Abstract
Objective
Defects in insulin signaling are associated with abnormal endothelial cell function, which occurs commonly in cardiovascular disease. Targets of insulin signaling in endothelial cells are incompletely understood. Protein S-palmitoylation, the reversible modification of proteins by the lipid palmitate, is a post-translational process relevant to cell signaling, but little is known about the role of insulin in protein palmitoylation.
Approach and Results
To test the hypothesis that insulin alters protein palmitoylation in endothelial cells, we combined acyl biotin exchange chemistry with SILAC to perform quantitative proteomic profiling of human endothelial cells. We identified ~380 putative palmitoylated proteins, of which >200 were not known to be palmitoylated. Less than 10% of the putative palmitoylated proteins were induced or suppressed by insulin. Of those potentially affected by insulin, <10 have been implicated in vascular function. For one, PAFAH1b3 (not previously known to be palmitoylated), we confirmed that insulin stimulated palmitoylation without affecting PAFAH1b3 protein abundance. Chemical inhibition of palmitoylation prevented insulin-induced angiogenesis in vitro; knockdown of PAFAH1b3 had the same effect. PAFAH1b3 knockdown also disrupted cell migration. Mutagenesis of cysteines at residues 56 and 206 prevented palmitoylation of PAFAH1b3, abolished its capacity to stimulate cell migration, and inhibited its association with detergent-resistant membranes (DRMs), which are implicated in cell signaling. Insulin promoted the association of wild type PAFAH1b3 with detergent-resistant membranes.
Conclusions
These findings provide proof of principle for employing proteomics to identify novel insulin-inducible palmitoylation targets relevant to endothelial function.
Keywords: Palmitoylation, insulin signaling, endothelial cell migration, angiogenesis
INTRODUCTION
Insulin resistance, usually reflecting decreased insulin-dependent glucose transport in peripheral tissues and decreased insulin-dependent suppression of endogenous glucose production, can occur independent of hyperglycemia if compensatory insulin secretion is sufficiently robust. However, sustained insulin resistance can have pleiotropic effects that are associated with cardiovascular complications 1. Optimal management to minimize the risk of these complications is unresolved 2–5. Insulin is an important mediator of endothelial function 6, and inactivation of the endothelial cell insulin receptor in mice increases atherosclerosis independent of traditional risk factors 7. However, the molecular mediators of insulin signaling in endothelial cells remain poorly understood. Identifying novel endothelial cell targets of insulin treatment could provide insight into the relationship between metabolism and inflammation that occurs in the setting of insulin resistance.
Lipids are involved in insulin signaling and impact endothelial cell function. Lipid molecules can integrate information to alter homeostasis through well-characterized mechanisms including the activation of nuclear receptors 8 and the complex network of lipid-modifying enzymes 9. Less is known about the how lipids affect cellular homeostasis through protein modifications such as prenylation, myristoylation and palmitoylation 10. Unlike other lipidation reactions, protein S-palmitoylation is reversible and posttranslational, making it inherently suitable (serving as an on/off switch based on the presence or absence of palmitate) for regulating function. G-proteins, scaffold proteins, kinases, vesicle proteins, and others utilize palmitoylation to modulate growth, differentiation, embryonic development, and cell-cell interactions 11, 12. Our recent observations point to an unexpected role for de novo lipogenesis in S-palmitoylation of eNOS in blood vessels 13 and mucin 2 in the intestine 14. Both of these palmitoylation events may be relevant to metabolic disorders since de novo lipogenesis is regulated by insulin.
Palmitoylated proteins have been identified in yeast, neurons, and certain membrane fractions15–17. Little is known about palmitoylation targets influenced by insulin. We tested the hypothesis that insulin alters the dynamics of protein palmitoylation in endothelial cells using SILAC (Stable Isotope Labeling by Amino acids in Cell culture). With this technique, proteomes are distinguished based on isotopes that correspond to experimental conditions 18, in this case the presence or absence of insulin treatment. We identified several novel palmitoylation targets regulated by insulin and demonstrated that one of these, PAFAH1b3 (Platelet-Activating Factor Acetylhydrolase IB subunit gamma), is likely to be important for endothelial cell function.
MATERIALS AND METHODS
Materials and Methods are available in the online-only Supplement.
RESULTS
Global identification of palmitoylation candidates
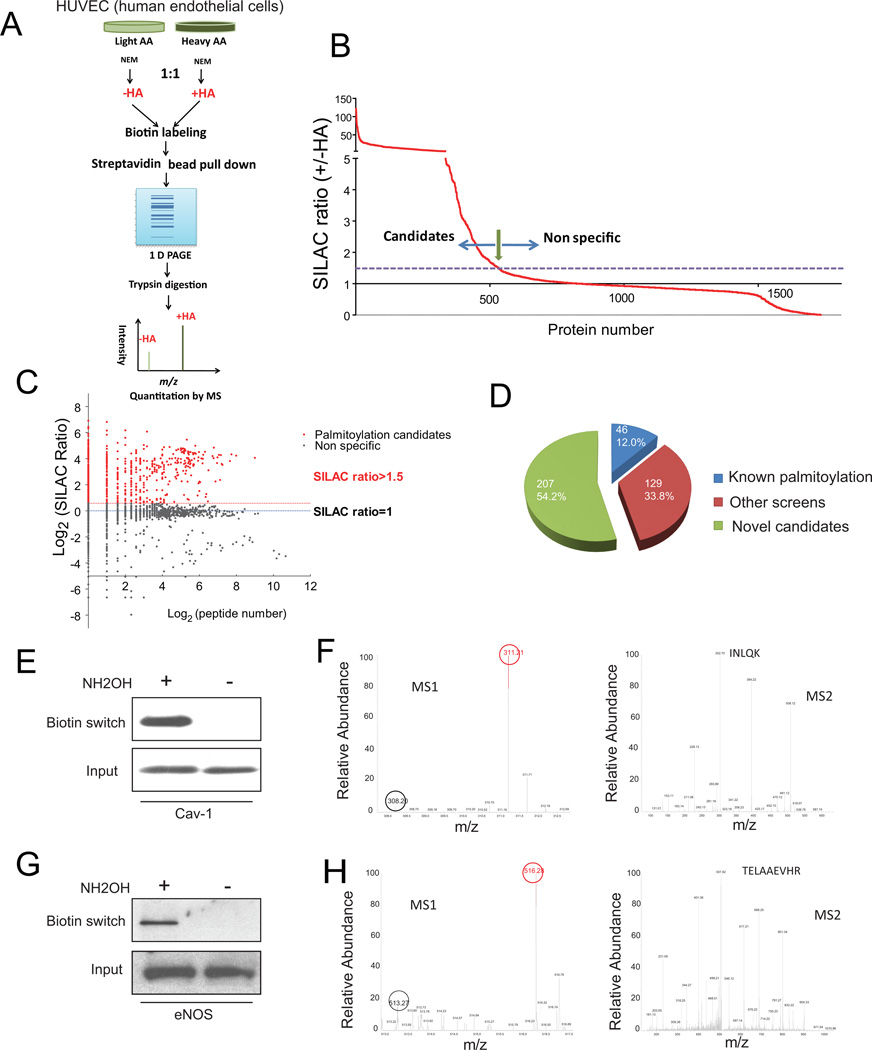
To first establish the feasibility of a quantitative proteomic strategy utilizing SILAC and focusing on palmitoylation, we screened HUVECs (Figure 1A). The screen utilized acyl-biotin exchange chemistry adapted for large-scale proteomics. With this technique, proteins are treated with N-ethylmaleimide (NEM in Figure 1A) to modify free thiols thus preventing their subsequent biotinylation. Subsequent treatment with hydroxylamine (HA in Figure 1A) cleaves the thioester bond between palmitate and cysteines, leaving cysteines susceptible to biotinyl-labeling. Cells cultured with “light” stable isotope-labeled amino acids were not treated with hydroxylamine and represent controls. The potential palmitoylated proteins should be enriched in the “heavy” samples (+HA), thus having higher SILAC ratios (H/L, which also represents +HA/-HA). Of the ~1700 proteins identified by acyl-biotin exchange and mass spectrometry, ~ 500 had a SILAC ratio of >1.5 (a threshold determined by the presence of known palmitoylated proteins) and are potential palmitoylated proteins (Figure 1B). More than 72% of these proteins were identified based on the presence of more than one peptide (Log2>=1, Figure 1C). These include G protein associated regulators, scaffold proteins, vesicular or membrane trafficking proteins, and cell adhesion or ECM interaction molecules (grouped based on functional characteristics in Supplemental Table I in the online-only Data Supplement). Among high confidence candidates (382 proteins with >=2 peptides identified, Figure 1D), 175 were either known palmitoylated proteins (46 or 12%) or previously identified 16, 17, 19 by palmitoylation screens (129 or 34%). The remaining 207 candidates (54%) represent potentially novel palmitoylated proteins. A complete list with protein descriptions and comparisons with prior screens is presented in Supplemental Table II in the online-only Data Supplement.
Figure 1.
Systematic identification of palmitoylated proteins by SILAC. A, SILAC strategy utilizing acyl biotin exchange. B, SILAC ratio of the proteome identified by mass spectrometry. C, Scatter plot of SILAC ratio vs. peptide number in log scale. D, Pie chart of identified palmitoylation candidates. E, Biotin switch assay for Cav-1. F, Mass spec demonstrating isotope enrichment (left) and peptide sequence (right) for Cav-1. G, Biotin switch assay for eNOS. H, Mass spec demonstrating isotope enrichment (left) and peptide sequence (right) for eNOS.
We verified our MS-based approach using caveolin-1 (Cav-1) and endothelial nitric oxide synthase (eNOS), known palmitoylated HUVEC proteins. For Cav-1 (Figure 1E) and eNOS (Figure 1G), the biotin switch signal was hydroxylamine-dependent, characteristic of palmitoylation of cysteines. “Heavy” peptide Cav-1, denoted by the red circle in the SILAC precursor scan (MS1), was more abundant than “light” peptide, denoted by the black circle, with the tandem spectrum (MS2) documenting the sequence of the peptide (Figure 1F). Similar results were seen for eNOS (Figure 1H). These results suggest that this SILAC-based approach is suitable for identifying palmitoylation candidates.
Detection of palmitoylated proteins in response to insulin
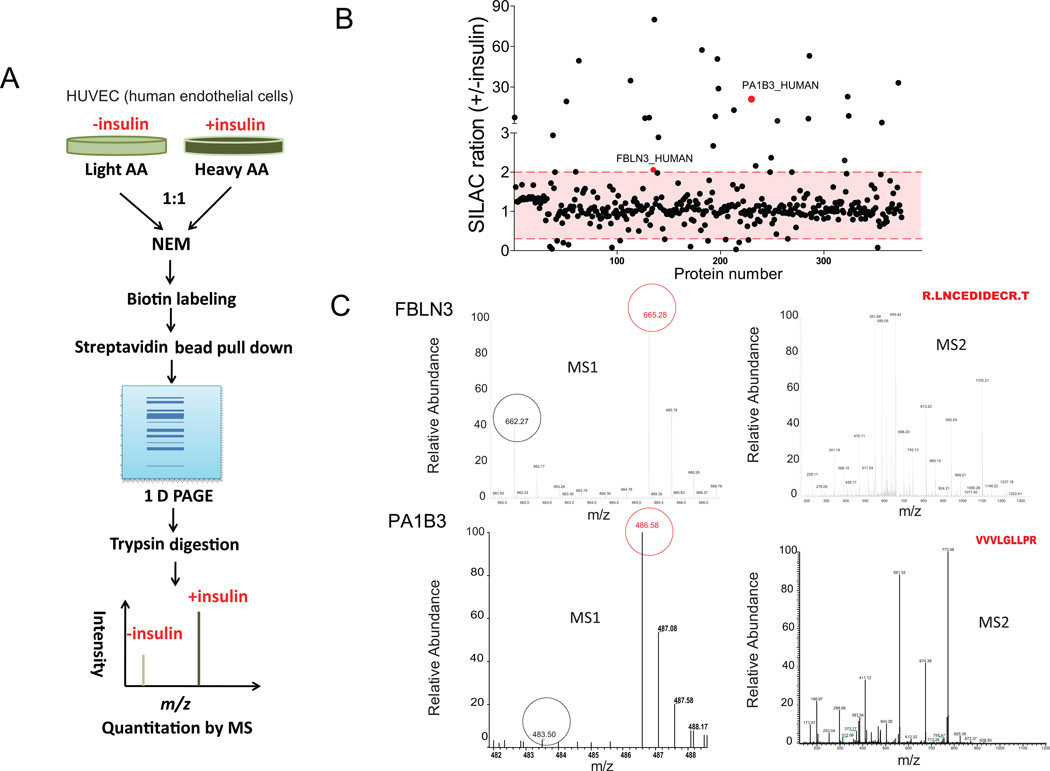
To pursue how insulin affects palmitoylation dynamics, “heavy” samples were treated with insulin (100 ng/ml) for 6 h, and “light” samples were treated with vehicle (Figure 2A). Since the median half life of mammalian proteins is estimated at ~46 h 20, steady state protein levels would be unlikely to be affected within 6 h and changes detected by SILAC with acyl biotin exchange (ratio of H/L) should reflect differential palmitoylation by insulin (+insulin/-insulin). We detected a palmitoylation proteome consisting of 375 high confidence (>=2 peptides identified) proteins (Figure 2B), of which 303 were also identified as palmitoylation candidates in our first screen, a finding that suggests reproducibility of our approach. Most candidates were unaffected by insulin treatment (>85% with SILAC ratios close to 1, Figure 2B). Less than 10% were either increased (>2 fold) or reduced (to <0.3) by insulin in HUVECs (Figure 2B). Fold changes and P values for these 35 proteins meeting our arbitrary induction or reduction criteria are shown in Supplemental Table III in the online-only Data Supplement. Mass spec verifications of two proteins identified in Figure 2B, FBLN3 (2.29 fold increase, P<0.0001) and PA1B3 (also known as PAFAH1b3, 21 fold increase, P=0.0083),are shown in Figure 2C. The SILAC precursor scan (MS1) demonstrated an increased m/z signal suggestive of insulin-induced palmitoylation and tandem spectrum (MS2) verified the sequences for each protein. A complete list of proteins detected in this screen is presented in Supplemental Table IV in the online-only Data Supplement.
Figure 2.
Effects of insulin on palmitoylation. A, SILAC strategy. B, SILAC ratios of palmitoylated proteins regulated by insulin. Several signals induced by insulin, including PAFAH1b3 (PA1B3), are labeled. C, Isotope enrichment and peptide sequencing are shown for two of the candidates labeled in panel B.
Palmitoylation is required for insulin effects on endothelial cell migration and angiogenesis
We focused on protein signals upregulated by insulin (SILAC ratio 2 or greater in Table 2) that might be relevant to vascular function. Membrane-associated proteins that utilize palmitoylation to increase hydrophobicity for impacting cell motility or membrane trafficking could be suitable targets for novel therapeutics in metabolic syndrome and type 2 diabetes. We identified several appropriate candidates including FBLN3 (fibulin-3), which associates with the extracellular matrix and promotes cell migration 21, 22, LYVE1 (Lymphatic vessel endothelial hyaluronic acid receptor 1 23, BST2 (Bone marrow stromal antigen 2) 24, and PAFAH1b3 25, 26 (pursued in detail below).
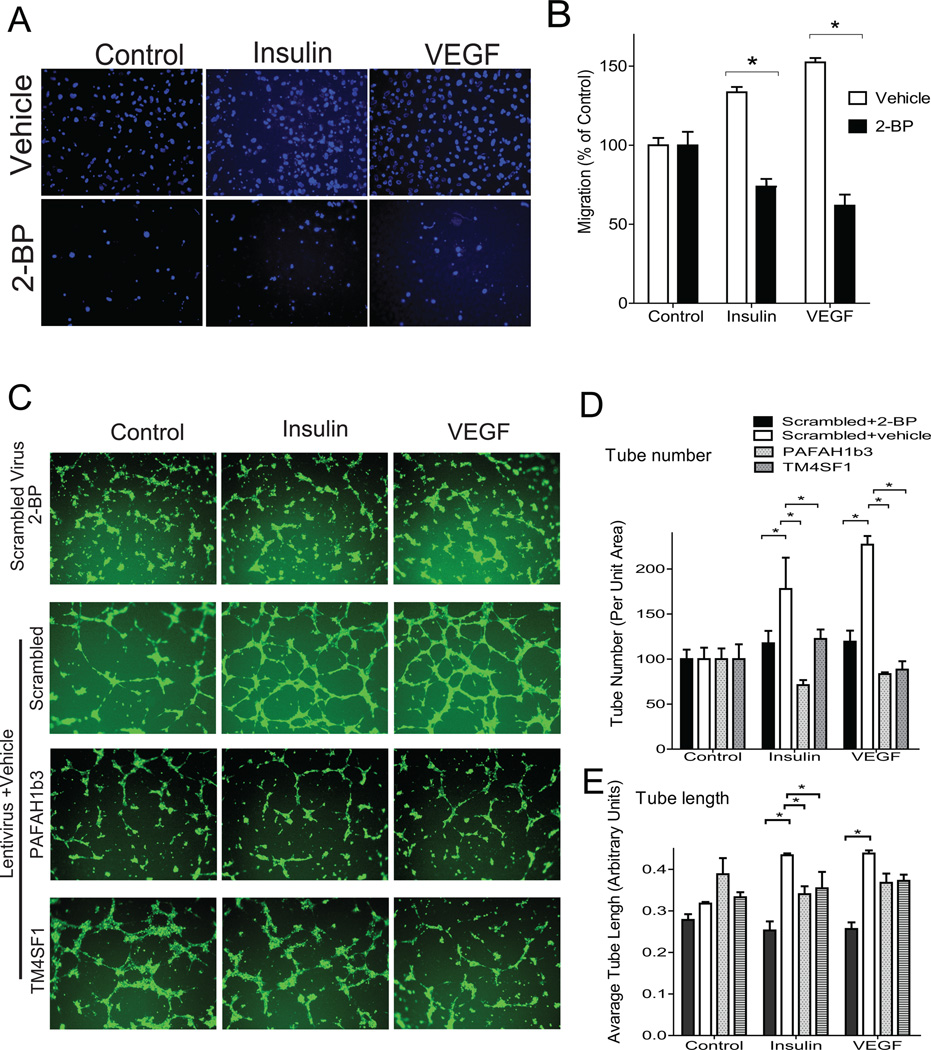
Since several of these palmitoylated proteins may be involved in cell migration, we addressed the potential role of global palmitoylation in migration using chamber transwell assays. Treating HUVECs with 2-bromopalmitate (2-BP, 20 μM), a palmitoylation inhibitor 27–29, blocked the ability of insulin or VEGF to stimulate migration through a semi-permeable membrane (Figure 3A,B). Since cell migration is an important component of angiogenesis, a vascular effect of insulin 30, we addressed the effects of palmitoylation on a surrogate of the angiogenic response, in vitro tube formation. Treatment with 2-BP blocked the ability of insulin or VEGF to stimulate tube formation (compare the top two rows in Figure 3C—results are quantified in Figure 3D,E). To determine if interfering with palmitoylation status has a similar effect in cells from an arterial source, we compared tube formation in bovine aortic endothelial cells and HUVECs. In both BAECs and HUVECs, 2-BP inhibited tube formation in a dose-dependent manner (Figure I in the online-only Data Supplement).
Figure 3.
Chemical inhibition of palmitoylation decreases insulin-induced and VEGF-induced cell migration and in vitro tube formation. A, Representative images (3 independent experiments) from cell transwell migration assays in the presence and absence of the palmitoylation inhibitor 2-bromopalmitate (2-BP, 20 μM). B, Quantified data for cell migration. C, Cells were infected with lentiviruses then treated with vehicle or 2-BP (20 μM) followed by plating on Matrigel. Representative images (3 independent experiments) are shown in the presence of control, insulin, or VEGF. D, Quantification of tube number (relative to controls) with different interventions. E, Quantification of tube length with different interventions. *indicates P<0.05 by ANOVA.
PAFAH1b3 was of particular interest because differential palmitoylation was enhanced more than 20 fold by insulin, and the protein is thought to connect lipid remodeling with Golgi-endosomal cargo transport 25, 26, which may be required for endothelial cells to migrate and form tube structures 31, 32. To our knowledge, palmitoylation of PAFAH1b3 has not been reported and this protein has not been implicated in endothelial function. As a positive control, we also studied TM4SF1 (Transmembrane 4 L6 family member 1), which was detected in each of our palmitoylation screens (Supplemental Tables I and IIII in the online-only Data Supplement). TM4SF1 is not known to be palmitoylated, but has been shown to be required for both endothelial cell migration 33 and VEGF-induced angiogenesis 34. Knockdown of PAFAH1b3 and TM4SF1 (extent of knockdown is shown in Figure II in the online-only Data Supplement) inhibited tube formation in response to insulin or VEGF (compare the bottom three rows in Figure 3C—results are quantified in Figure 3D,E). For each knockdown, effects on tube number were comparable with 2-BP, while effects on tube length were less than those seen with 2-BP. These data suggest that palmitoylation is required for insulin-activated endothelial migration and tube formation.
Insulin regulates PAFAH1b3 palmitoylation
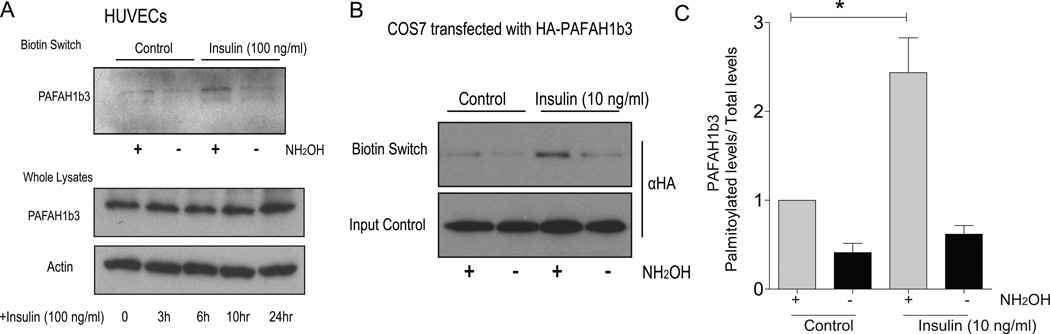
When the biotin switch assay was repeated in HUVECs, endogenous palmitoylated PAFAH1b3 was detected and shown to be induced by insulin (Figure 4A, top panel). This induction was not due to effects of insulin on PAFAH1b3 protein abundance since PAFAH1b3 levels determined by Western blotting using a PAFAH1b3 antibody were not increased with insulin incubation for up to 24 h (Figure 4A, bottom panel). To confirm this observation in an independent experimental system with an antibody to an easily detectable tag, mouse PAFAH1b3 cDNA was cloned into an expression vector with an N-terminal hemagglutinin epitope (HA-PAFAH1b3). When expressed in COS cells, the recombinant protein showed increased palmitoylation in response to insulin at a concentration of 10 ng/ml as shown in Figure 4B. Quantified results from this experiment demonstrating a significant increase in palmitoylation with 10 ng/ml insulin are shown in Figure 4C.
Figure 4.
PAFAH1b3 is a novel palmitoylated protein regulated by insulin. A, Top-Biotin switch assay for palmitoylation of endogenous PAFAH1b3 protein. Bottom-Effects of insulin on total protein abundance. B, Effects of insulin on palmitoylation of tagged PAFAH1b3. C, The quantification of biotin switch assay for palmitoylation of HA-PAFAH1b3 protein in transfected cells treated with control or insulin (3 independent experiments). * p<0.05 by two-way ANOVA.
To implicate an insulin receptor-dependent process in the stimulation of PAFAH1b3 palmitoylation by insulin, we performed biotin switch assays in cells after knocking down the insulin receptor. Insulin (10 ng/ml) increased PAFAH1b3 palmitoylation in control (Scrambled) cells, but this effect was diminished in cells treated with an shRNA to the insulin receptor (Figure III in the online-only Data Supplement).
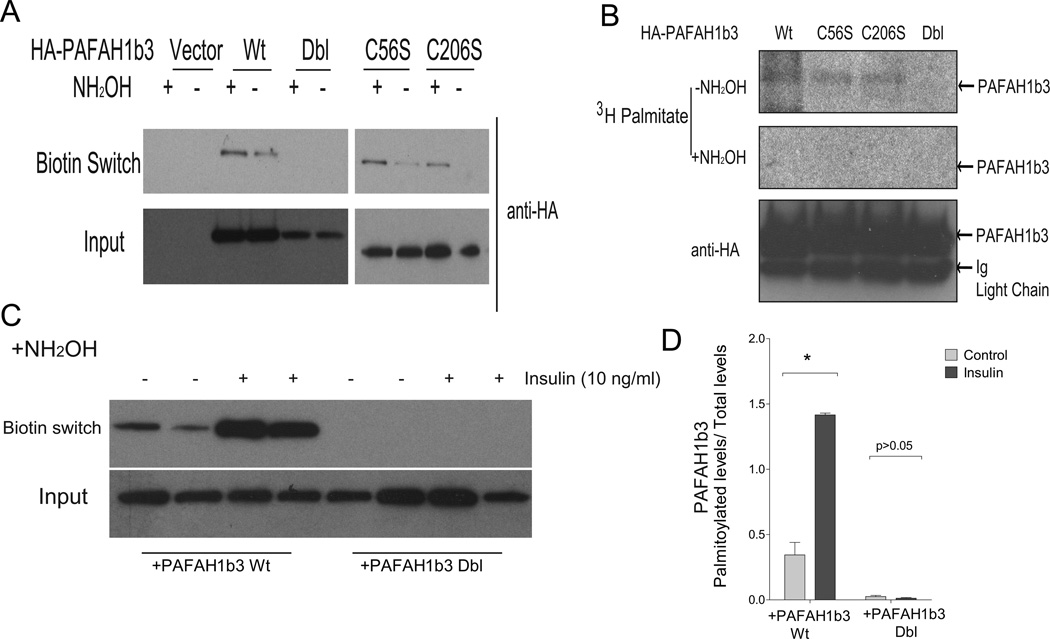
Since there are only two cysteine residues (56 and 206 in mouse) in PAFAH1b3, we introduced conservative (cysteine to serine) mutations at both of these residues and generated recombinant proteins with each single (C56S and C206S) mutation as well as another with both mutations (double or dbl, C56S/C206S). In biotin switch palmitoylation assays, each single mutant was palmitoylated but the double mutant was not (Figure 5A). To confirm this observation with an independent assay, cells were labeled with [3H]-palmitate, then tagged recombinant proteins were immunoprecipitated and subjected to autoradiography. Radiolabeled bands disappeared with hydroxylamine (NH2OH), demonstrating thioester bond dependence of the association between the label and the protein PAFAH1b3 (Figure 5B), which is characteristic of palmitoylation. Consistent with a role for both cysteines in palmitoylation, the radiolabeled signal was absent when the double mutant was studied despite the presence of similar amounts of protein as compared to cells expressing the wild type and single mutant constructs. These results suggest that both PAFAH1b3 cysteine residues are palmitoylated. To provide additional evidence that these cysteines are required for insulin induction of palmitoylation, the biotin switch assay was repeated in several experiments with wild type and double cysteine mutation PAFAH1b3 in the presence and absence of insulin. Insulin (10 ng/ml) increased palmitoylation in the wild type but not the mutated protein as shown in Figure 5C with quantification of the results provided in Figure 5D.
Figure 5.
Identification of palmitoylation sites of PAFAH1b3 protein. A, Palmitoylation status of mutant and wild type PAFAH1b3 protein by biotin switch assay. B, Confirmation of palmitoylation with a radiolabel assay. [3H]-palmitic acid associated with PAFAH1b3 was detected by autoradiography. C-D, Representative blots (C) and quantification of biotin switch assay (D) for palmitoylation of HA-PAFAH1b3 protein in the absence or presence of insulin. Wild type (Wt) or palmitoylation-deficient (Dbl for double cysteine mutation) PAFAH1b3 were assayed (n=4).
Palmitoylation of PAFAH1b3 is required to promote cell migration
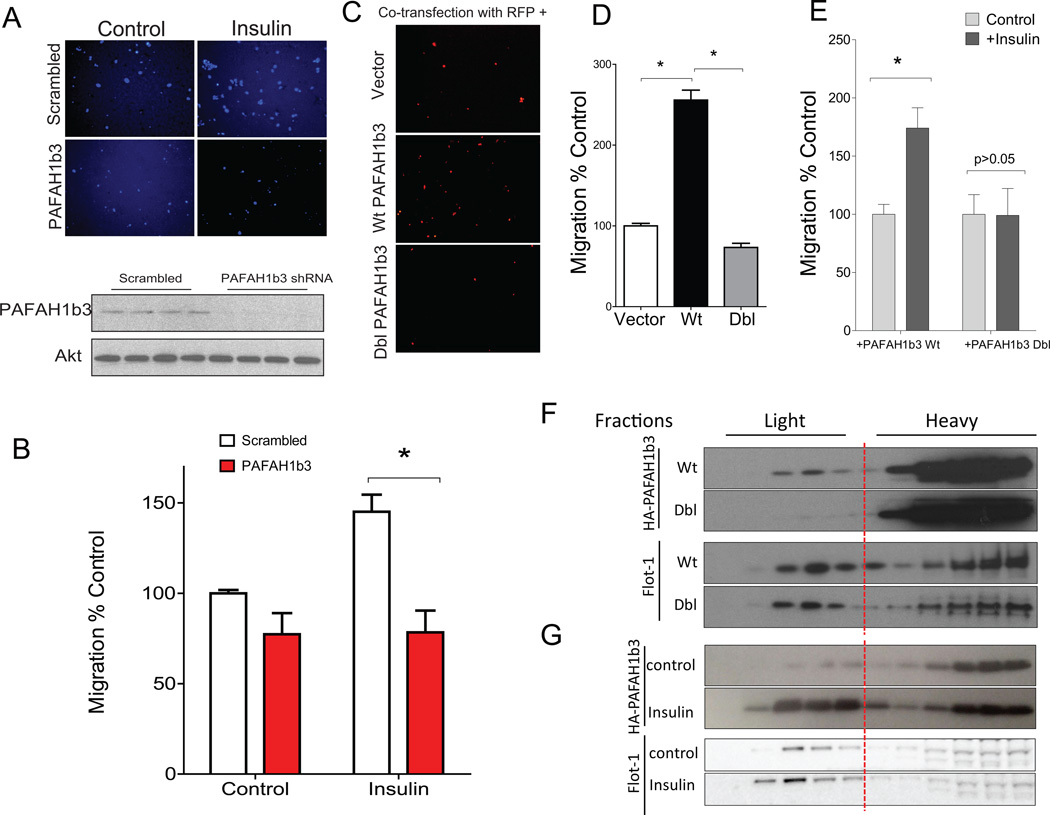
Knockdown of PAFAH1b3 abrogates the ability of insulin (and VEGF) to stimulate angiogenesis in vitro (Figure 3). Angiogenesis requires cell migration. Consistent with the effects of PAFAH1b3 knockdown on angiogenesis, PAFAH1b3 knockdown also eliminated the stimulatory effect of insulin on cell migration (Figure 6, representative images in A, quantified results in B). Overexpression of wild type PAFAH1b3 in HEK 293 cells promoted cell migration, but this effect was lost with overexpression of the C56S/C206S mutant (Dbl) (Figure 6, representative images in C, quantified results in D). Insulin treatment promoted cell migration in cells expressing wild type PAFAH1b3 but did not promote migration in cells expressing the C56S/C206S mutant (Dbl); quantified results are shown in Figure 6E. Since the C56S/C206S mutant cannot be palmitoylated (Figure 5), these findings suggest that the cell migratory effects of PAFAH1b3 require its modification by palmitoylation.
Figure 6.
Palmitoylation of PAFAH1b3 is required to promote cell migration and DRM localization. A, Representative images (3 independent experiments) from cell transwell migration assays in HUVECs infected with either scrambled or PAFAH1b3 shRNA lentiviruses. Demonstration of the knockdown is shown in the bottom panel. B, Quantification of migration data for experiments of Panel A. C, Representative images (3 independent experiments) from cell transwell migration assays of 293 cells transfected with control vector, or vector expressing wild type (Wt) or palmitoylation-deficient (Dbl for double cysteine mutation) PAFAH1b3. Cells were co-transfected with RFP for fluorescent imaging. D, Quantification of migration data for experiments of Panel C. E, Quantification of cell transwell migration assays of 293 cells transfected with wild type (Wt) or palmitoylation-deficient (Dbl) PAFAH1b3 in the absence or presence of insulin (4 independent experiments). F, Detergent-Resistant Membrane localization of wild type or mutant PAFAH1b3. G, Insulin stimulates PAFAH1b3 association with Detergent-Resistant Membranes. *, p<0.05 by ANOVA.
Palmitoylation-dependent localization of PAFAH1b3 in Detergent-Resistant Membranes (DRMs)
Palmitoylation, by altering interactions with membranes or proteins, can determine subcellular localization. Because palmitoylation would be expected to increase protein hydrophobicity and promote interaction with certain organized membrane domains, we prepared Detergent-Resistant Membranes (DRMs), which are associated with proteins important for cell signaling 35. A proportion of wild type PAFAH1b3 was found in DRMs but this association was reduced in cells expressing the palmitoylation-deficient C56S/C206S mutant (Dbl) as shown in the top panels of Figure 6F. Expression of the mutant had no effect on DRM localization of the control protein flotillin-1 (Flot-1) as shown in bottom panels of Figure 6F. Insulin (10 ng/ml) treatment of wild type PAFAH1b3, which increases its palmitoylation (Figures 4 and 5), also increases its association with DRMs as shown in Figure 6G. When skeletal muscle (a tissue that is relatively enriched in endothelial cells) was isolated from control mice and mice rendered insulin-deficient by treatment with streptozotocin, PAFAH1b3 association was decreased in DRMs from streptozotocin-treated as compared to control mice (Figure IV in the online-only Data Supplement).
DISCUSSION
Endothelial dysfunction is characteristic of cardiovascular disease 36. With a goal of identifying potentially novel targets that might link insulin, insulin resistance, and cardiovascular disease, we focused on the potential relationship between insulin signaling and the posttranslational process of protein S-palmitoylation in endothelial cells.
Using a mass spec-based proteomic screen to quantify differential palmitoylation, we identified several putative novel targets of insulin signaling in endothelial cells (Figure 2, Supplemental Tables III and IV in the online-only Data Supplement). Global inhibition of palmitoylation by chemical means (Figure 3, Figure I in the online-only Data Supplement) and genetic inactivation of the insulin-induced palmitoylated protein PAFAH1b3 (Figures 3,6) disrupted endothelial cell migration/tube formation. Insulin has pleiotropic effects, and it alters the abundance of many proteins. However, cells were exposed to insulin for only 6 h, and since most intracellular proteins have half-lives considerably longer than 6 h 20, it is likely that our differential palmitoylation screen selected for proteins differing predominantly by palmitoylation status instead of protein abundance. We verified that for PAFAH1b3, a novel insulin-responsive palmitoylated protein required for insulin-induced angiogenesis (Figure 3) and cell migration (Figure 6), palmitoylation is increased by insulin without affecting PAFAH1b3 protein levels (Figure 4). Moreover, we identified the palmitoylation sites in PAFAH1b3 using two independent techniques (Figure 5), and demonstrated that preventing palmitoylation through site-directed mutagenesis disrupts the capacity for PAFAH1b3 to stimulate cell migration (Figure 6). Collectively, these results suggest that protein palmitoylation may participate in insulin-activated endothelial function.
Protein palmitoylation involves the addition of palmitate (C16:0) to cysteine residues through a thioester bond. Our screen for palmitoylated proteins utilized acyl biotin exchange chemistry, an approach notable for the generation of false positive results 15, 37. For example, enzymes with active site cysteines, such as ubiquitin conjugating enzymes (of which several were identified as shown in the Supplemental Tables in the online-only Data Supplement), might be detected through biotinylation at the active site cysteine. However, it is possible that such enzymes might also be palmitoylated at cysteines distinct from the active site, so we elected not to dismiss these and related types of enzymes as false positives in the absence of direct characterization of their capacity for lipidation at regions distinct from their active sites.
While this manuscript was in preparation, a rigorously characterized palmitoylation proteome was reported in endothelial cells 38. That proteome did not address insulin-responsive palmitoylated proteins, the focus of the current work. Our initial screen found many of the same palmitoylated proteins identified by Marin and colleagues 38 including PECAM1, calnexin, Cav-1, and eNOS (Supplemental Table II in the online-only Data Supplement and Figure 1). There were also discrepancies, notably our inability to detect SOD-1, perhaps related to the fact that the current work profiled HUVECs while Marin et al. profiled an immortalized endothelial cell line.
Our study has limitations. We used 2-bromopalmitate to assess the possible role of global palmitoylation on endothelial function. While 2-bromopalmitate is known to inhibit palmitoylation 27–29, this chemical also has other well-characterized effects on fatty acid metabolism 39. However, tube formation was similarly affected by this chemical and knockdown of either PAFAH1b3 or TM4SF1 (palmitoylated proteins identified in this study), consistent with the notion that palmitoylation affects endothelial function. We characterized PAFAH1b3 (Figures 3–6 and Figures III-IV in the online-only Data Supplement), a protein not previously known to be palmitoylated, that is a subunit of an enzyme complex implicated in lipid remodeling, Golgi structure, and endocytic recycling 25–28. Our results demonstrate that insulin promotes palmitoylation of PAFAH1b3, but the mechanism by which this occurs is unknown. Insulin might increase palmitoylation 29 by promoting the activity of one of the more than 20 known acyltransferases, inhibiting acyl thioesterase activity, increasing palmitate accessibility, altering protein trafficking to sites better suited for acylation, or impacting any of several other processes related to posttranslational modification of proteins. However, our work does provide insight into the mechanism by which palmitoylation affects the cell biology of endothelial cells. For PAFAH1b3, we demonstrate that palmitoylation is required for localization of this protein to DRMs (Figure 6), and that insulin both increases palmitoylation of this protein (Figures 4–5) as well as the association of this protein with DRMs (Figure 6). While DRMs do not equate with caveolae or lipid rafts, many proteins isolated by this technique are involved in endothelial function and cell signaling 40.
Protein lipidation can affect cell signaling and is an important contributor to disease processes 41. Our demonstration of defective insulin-stimulated angiogenesis in cells treated with a palmitoylation inhibitor or deficient in discrete palmitoylated proteins suggests that altered palmitoylation could contribute to endothelial dysfunction. These results raise the possibility that promoting palmitoylation or promoting chemical modifications of proteins that mimic the effects of acylation could improve endothelial function.
Supplementary Material
SIGNIFICANCE.
Endothelial dysfunction is common in cardiovascular disease, which complicates metabolic disorders associated with insulin resistance. Insulin affects endothelial function but the protein targets of insulin in endothelial cells are incompletely defined. Proteins can be modified by palmitoylation, the reversible process of forming a thioester bond between a cysteine residue and the saturated fatty acid palmitate. Here we provide an initial comprehensive evaluation of palmitoylated proteins regulated by insulin in endothelial cells. For one such protein not previously known to be palmitoylated, we utilize biochemical techniques, site-directed mutagenesis, and cell biology assays to demonstrate that this modification is critical for cell migration and angiogenesis. This work thus identifies candidate proteins for pharmacologic targeting to improve endothelial function.
ACKNOWLEDGMENTS
None.
Sources of Funding: This work was supported by National Institutes of Health grants DK076729, DK088083, RR00954, DK52574, DK20579, and DK56341.
Abbreviations
- SILAC
Stable Isotope Labeling by Amino acids in Cell culture
- PAFAH1b3
Platelet-Activating Factor Acetylhydrolase IB subunit gamma
Footnotes
Disclosures: All authors state no actual or perceived conflicts of interests.
REFERENCES
- 1.Razani B, Chakravarthy MV, Semenkovich CF. Insulin resistance and atherosclerosis. Endocrinol Metab Clin North Am. 2008;37:603–621. doi: 10.1016/j.ecl.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerstein HC, Miller ME, Byington RP, Goff DC, Jr, Bigger JT, Buse JB, Cushman WC, Genuth S, Ismail-Beigi F, Grimm RH, Jr, Probstfield JL, Simons-Morton DG, Friedewald WT. Effects of intensive glucose lowering in type 2 diabetes. N Engl J Med. 2008;358:2545–2559. doi: 10.1056/NEJMoa0802743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currie CJ, Peters JR, Tynan A, Evans M, Heine RJ, Bracco OL, Zagar T, Poole CD. Survival as a function of HgbA(1c) in people with type 2 diabetes: A retrospective cohort study. Lancet. 2010;375:481–489. doi: 10.1016/S0140-6736(09)61969-3. [DOI] [PubMed] [Google Scholar]
- 4.Cushman WC, Evans GW, Byington RP, et al. Effects of intensive blood-pressure control in type 2 diabetes mellitus. N Engl J Med. 2010;362:1575–1585. doi: 10.1056/NEJMoa1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Accord Study Group. Ginsberg HN, Elam MB, et al. Effects of combination lipid therapy in type 2 diabetes mellitus. N Engl J Med. 2010;362:1563–1574. doi: 10.1056/NEJMoa1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rask-Madsen C, Kahn CR. Tissue-specific insulin signaling, metabolic syndrome, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2012;32:2052–2059. doi: 10.1161/ATVBAHA.111.241919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rask-Madsen C, Li Q, Freund B, et al. Loss of insulin signaling in vascular endothelial cells accelerates atherosclerosis in apolipoprotein e null mice. Cell Metab. 2010;11:379–389. doi: 10.1016/j.cmet.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–477. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 9.Wymann MP, Schneiter R. Lipid signalling in disease. Nature Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 10.Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol. 2010;6:498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 11.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006;2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 12.Linder ME, Deschenes RJ. Palmitoylation: Policing protein stability and traffic. Nature Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 13.Wei X, Schneider JG, Shenouda SM, Lee A, Towler DA, Chakravarthy MV, Vita JA, Semenkovich CF. De novo lipogenesis maintains vascular homeostasis through endothelial nitric-oxide synthase (eNOS) palmitoylation. J Biol Chem. 2011;286:2933–2945. doi: 10.1074/jbc.M110.193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei X, Yang Z, Rey FE, Ridaura VK, Davidson NO, Gordon JI, Semenkovich CF. Fatty acid synthase modulates intestinal barrier function through palmitoylation of mucin 2. Cell Host Microbe. 2012;11:140–152. doi: 10.1016/j.chom.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, Green WN, Yates JR, 3rd, Davis NG, El-Husseini A. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 21.Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: Physiological and disease perspectives. EMBO Rep. 2003;4:1127–1131. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, Thirtamara-Rajamani KK, Sim H, Viapiano MS. Fibulin-3 is uniquely upregulated in malignant gliomas and promotes tumor cell motility and invasion. Mol Cancer Res. 2009;7:1756–1770. doi: 10.1158/1541-7786.MCR-09-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foskett AM, Ezekiel UR, Trzeciakowski JP, Zawieja DC, Muthuchamy M. Hypoxia and extracellular matrix proteins influence angiogenesis and lymphangiogenesis in mouse embryoid bodies. Front Physiol. 2011;2:103. doi: 10.3389/fphys.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo H, Park SH, Ye SK, Kim M. IFN-gamma-induced BST2 mediates monocyte adhesion to human endothelial cells. Cell Immunol. 2011;267:23–29. doi: 10.1016/j.cellimm.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Bechler ME, Doody AM, Ha KD, Judson BL, Chen I, Brown WJ. The phospholipase a enzyme complex PAFAH 1b mediates endosomal membrane tubule formation and trafficking. Mol Biol Cell. 2011;22:2348–2359. doi: 10.1091/mbc.E09-12-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bechler ME, Doody AM, Racoosin E, Lin L, Lee KH, Brown WJ. The phospholipase complex PAFAH 1b regulates the functional organization of the golgi complex. J Cell Biol. 2010;190:45–53. doi: 10.1083/jcb.200908105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho YS, Swenson L, Derewenda U, Serre L, Wei Y, Dauter Z, Hattori M, Adachi T, Aoki J, Arai H, Inoue K, Derewenda ZS. Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer. Nature. 1997;385:89–93. doi: 10.1038/385089a0. [DOI] [PubMed] [Google Scholar]
- 28.Yan W, Assadi AH, Wynshaw-Boris A, Eichele G, Matzuk MM, Clark GD. Previously uncharacterized roles of platelet-activating factor acetylhydrolase 1b complex in mouse spermatogenesis. Proc Natl Acad Sci U S A. 2003;100:7189–7194. doi: 10.1073/pnas.1236145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linder ME, Deschenes RJ. New insights into the mechanisms of protein palmitoylation. Biochemistry. 2003;42:4311–4320. doi: 10.1021/bi034159a. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Petreaca M, Martins-Green M. Cell and molecular mechanisms of insulin-induced angiogenesis. J Cell Mol Med. 2009;13:4492–4504. doi: 10.1111/j.1582-4934.2008.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nature Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu VW, Bai M, Li J. Getting active: Protein sorting in endocytic recycling. Nature Rev Mol Cell Biol. 2012;13:323–328. doi: 10.1038/nrm3332. [DOI] [PubMed] [Google Scholar]
- 33.Zukauskas A, Merley A, Li D, Ang LH, Sciuto TE, Salman S, Dvorak AM, Dvorak HF, Jaminet SC. Tm4sf1: A tetraspanin-like protein necessary for nanopodia formation and endothelial cell migration. Angiogenesis. 2011;14:345–354. doi: 10.1007/s10456-011-9218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih SC, Zukauskas A, Li D, Liu G, Ang LH, Nagy JA, Brown LF, Dvorak HF. The l6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer research. 2009;69:3272–3277. doi: 10.1158/0008-5472.CAN-08-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- 36.Bonetti PO, Lerman LO, Lerman A. Endothelial dysfunction: A marker of atherosclerotic risk. Arterioscler Thromb Vascular Biol. 2003;23:168–175. doi: 10.1161/01.atv.0000051384.43104.fc. [DOI] [PubMed] [Google Scholar]
- 37.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: Purification and identification. Nat Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 38.Marin EP, Derakhshan B, Lam TT, Davalos A, Sessa WC. Endothelial cell palmitoylproteomic identifies novel lipid-modified targets and potential substrates for protein acyl transferases. Circ Res. 2012;110:1336–1344. doi: 10.1161/CIRCRESAHA.112.269514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman RA, Rao P, Fogelsong RJ, Bardes ES. 2-bromopalmitoyl-CoA and 2- bromopalmitate: Promiscuous inhibitors of membrane-bound enzymes. Biochim Biophys Acta. 1992;1125:203–209. doi: 10.1016/0005-2760(92)90046-x. [DOI] [PubMed] [Google Scholar]
- 40.Guo Y, Singleton PA, Rowshan A, Gucek M, Cole RN, Graham DR, Van Eyk JE, Garcia JG. Quantitative proteomics analysis of human endothelial cell membrane rafts: Evidence of MARCKS and MRP regulation in the sphingosine 1-phosphate-induced barrier enhancement. Mol Cell Proteomics. 2007;6:689–696. doi: 10.1074/mcp.M600398-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Resh MD. Targeting protein lipidation in disease. Trends Mol Med. 2012;18:206–214. doi: 10.1016/j.molmed.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.