-life of Rad4 to be 4 hours. Samples were examined and photographed on a Zeiss Imager M1 AXIO fluorescence microscope paired with a Photometrics Coolsnap HQ2camera. UV exposures were done in triplicate, and greater than 150 cells were counted and analyzed for each treatment.
3. Results
3.1. Mutation in CDC20 Results in Increased Sensitivity to Ultraviolet Radiation in Δrad4 Backgrounds
The Δrad4/cdc20-1 double mutant was initially identified as displaying a synthetic sick phenotype in a synthetic genome analysis using a query strain harboring the cdc20-1 temperature sensitive allele (Figure 1(a)). The UV sensitivity of the Δrad4 strain was increased approximately 10-fold by the point mutation in cdc20-1 (Figure 1(b)). When CDC20 under the control of the gal1 promoter was reintroduced into the double mutant strain and then irradiated, UV resistance comparable to that seen in Δrad4 strains was restored (Figure 1(c)). This was not observed when cdc20-1 was transformed into the double mutant. When cdc20-1 was introduced into the Δrad4 strain survival at 10 J/m2was less than that caused when CDC20 was overexpressed in the same strain, indicating a dominant negative phenotype of the cdc20-1 mutant allele. The sensitivities of neither the single nor double mutant to hydroxyurea, MMS, phleomycin, and benomyl were significantly different as compared to wild-type or the cdc20-1 strains alone (Figure 1(d)).
Figure 1.
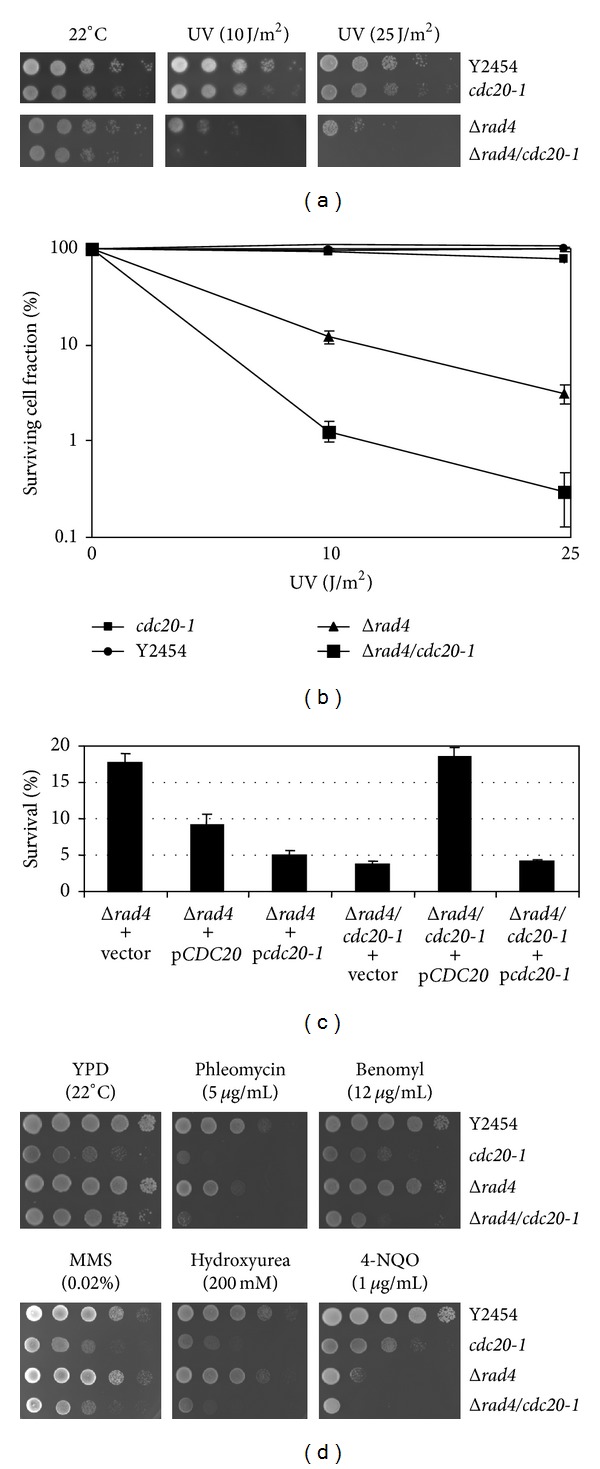
UV resistance of Δrad4 mutants was diminished by the introduction of temperature-sensitive cdc20-1 allele. (a) The optical density A 600 of overnight cultures was normalized to 1.0, and a 1 : 10 serial dilution was prepared. Onto YPD, 5 μL of each dilution was spotted and plates were exposed to UV radiation at energy levels corresponding to 10 and 25 J/m2. Plates were then incubated at 22°C for 3 days. The growth of Δrad4 harboring cdc20-1 was greatly reduced relative to the Δrad4 single mutant or the wild-type strain (Y2454). (b) Diluted cultures of yeast were plated on YPD and then exposed to UV at energy levels of 10 and 25 J/m2, or none at all. Following 3 days of incubation at 22°C, colonies were counted and percent survival was calculated as the total number of colonies on the treated plates divided by the number of colonies on the untreated plates. The Δrad4/cdc20-1 mutant exhibits a tenfold decrease in survivability relative to the Δrad4 strain at both energy levels tested. (c) Plasmids were constructed in which either CDC20 or cdc20-1 was placed under the control of the gal1 promoter. These plasmids, in addition to the empty vector, were transformed into yeast strains containing Δrad4 and Δrad4/cdc20-1. Survivability was determined as described in (c), with cells plated on SCD lacking uracil rather than YPD. Overexpression of CDC20 (pCDC20) resulted in increased survivability of the Δrad4/cdc20-1 strain, while overexpression of cdc20-1 (pcdc20-1) did not. (d) Yeast cells were prepared as in (a) and spotted onto YPD media containing 5 μg/mL phleomycin, 12 μg/mL benomyl, 0.02% MMS, 200 mM hydroxyurea, and 1 μg/mL 4-NQO. The Δrad4/cdc20-1 strain exhibited decreased growth on 4-NQO relative to Δrad4 but was otherwise similar in growth patterns to the cdc20-1 mutant.
3.2. Δrad4 Enhances RNR3:lacZ Induction While Δrad4/cdc20-1 Compromises Induction upon Exposure to UV Irradiation and 4-NQO
It has been reported that inactivation of certain DNA repair pathways in S. cerevisiae may diminish or enhance gene expression in response to DNA damage [46]. In this assay, plasmid with RNR3 fused to lacZ was transformed into Δrad4/cdc20-1, Δrad4, cdc20-1, and wild-type strains. RNR3 is the large subunit of the ribonucleotide reductase complex, which catalyzes dNTP synthesis and is subject to both DNA replication and repair checkpoints. We examined the response of Δrad4 and Δrad4/cdc20-1 strains to both UV radiation and 4-NQO. Deletion of RAD4 enhanced RNR3-lacZ induction by 4-NQO 3.5- and 2.5-fold at concentrations of 0.2 and 0.5 μg/mL (Figure 2(a)). UV radiation at energy levels of 10 and 25 J/m2 enhanced this induction by 2- and 7-fold, respectively (Figure 2(b)). With the introduction of cdc20-1 into the Δrad4 background, however, the fold increase was diminished and not different than either wild-type or cdc20-1 strains alone. These data are not consistent with the values for survivability following UV exposure and indicate an altered capacity for DNA repair that is not related to cell survival in strains harboring both Δrad4 and cdc20-1 simultaneously.
Figure 2.
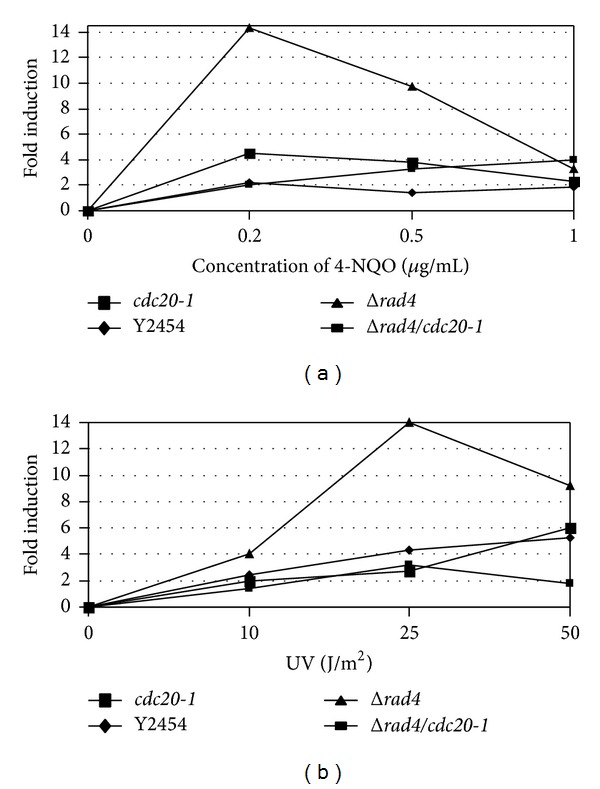
DNA damage-induced RNR3-lacZ expression is reduced in Δrad4/cdc20 mutants that have been (a) exposed to 4-NQO or (b) UV irradiated. Strains were transformed with pZZ2 (gift from S. Elledge), which contains the large subunit of the ribonucleotide reductase complex (RNR3) fused in-frame to lacZ. Cultures were exposed to UV or 4-NQO, incubated for 4 hours, and assayed for β-galactosidase activity. Fold induction was calculated as the ratio of β-gal activity of treated cultures cells to that of untreated cultures. In both treatments, Δrad4 strains were capable of evoking RNR3-lacZ expression, while Δrad4/cdc20-1 strains did not significantly affect this activity relative to wild-type (Y2454) or cdc20-1 strains.
3.3. Δrad4/cdc20-1 Mutant Strains Are Defective in Plasmid Rejoining In Vivo
To determine whether introduction of cdc20-1 affected the cell's ability to repair double strand breaks, a plasmid end-joining assay was performed [47, 48]. Briefly, strains of S. cerevisiae were transformed with a linearized plasmid (pYES.2) containing the URA3 marker. To normalize for differences in transformation efficiencies between strains, an uncut version of the same plasmid was transformed, in parallel. Since the plasmid must be recircularized in order to revert the ura − phenotype of the selected strains, the number of transformants obtained with the linearized plasmid normalized to the number obtained with the uncut plasmid provides a quantitation of the DSB-repair ability of the yeast strains. As shown in Figure 3, Δrad4 strains were able to repair DSBs, although not as efficiently as wild type. The double mutant, however, had significantly diminished ability to repair these defects as compared with the single deletion alone. These results indicate that introduction of cdc20-1 affects the cells ability to repair damaged DNA in an NER-defective strain.
Figure 3.
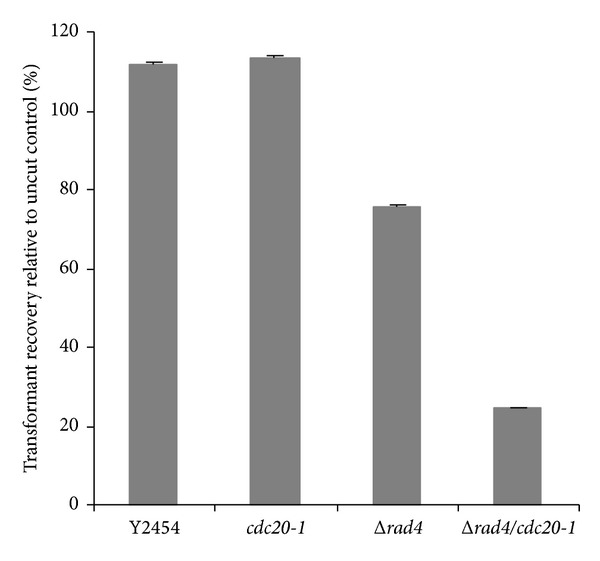
Activity of Cdc20 contributes to the ability of Δrad4 strains to carry out double strand break repair. Plasmid DNA (pYES.2/URA3 +) was linearized with PvuII, and aliquots from the same pool of digested DNA were used to transform Δrad4, Δrad4/cdc20-1, cdc20-1, or a wild-type strain (Y2454). Following incubation at 22°C for 4 days, colonies were counted, and transformation efficiencies were calculated. The fold changes (linearized/uncut) are represented.
3.4. Microscopic Analysis of Rad4-GFP/cdc20-1 Mutants Reveals Altered Rad4 Fluorescence upon UV Irradiation
To determine whether cdc20-1 may have an effect on expression or stability of RAD4 we utilized a strain of yeast harboring a C-terminal GFP tag on RAD4, with or without a chromosomal copy of cdc20-1. Liquid cultures were irradiated at 25 and 100 J/m2 and examined four hours after exposure. Fluorescence was observed in strains carrying the tagged copy of RAD4 in an otherwise wild-type background, regardless of treatment or time of outgrowth. When cdc20-1 was present, however, fluorescence was diminished following UV irradiation such that around 60% of the cells had visible fluorescence (Figure 4). Since strains with cdc20-1 were capable of survival on solid media at these energy levels, we do not attribute decreased fluorescence to cell death.
Figure 4.
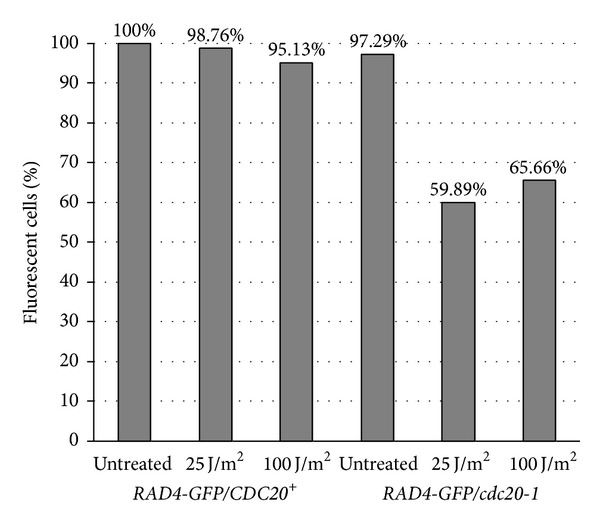
The cdc20-1 mutant allele contributes to diminished expression of Rad4-GFP upon UV irradiation. Strains harboring cdc20-1 show less fluorescence of Rad4-GFP. Cultures were grown overnight at 22°C and then UV irradiated using a Stratagene UV Stratalinker 1800. An outgrowth of four hours was then allowed. Three replicates were performed, and percentages were determined on a sample size of greater than 150 cells. All images were captured using a Zeiss Imager M1 AXIO paired with a Photometrics Coolsnap HQ2camera.
4. Discussion
Here we demonstrate that Δrad4 and cdc20-1 exhibit a synthetic growth defect upon UV exposure in Saccharomyces cerevisiae. We examined strains carrying a deletion of RAD4 and the conditional cdc20-1 allele and found that the two mutations in combination resulted in extreme UV sensitivity. Overexpression of CDC20 in Δrad4/cdc20-1 restored the slight UV resistance observed in the single mutant Δrad4. Microscopic analyses indicate that in the presence of cdc20-1, Rad4-GFP fluorescence is significantly diminished relative to wild type after a four outgrowth following UV irradiation. When introduced into a NER-defective strain (Δrad4), survival following UV irradiation is severely compromised. Our data demonstrate that this UV sensitivity may be the result of a reduced capability to repair double strand breaks, as the Δrad4/cdc20-1 strain had a diminished capacity to anneal DSBs in a plasmid end-joining assay.
We propose that Cdc20 may act to indirectly modulate the nucleotide excision repair pathway by way of its role in protein degradation. In a report by Gillette et al. [9] a novel cullin-based E3 ubiquitin ligase (ECS ligase) was identified that ubiquitinates Rad4 following UV exposure. If Cdc20 has a role in regulation of ECS ligase activity, the ubiquitination of Rad4 could be affected indirectly; however, Cdc20 may also directly regulate its ubiquitination through the APC/C. Rad4 contains a putative cyclin destruction box (D box) in its amino terminus, although its expression does not exhibit cell cycle periodicity. D box motifs contain two conserved residues (RXXL) and several other more moderately conserved residues. They are a highly conserved sequence common to substrates of the APC/C [49–52]. Previous studies have shown that Rad4 is stable, with a half-life of approximately 4 hours [5]. Upon UV exposure, these levels decrease as the protein is proteolyzed by the 26S-proteasome. The ubiquitination, not the stabilization, of Rad4 in response to UV irradiation via the ECS ligase regulates NER in part [9]. Without an examination of Rad4 ubiquitination in cdc20-1 mutants, however, it is impossible for us to state that this is indeed the case, although microscopic examinations lend proof to this argument.
Despite the strong genetic evidence that Cdc20 affects the ability of Saccharomyces cerevisiae to carry out NER in the absence of RAD4 we have yet to demonstrate biochemically how Cdc20 acts in this pathway. There are a number of NER proteins that contain putative D boxes and many other candidate proteins that may be targeted for ubiquitination via the APC/CCdc20 or acted upon by Cdc20 alone. We do not exclude the possibility that unidentified protein is a member of the chromatin remodeling complex as Rad4/Rad23 complex is known to have a role in maintenance of heterochromatin structure [53, 54]. In addition, recent studies have shown that Rad4 can bind to the proteasome without affecting overall protein degradation ability of the complex [36], and studies have yet to elucidate mediators of this direct binding. Further research is necessary to dissect this interaction and to understand the roles of each component. This study is the first, however, to present evidence that Cdc20 may play a role in the degradation of proteins involved in nucleotide excision repair.
Acknowledgments
The authors wish to thank Dr. Steven Elledge for the RNR3:lacZ plasmid pZZ2 and Dr. Dan Burke for strain 405-1-1 that contained the cdc20-1 allele. They would also like to thank Elizabeth Charlotin, Sarah Armstrong, and Sarah Lea McGuire for helpful discussion and critical reading of the paper, as well as Glen Shearer, George Santangelo, Kristine Willis, and Mohamed Elasri for technical advice and support. This work was supported by the NIH/NCRR INBRE Program Grant RR016476 and the Science Department at Dominican College.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Molecular and Cellular Biology. 2000;20(21):8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan F, Jin S, Amundson SA, et al. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene. 2002;21(49):7488–7496. doi: 10.1038/sj.onc.1205896. [DOI] [PubMed] [Google Scholar]
- 3.Sabbioneda S, Bortolomai I, Giannattasio M, Plevani P, Muzi-Falconi M. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair. 2007;6(1):121–127. doi: 10.1016/j.dnarep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(21):11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lommel L, Ortolan T, Chen L, Madura K, Sweder KS. Proteolysis of a nucleotide excision repair protein by the 26S proteasome. Current Genetics. 2002;42(1):9–20. doi: 10.1007/s00294-002-0332-9. [DOI] [PubMed] [Google Scholar]
- 6.Schauber C, Chen L, Tongaonkar P, et al. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391(6668):715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 7.Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nature Cell Biology. 2000;2(9):601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 8.Ng JM, Vermeulen W, van der Horst GTJ, et al. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes & Development. 2003;17(13):1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillette TG, Yu S, Zhou Z, Waters R, Johnston SA, Reed SH. Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. The EMBO Journal. 2006;25(11):2529–2538. doi: 10.1038/sj.emboj.7601120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi S, Loda M. The role of the ubiquitination-proteasome pathway in breast cancer: use of mouse models for analyzing ubiquitination processes. Breast Cancer Research. 2003;5(1):16–22. doi: 10.1186/bcr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutcliffe JS, Jiang Y-H, Galjaard R-J, et al. The E6-AP ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Research. 1997;7(4):368–377. doi: 10.1101/gr.7.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohh M. Ubiquitin pathway in VHL cancer syndrome. Neoplasia. 2006;8(8):623–629. doi: 10.1593/neo.06442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe D, Reiner T, Keeley JL, Pizzini M, Keil RL. Ubiquitin metabolism affects cellular response to volatile anesthetics in yeast. Molecular and Cellular Biology. 1999;19(12):8254–8262. doi: 10.1128/mcb.19.12.8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foury F. Human genetic diseases: a cross-talk between man and yeast. Gene. 1997;195(1):1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 15.Hershko A, Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 16.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae . Genetics. 2012;192(2):319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickart CM. Ubiquitin enters the new millennium. Molecular Cell. 2001;8(3):499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 18.Peters J-M. The anaphase-promoting complex: proteolysis in mitosis and beyond. Molecular Cell. 2002;9(5):931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 19.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes & Development. 2002;16(17):2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 20.Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Current Opinion in Cell Biology. 2002;14(6):706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- 21.Ross KE, Cohen-Fix O. The role of Cdh1p in maintaining genomic stability in budding yeast. Genetics. 2003;165(2):489–503. doi: 10.1093/genetics/165.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marston A. Chromosome segregation in budding yeast: sister chromatid cohesion and related mechanisms. Genetics. 2014;196(1):31–63. doi: 10.1534/genetics.112.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan J, Chen R-H. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae . Genes & Development. 2004;18(12):1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke DJ, Segal M, Andrews CA, et al. S-phase checkpoint controls mitosis via an APC-independent Cdc20p function. Nature Cell Biology. 2003;5(10):928–935. doi: 10.1038/ncb1046. [DOI] [PubMed] [Google Scholar]
- 25.Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters J-M. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(20):11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes & Development. 1999;13(7):768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 27.Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutation Research. 2000;451(1-2):13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 28.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends in Genetics. 2012;28(1):566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Hanawalt PC. Controlling the efficiency of excision repair. Mutation Research. 2001;485(1):3–13. doi: 10.1016/s0921-8777(00)00071-9. [DOI] [PubMed] [Google Scholar]
- 30.de Feraudy S, Revet I, Bezrookove V, Feeney L, Cleaver JE. A minority of foci or pan-nuclear apoptotic staining of γH2AX in the S phase after UV damage contain DNA double-strand breaks. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(15):6870–6875. doi: 10.1073/pnas.1002175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braithwaite E, Wu X, Wang Z. Repair of DNA lesions: mechanisms and relative repair efficiencies. Mutation Research. 1999;424(1-2):207–219. doi: 10.1016/s0027-5107(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 32.Kusumoto R, Masutani C, Sugasawa K, et al. Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro . Mutation Research. 2001;485(3):219–227. doi: 10.1016/s0921-8777(00)00082-3. [DOI] [PubMed] [Google Scholar]
- 33.Cadet J, Bellon S, Douki T, et al. Radiation-induced DNA damage: formation, measurement, and biochemical features. Journal of Environmental Pathology, Toxicology and Oncology. 2004;23(1):33–43. doi: 10.1615/jenvpathtoxoncol.v23.i1.30. [DOI] [PubMed] [Google Scholar]
- 34.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Molecular and Cellular Biology. 2002;22(13):4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Yan J, Kim I, Liu C, Huo K, Rao H. Rad4 regulates protein turnover at a postubiquitylation step. Molecular Biology of the Cell. 2010;21(1):177–185. doi: 10.1091/mbc.E09-04-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q-E, Han C, Zhang B, Sabapathy K, Wani AA. Nucleotide excision repair factor XPC enhances DNA damage-induced apoptosis by downregulating the antiapoptotic short isoform of caspase-2. Cancer Research. 2012;72(3):666–675. doi: 10.1158/0008-5472.CAN-11-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Z, Liu S, Zhang Y, Wang Z. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Research. 2004;32(20):5981–5990. doi: 10.1093/nar/gkh934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazón G, Lam AF, Kwen Ho C, Kupiec M, Symington LS. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nature Structural & Molecular Biology. 2012;19:964–971. doi: 10.1038/nsmb.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guzder SN, Sung P, Prakash L, Prakash S. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. The Journal of Biological Chemistry. 1998;273(47):31541–31546. doi: 10.1074/jbc.273.47.31541. [DOI] [PubMed] [Google Scholar]
- 41.Jansen LE, Verhage RA, Brouwer J. Preferential binding of yeast Rad4·Rad23 complex to damaged DNA. The Journal of Biological Chemistry. 1998;273(50):33111–33114. doi: 10.1074/jbc.273.50.33111. [DOI] [PubMed] [Google Scholar]
- 42.Birrell GW, Giaever G, Chu AM, Davis RW, Brown JM. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(22):12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortolan TG, Chen L, Tongaonkar P, Madura K. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Research. 2004;32(22):6490–6500. doi: 10.1093/nar/gkh987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie Z, Liu S, Zhang Y, Wang Z. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Research. 2004;32(20):5981–5990. doi: 10.1093/nar/gkh934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11(4):355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 46.Jia X, Xiao W. Compromised DNA repair enhances sensitivity of the yeast RNR3-lacZ genotoxicity testing system. Toxicological Sciences. 2003;75(1):82–88. doi: 10.1093/toxsci/kfg158. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki K, Imai Y, Yamashita I, Fukui S. In vivo ligation of linear DNA molecules to circular forms in the yeast Saccharomyces cerevisiae . Journal of Bacteriology. 1983;155(2):747–754. doi: 10.1128/jb.155.2.747-754.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Research. 1996;24(23):4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sudakin V, Ganoth D, Dahan A, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Molecular Biology of the Cell. 1995;6(2):185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285(5426):418–421. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 51.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae . The EMBO Journal. 1998;17(5):1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen BO, Wagener C, Marinoni F, et al. Cell cycle-and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes & Development. 2000;14(18):2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong F, Fahy D, Smerdon MJ. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nature Structural & Molecular Biology. 2006;13(10):902–907. doi: 10.1038/nsmb1152. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Chen H, Bi X, Gong F. Detection of an altered heterochromatin structure in the absence of the nucleotide excision repair protein Rad4 in Saccharomyces cerevisiae . Cell Cycle. 2013;12(15):2435–2442. doi: 10.4161/cc.25457. [DOI] [PMC free article] [PubMed] [Google Scholar]


