Abstract
Phenotypic screening is making a comeback in drug discovery as the maturation of chemical proteomics methods has facilitated target identification for bioactive small molecules. A limitation of these approaches is that time-consuming genetic methods or other means is often required to determine the biologically relevant target(s) from among multiple protein-compound interactions that are typically detected. Here, we have combined phenotypic screening of a directed small-molecule library with competitive activity-based protein profiling to map and functionally characterize the targets of screening hits. Using this approach, we identify carboxylesterase 3 (Ces3 or Ces1d) as a primary molecular target of bioactive compounds that promote lipid storage in adipocytes. We further show that Ces3 activity is dramatically elevated during adipocyte differentiation. Treatment of two mouse models of obesity-diabetes with a Ces3 inhibitor ameliorates multiple features of metabolic syndrome, illustrating the power of the described strategy to accelerate the identification and pharmacologic validation of new therapeutic targets.
Phenotypic screening is seeing a renaissance in drug discovery, as the target-focused approaches that have dominated the industry over the past several decades have yet to translate into a greater number of new medicines.1 A recent survey of all first-in-class small-molecule medications approved between 1999 and 2008 has shown that, only 17 (34%) came from target-based approaches, as compared to 28 (56%) derived from phenotypic screens.2 In a target-oriented campaign, molecules are optimized on a single protein that is hypothesized to play a critical role in the disease in question. The advantages of this approach are clear: screens with greater throughput can be developed, and concrete measures of target modulation provide a rational path to optimize leads. The drawbacks of a target-centric strategy are that interaction of a compound with a single protein may not be sufficient to elicit a therapeutic response in vivo, and that insufficient knowledge of the mechanism of action can result in unexpected toxicity or lack of efficacy for the optimized drug candidate. These limitations are particularly evident in metabolic disease indications, where therapeutic outcome reflects a complex interplay across multiple organs.
In contrast, phenotypic screening aims to identify compounds that induce a desirable biological response without making a priori assumptions about the underlying molecular target(s). Phenotypic screening thus provides a less biased approach to chemically interrogate the proteome in its native context and increases the likelihood of uncovering new biology, as well as small-molecules that modulate targets that are part of dynamic complexes and signaling pathways. As a result, bioactive molecules isolated in phenotypic screens could be viewed as being more likely to have therapeutic impact in vivo. Phenotypic screening hits may also constitute better starting points for optimization, since they must be cell-permeable and engage their targets with sufficient affinity to displace endogenous interacting metabolites or proteins. Some of the disadvantages of phenotypic screening are that the assays have lower throughput and must serve as robust surrogates for the desired outcome in vivo. However, the greatest challenge of phenotypic screens is arguably the identification of the molecular targets of bioactive small-molecules. Without a target(s), optimization and pre-clinical development of phenotypic hits are challenging tasks.
Advances in chemical proteomics, genomics, and informatics are beginning to provide tools to overcome the hurdle of target identification for phenotypic screening hits.3-7 Traditional affinity chromatography, in which the bioactive molecule is coupled to a solid matrix and used to pull down interacting proteins, has benefitted greatly by the development of more sensitive and quantitative proteomic approaches (e.g., stable isotope labeling by amino acids in cell culture, SILAC).8,9 Nonetheless, validation of the molecular target from a list of interacting proteins still requires further genetic, chemical, and/or biophysical methods.
Activity-based protein profiling (ABPP) is a chemical proteomics approach that exploits the conserved mechanistic and/or structural features of large enzyme families to develop chemical probes that irreversibly bind their active sites.10,11 An activity-based probe consists of a reactive group that interacts with the active sites of proteins coupled to a reporter tag that can be used to visualize probe-labeled enzymes by SDS-PAGE (e.g., rhodamine) or to enrich and identify these proteins using avidin-biotin and mass spectrometry-based proteomics methods, respectively. ABPP has been used to comparatively profile enzyme activities across different types of cells, tissues, and disease states.10 When performed in a competitive mode, ABPP can also identify the target(s) to which a small-molecule inhibitor binds directly in native cell and tissue proteomes.12 This feature can lead to the rapid identification of the molecular target of bioactive compounds emerging from phenotypic screens.
Here, we have tested whether competitive ABPP could hasten the identification of targets of small-molecules that show activity in a cell-based assay measuring differentiation and lipid accumulation in adipocytes, readouts that can serve to identify molecules of potential value as insulin sensitizers in vivo.13 Insulin resistance and type 2 diabetes are characterized by increased levels of plasma free fatty acids and ectopic lipid deposition.14 Thus, agents that restore normal tissue lipid partitioning often enhance insulin sensitivity. To increase the probability of success, we employed a small-molecule library and competitive ABPP probes directed to interact with serine hydrolases, a large and diverse class of enzymes that plays important roles in mammalian metabolism and includes members with validated therapeutic potential in diabetes and obesity.15,16 A subset of bioactive compounds identified in the screen was found by competitive ABPP to target the serine hydrolase Ces3 (or Ces1d) in mouse adipocytes. Administration of one of these Ces3 inhibitors to high-fat fed or obese-diabetic db/db mice protected them from weight gain, improved blood lipid levels, and increased insulin sensitivity and glucose tolerance. We also found that the activity of human CES1 (the orthologue of mouse Ces3) is elevated in adipose tissue of humans with obesity and type 2 diabetes. These data show that phenotypic screening of directed small-molecule libraries paired with cognate probes for competitive ABPP can facilitate rapid identification and in vivo validation of the molecular target of bioactive compounds of potential therapeutic relevance to metabolic disease.
RESULTS
Profile of serine hydrolase activity during adipogenesis
To pilot the integration of cell-based screening with competitive ABPP as an approach to discover new metabolic drug targets, we chose to screen a focused library of serine hydrolase inhibitors in a phenotypic screen for cellular adipogenesis and lipid storage. Serine hydrolases play important roles in many physiological and disease processes, including lipid metabolism and adipocyte function (e.g., hormone sensitive lipase), and diabetes (e.g., DPPIV).15,17 Previous studies have inventoried serine hydrolase activities in adipocytes using phosphonate probes, but, in these experiments, a comparison to predifferentiated cells was not performed.18-20 With this goal in mind, we profiled serine hydrolase activities in predifferentiated and differentiated C3H10T1/2 (10T1/2) and 3T3-L1 cells using reporter-tagged fluorophosphonates (FPs), which have been shown to serve as near-universal activity-probes for mammalian serine hydrolases.21 Proteomes from undifferentiated and differentiated 10T1/2 and 3T3-L1 cells were incubated with either a fluorescent FP probe (FP-rhodamine) to visualize serine hydrolase activity by SDS-PAGE and in-gel fluorescence scanning, or with a biotinylated FP probe (FP-biotin) for affinity enrichment, identification, and quantitation of active serine hydrolases using avidin chromatography coupled with multidimensional liquid chromatography-MS/MS (ABPPMudPIT).22 We found that adipogenesis was accompanied by the suppression of a handful of serine hydrolases that were primarily active in the predifferentiated state, and by the robust induction of many other serine hydrolase activities that were elevated in mature adipocytes (Fig. 1a; Supplementary Results, Supplementary Table 1 and Supplementary Datasets 1 and 2). Serine hydrolase activities enriched in adipocytes include enzymes previously associated with lipid metabolism in fat cells (e.g., FAS, HSL, LPL), but also many poorly annotated proteins with no prior link to adipogenesis (e.g., ABHD11, ABHD6, Serhl). The extensive induction of serine hydrolase activities during adipogenesis emphasizes the importance of these enzymes in adipocyte physiology.
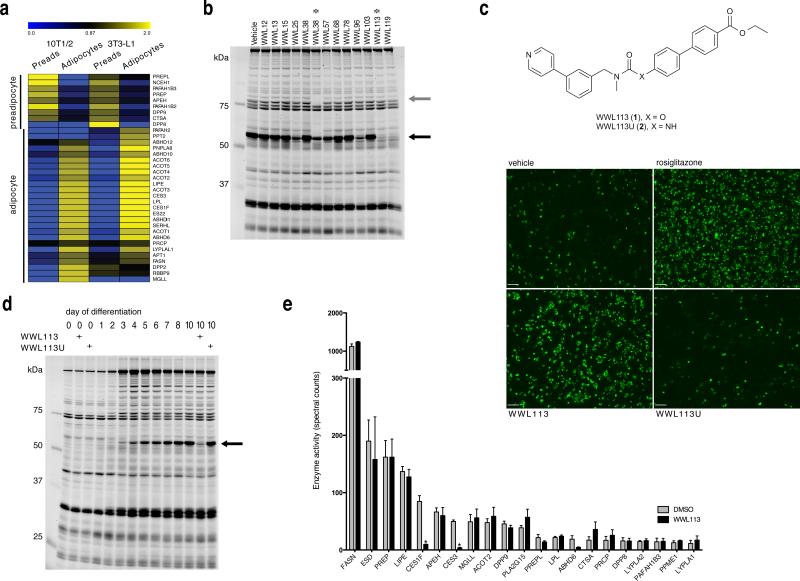
Figure 1. Isolation of serine hydrolase inhibitors with adipogenic activity and identification of their molecular target.
(a) Hierarchical cluster analysis of serine hydrolase signals detected by ABPP-MudPIT in 3T3-L1 and 10T1/2 preadipocyte and adipocyte proteomes. Data represent the normalized mean of three independent experiments. (b) Gel-based competitive ABPP analysis of adipocyte 10T1/2 cells labeled in situ with carbamates that promote differentiation and lipid accumulation in fat cells. A ~60 kDa serine hydrolase (black arrow) is inhibited by multiple proadipogenic carbamates. WWL38 inhibits HSL (grey arrow); WWL113 appears specific for the 60kDa activity. Image is representative of 2 independent experiments. (c) Structure of WWL113 and its urea derivative (WWL113U). WWL113, but not WWL113U, promotes adipocyte formation and lipid storage in 10T1/2 cells. Green fluorescence corresponds to Nile red staining. Images are representative of 10 independent experiments. Scale bar = 250 μm. (d) Competitive ABPP shows that WWL113 specifically targets a ~60 kDa serine hydrolase activity that is highly induced during differentiation of 10T1/2 adipocytes; WWL113U has no effect. Image is representative of 3 independent experiments. (e) Competitive ABPP-MudPIT analysis of proteomes from 10T1/2 adipocytes incubated with WWL113 reveals that this compound is a Ces3/Ces1f inhibitor. Error bars represent s.d. (n = 3).
Screen to discern important adipocyte serine hydrolases
The existence of a large number of uncharacterized serine hydrolase activities in mature fat cells highlights the opportunity to elucidate new enzymatic pathways involved in adipose tissue biology. We sought to identify those most relevant to fat cell physiology by screening a library of serine hydrolase-directed inhibitors. Carbamates with an activated leaving group serve as a privileged scaffold for serine hydrolase inhibitors.23 A recent in vitro screen of 140+ carbamates against a panel of 72 serine hydrolases21 has led to the discovery of lead inhibitors for >30 serine hydrolases, underscoring the potential to identify pharmacological probes for these enzymes using a modestly-sized library. We screened this carbamate library21 in a phenotypic assay for adipocyte formation and lipid storage; compounds that score as hits in this screen can have potential as insulin sensitizers in vivo.13 10T1/2 preadipocytes were induced to differentiate into adipocytes in the presence of either vehicle (DMSO) or one of the carbamate compounds (10 μM). The PPARγ ligand rosiglitazone served as a positive control. Cells were incubated with compounds for 8 days and the extent of adipocyte differentiation was evaluated using a fluorescent lipid dye (Nile red). This screen yielded a number of carbamates that promoted a substantial increase of lipid accumulation in differentiating adipocytes. These molecules had similar effects in 3T3-L1 cells (Supplementary Fig. 1).
We next used competitive ABPP to identify the molecular targets of pro-adipogenic carbamates. In this modality of ABPP, cells or proteomes are exposed to the bioactive carbamate prior to labeling with an FP probe. Serine hydrolases that react with a carbamate inhibitor are detected by a loss in their FP probe labeling. 10T1/2 adipocytes were treated for 4 hr with each hit (10 μM) from the phenotypic screen, proteomes harvested, and serine hydrolase activity evaluated using the FP-rhodamine probe (Fig. 1b). Some of the carbamates (e.g., WWL38) were found to target an ~80-95 kDa serine hydrolase doublet that we surmised (and later confirmed by ABPP-MudPIT; Supplementary Fig. 2a; Supplementary Dataset 2) to represent hormone sensitive lipase (HSL, Lipe), an enzyme known to regulate neutral lipid catabolism in adipocytes.24 Another set of pro-adipogenic carbamates, however, inhibited a distinct ~60 kDa serine hydrolase activity (Fig. 1b). We selected one of these carbamates, WWL113 (1) (Fig. 1c), which appeared to show excellent selectivity for the 60 kDa serine hydrolase(s) for further characterization.
To gain confidence that the pro-adipogenic ability of WWL113 was due to serine hydrolase inhibition, we tested the properties of a urea analogue (WWL113U (2); Fig. 1c), which should be impaired in its capacity to inhibit serine hydrolases due to reduced reactivity. Only WWL113, but not WWL113U, showed activity in the original phenotypic assay (Fig. 1c). Cells treated with WWL113, but not WWL113U, showed increased expression of fat cell markers (e.g, the transcription factors PPARγ and C/EBPα, the hormone adiponectin) indicating that they are functional adipocytes (Supplementary Fig. 2b). Importantly, the ~60 kDa target(s) of WWL113 was not inhibited by WWL113U (Fig. 1d), confirming the utility of this agent as a negative-control probe. The putative ~60 kDa target(s) of WWL113 was also strongly elevated during adipocyte differentiation, with the activity being undetectable in preadipocytes and increasing steadily from day 3 to day 10 of differentiation (Fig. 1d). WWL113 is not a PPARγ ligand, as evaluated using transfections and a cell-free binding assay, excluding this explanation for its effect in adipocytes (Supplementary Fig. 3).
WWL113 is a Ces3/Ces1f inhibitor
We next used ABPP-MudPIT to identify the target(s) of WWL113. 10T1/2 adipocyte proteome was incubated with either DMSO or 10 μM WWL113 for 45 min and then treated with FP-biotin (5 μM, 2 hr). Active serine hydrolases (i.e. FP-biotin labeled) were enriched using avidin beads, subjected to tryptic digest, and analyzed by multidimensional LC-MS/MS (ABPP-MudPIT22). Serine hydrolase activities were quantified by spectral counting, which revealed that WWL113 inhibited two carboxylesterase enzymes – Ces3 (or Ces1d) and Ces1f (or CesML1) – with molecular weights (61,788 and 61, 612 Da, respectively) that match the ~60 kDa activity observed by gel-based ABPP (Fig. 1e; Supplementary Dataset 3). WWL113 treatment also reduced the activity signals for ABHD6, although these data did not reach statistical significance (p = 0.08). Ces3 and Ces1f are endoplasmic reticulum glycoproteins that have been previously associated with adipocyte lipolysis.25-28 Importantly, WWL113 did not inhibit any of the other serine hydrolases detected by ABPP-MudPIT, including several known to be involved in adipocyte lipolysis, such as hormone sensitive lipase (Lipe) and monoglyceride lipase (Mgll).
Acute treatment of differentiated adipocytes with WWL113, but not WWL113U, diminished basal and, to a lesser extent, hormone-induced lipolysis, suggesting a functional role for the target of this compound in the regulation of lipid breakdown in fat cells (Supplementary Fig. 2c). Consistent with this premise, untargeted metabolite profiling of 10T1/2 adipocytes differentiated in the presence of 10 μM WWL113 identified several elevated triacylglycerol (TAG) species (Supplementary Fig. 4). Some of these TAGs were also increased in cells differentiated in the presence of 1 μM rosiglitazone, while others, in particular long chain, polyunsaturated TAGs appeared to accumulate to a greater degree in WWL113-treated cells (Supplementary Fig. 4). None of these changes were observed in cells treated with WWL113U.
Discovery of a more selective, distinct Ces3 inhibitor
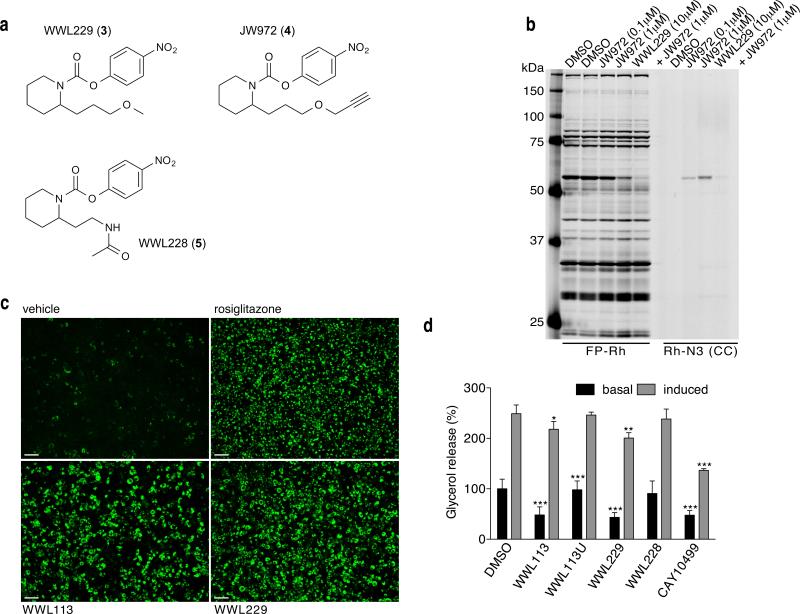
We confirmed that WWL113, but not WWL113U is a potent inhibitor of Ces3 and Ces1f by competitive ABPP of enzymes recombinantly expressed in HEK293T cells (IC50 value of ~0.1 μM for each enzyme; Supplementary Fig. 5). We also established that WWL113 inhibits recombinantly expressed ABHD6 (Supplementary Fig. 6a). We next used gel-based competitive ABPP to identify a second, structurally distinct carbamate (WWL229 (3); Fig. 2a) that inhibited recombinant Ces3, but not Ces1f, ABHD6, or other tested serine hydrolases (Supplementary Fig. 6). WWL229 also selectively inhibited Ces3, but not Ces1f, ABHD6, other serine hydrolases, in adipocyte proteomes as determined by ABPP-MudPIT (Supplementary Fig. 7; Supplementary Dataset 4). We examined whether WWL229 reacts with other proteins in adipocyte proteomes beyond serine hydrolases by creating an alkyne-derivatized analogue JW972 (4) (Fig. 2a; Supplementary Note), which enabled proteome-wide reactivity analysis by click chemistry (CC)-ABPP.29 Labeling of cultured adipocytes in situ with JW972, followed by copper-catalyzed conjugation to an azide-rhodamine reporter tag (click chemistry30), revealed a single ~60 kDa protein target across the adipocyte proteome, and the labeling of this target by JW972 was substantially blocked by pre-treatment with WWL229 (Fig. 2b). We also enriched proteins labeled by JW972 (1 μM) using an azide-biotin tag and ABPP-MudPIT methods, which identified only two JW972-labeled proteins across the entire adipocyte proteome – Ces3 and Ces1f (Supplementary Fig. 8; Supplementary Datasets 5 and 6). We suspect that Ces1f was also enriched by JW972, despite showing a much lower potency of inhibition compared to Ces3 (Supplementary Fig. 5), because only a small fraction of protein labeling may be needed to enable enrichment and identification by ABPP-MudPIT.
Figure 2. WWL229, a selective Ces3 inhibitor, recapitulates the effects of WWL113 in adipocytes.
(a) Structures of WWL229, WWL228, and JW972. (b) Gel-based ABPP analysis of proteomes from adipocytes incubated in situ for 4 hr with JW972 alone or in combination with WWL229, and subsequently labeled in vitro with FP-rhodamine (FP-Rh) or with rhodamine-azide (Rh-N3). The WWL229-click probe (JW972) labels an activity that is competed away by an excess of WWL229. Gel is representative of 3 independent experiments. (c) WWL229 (10 μM) promotes adipocyte formation and lipid storage in 10T1/2 cells to the same extent as WWL113. Green fluorescence (Nile red staining) was measured at day 8 of differentiation. Scale bar = 200 μm; images are representative of 5 independent experiments. (d) WWL229 and WWL113 block basal lipolysis in 10T1/2 adipocytes to an equivalent degree. WWL228 and WWL113U have no effect. CAY10499 is a promiscuous lipolysis inhibitor. Data are presented as mean ± s.d. (n = 4), *p <0.05, **p < 0.01, and ***p < 0.001 vehicle vs. treated cells.
Importantly, we found that WWL229 recapitulated all of the biological effects of WWL113 in cultured adipocytes to an equivalent extent, including the promotion of lipid storage in adipocytes (Fig. 2c) and the blockade of basal lipolysis (Fig. 2d). These effects were not observed with WWL228 (5), a structural analogue of WWL229 that does not inhibit Ces3 activity (Fig. 2; Supplementary Fig. 6).
WWL113 treatment ameliorates obesity-diabetes in mice
One attribute of carbamate-based irreversible inhibitors of serine hydrolases, especially when used in combination with competitive ABPP, is that they provide a relatively straightforward class of probes for in vivo studies that often do not require extensive phamacokinetic optimization.21 In this approach, animals are dosed with the carbamate of interest and tissues harvested and labeled with FP-probes to discern inhibited serine hydrolase activities at the tested dose of carbamate. Because pilot studies indicated that WWL113 exhibited significantly greater inhibitory activity in mice compared to WWL229, we chose WWL113 for in vivo experiments. To evaluate the potential of WWL113 as a tool compound for animal studies, mice were given a single 30 mg/kg oral dose of WWL113 and tissues analyzed 4 hr later. This dose was sufficient to inhibit Ces3 activity in liver (>90%) and adipose tissue (>75%; Supplementary Fig. 9a), the sites of highest Ces3 expression.31 ABPPMudPIT revealed that the activity of several other related Ces enzymes was affected by WWL113 treatment (Supplementary Fig. 9b,c; Supplementary Datasets 7 and 8). In white adipose tissue, Ces1f and Esterase 22 (Ces1e), two additional Ces enzymes with lower ABPP signals than Ces3 (4 and 14-fold less respectively), were also inhibited by WWL113. In liver, Ces1 (Ces1g), Esterase1 (Ces1c), and AADAC were also inhibited by WWL113. In vitro assays with recombinant proteomes indicated that Ces3, Ces1f, Ces1, and Ces1c are direct targets of WWL113; changes in the activity of other hydrolases (e.g., Es22) may reflect indirect regulation of these enzymes as a result of Ces3 inhibition (Supplementary Fig. 9d). Because multiple CES enzymes are capable of hydrolyzing TAGs (e.g., both Ces3 and Ces1f have this enzymatic activity27,28), we conclude that WWL113 may offer a valuable in vivo pharmacological probe to test the role that this group of enzymes plays in obesity and type 2 diabetes.
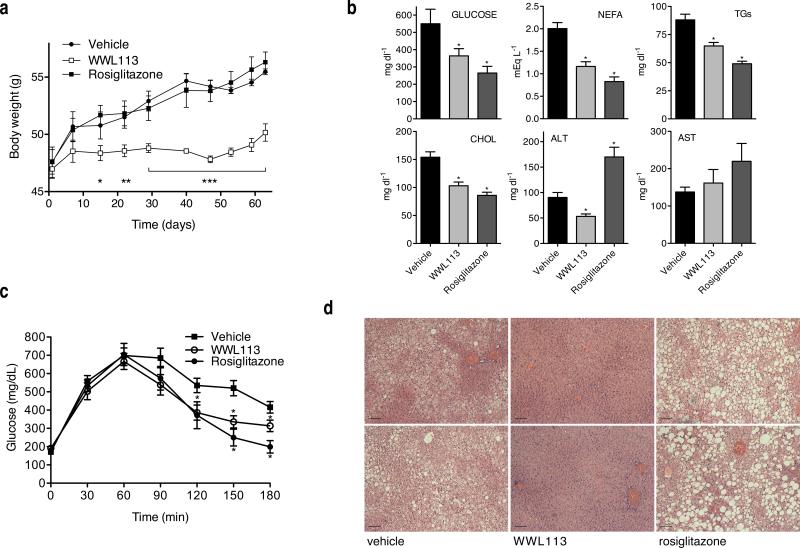
Daily oral administration of WWL113 (30 mg/kg) to obese-diabetic db/db mice for 9 weeks resulted in major improvement of multiple features of metabolic syndrome. Treated mice gained weight at a much slower rate than those receiving vehicle or rosiglitazone (4 mg/kg), a synthetic PPARγ ligand used as a control for enhanced insulin sensitivity (Fig. 3a). This effect was noted within 2 weeks of WWL113 administration and could not be ascribed to decreased food intake or intestinal lipid absorption (Supplementary Fig. 10a,b). Plasma analysis after 3 weeks of treatment showed dramatic improvement in glucose and lipid profiles, comparable to that brought about by the clinical agent rosiglitazone (Fig. 3b). Mice treated with WWL113 had lower levels of non-esterified free fatty acids (NEFA), triglycerides (TGs), total cholesterol, and fasted glucose, as well as enhanced glucose tolerance (not shown). To examine the possibility that improved glucose tolerance in WWL113-treated mice was due to their reduced weight, a second set of db/db mice was treated with WWL113 and the test was performed after 8 days of dosing, a point at which there were no weight differences between groups (Supplementary Fig. 10c). Glucose tolerance was enhanced in diabetic mice treated with WWL113 in this short regimen (Fig. 3c).
Figure 3. WWL113 treatment corrects multiple features of metabolic syndrome in db/db mice.
8-week-old db/db mice (n = 10 per group) were dose orally once a day with vehicle, 30 mg/kg WWL113, or 4 mg/kg rosiglitazone. (a) WWL113-treated db/db mice put on weight at a slower rate. (b) Blood chemistry after 3 weeks of WWL113 treatment. WWL113 treatment lowers circulating glucose, free fatty acids (NEFA), triglycerides (TGs), and total cholesterol. (c) WWL113 enhances glucose tolerance after 8 days of treatment in animals of equivalent weight. Glucose tolerance test (1 g/kg intraperitoneal injection), n = 10 per group. (d) Complete clearance of hepatic lipids in db/db mice treated with WWL113 for 3 months (Hematoxylin and Eosin staining showing two representative animals per group). Scale bar = 100 μm. In all cases, error bars represent s.e.m. and *p < 0.05, **p < 0.01, and ***p < 0.001 vs. vehicle-treated mice.
Much of the beneficial impact that WWL113 treatment had on metabolic parameters may have been due to its dramatic effect on hepatic lipid accumulation. Ectopic fat deposition in the liver is a common feature of obesity and type 2 diabetes that may render this organ less sensitive to insulin.32 In stark contrast to animals treated with the PPARγ ligand, which showed increased liver lipid accumulation, a side-effect of thiazolidinediones in rodents,33 obese-diabetic mice treated chronically with WWL113 displayed complete clearance of the excess lipids that normally accrue in the liver of these animals (Fig. 3d). Resolution of hepatic steatosis was accompanied by unchanged aspartate aminotransferase (AST) and decreased alanine aminotransferase (ALT) activity, markers of liver damage (Fig. 3b). Expression of liver lipogenic genes (e.g., SREBP-1c) was decreased in WWL113-treated animals, while expression of genes involved in fatty acid oxidation and ketogenesis was induced (e.g., PPARγ, Acadl, HMGCL) (Supplementary Fig. 10d). These changes were reflected in an increase in circulating ketone bodies, the end product of fatty acid oxidation, that became evident within 2 weeks of the start of WWL113 treatment (Supplementary Fig. 10e). These increases in oxidative gene expression were not due to direct activation of PPARγ by WWL113 (Supplementary Fig. 3a). Wild type mice treated for 30 days with the same dose of WWL113 showed no difference in weight or glycemia, indicating that the changes WWL113 elicited in diabetic mice are not due to toxic effects (Supplementary Fig. 11).
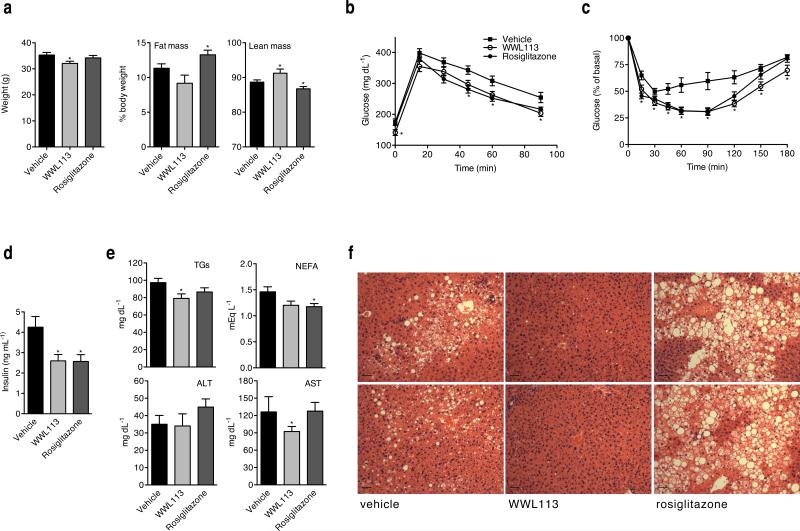
Treatment with WWL113 had similar effects in diet-induced obesity (DIO) mice, a non-genetic model that more closely resembles the development of obesity and insulin resistance in humans. C57BL/6J mice were fed a high-fat diet for 12 weeks prior to the start of 9 weeks of compound treatment (oral 50 mg/kg daily dose). DIO mice dosed with WWL113 were more resistant to weight gain than controls and showed enhanced glucose tolerance (Fig. 4a,b). WWL113-treated mice also had lower levels of plasma insulin, and increased whole-body insulin sensitivity as measured using insulin tolerance tests (Fig. 4c,d). Circulating triglycerides were significantly decreased, while NEFAs showed a downward trend that did not reach statistical significance (Fig. 4e). Although white adipocytes in the visceral and subcutaneous depots of WWL113-treated mice appeared larger than those in vehicle-treated animals, this difference was not significant. More striking were the changes seen in the liver. WWL113-treated animals showed complete absence of ectopic lipid deposition in the liver (Fig. 4f). Together, these data indicate that inhibition of Ces3/Ces1f activity has multiple beneficial effects in lipid and glucose homeostasis in genetic and diet-induced mouse models of obesity, insulin resistance, and type 2 diabetes.
Figure 4. WWL113 treatment enhances insulin sensitivity and glucose tolerance in a model of diet-induced obesity.
12-week-old C57BL/6J male mice fed a 60 kcal% fat diet since the time of weaning were treated orally once per day with vehicle, 50 mg/kg WWL113, or 4 mg/kg rosiglitazone for 50 days (n = 10 per group). (a) Body weight is decreased in DIO mice treated with WWL113 without a significant change in fat mass. (b) Glucose and (c) Insulin tolerance tests show that WWL113 enhances insulin sensitivity and glucose homeostasis. (d) WWL113 treatment decreases plasma insulin levels. (e) WWL113 treatment significantly reduces plasma triglyceride levels, and shows a tendency to decrease circulating free fatty acids. The levels of AST are also significantly reduced, perhaps as a consequence of the absence of liver steatosis in WWL113-treated mice. (f) Hematoxylin and Eosin staining showing two representative animals per group. Scale bar = 60 μm. In all cases, error bars represent s.e.m. and *p < 0.05 vs. vehicle-treated mice.
WWL113 treatment reduces hepatic diacylglycerol species
The dramatic effects of WWL113 treatment on liver lipid accumulation prompted us to explore the nature of the metabolite changes induced by this compound. Livers from db/db mice treated with WWL113 for 14 days were subjected to lipidomic analysis. Consistent with the histology, lipid profiling revealed that various TAG, DAG, and MAG species were significantly reduced in mice dosed with WWL113 (Table 2). Changes were not observed in the levels of other detected lipid species, such as phospholipids. To discern which of these changes were directly related to Ces3/Ces1f inhibition, as opposed to a consequence of the enhanced metabolic state of treated animals, wild-type mice were given a single dose of WWL113 and their livers analyzed 4 hr later. In this acute setting, the levels of bulk TAG species did not change, but several DAG and MAG species, including 16:0/20:4 DAG and 20:4 MAG, were significantly decreased (Table 1). These findings are consistent with the proposed role of Ces3 and Ces1f as ER-localized triacylglycerol hydrolases that act on limited TAG pools.31
Table 1.
Hepatic lipid levels in WWL113-treated mice
| db/db 14 days | wt 4 hr | |
|---|---|---|
| WWL113/control | WWL113/control | |
| monoacylglycerols (MAG) | ||
| C16:0 MAG | 0.6 | 0.8 |
| C18:0 MAG | 0.4* | 1.5 |
| C20:4 MAG | 0.7* | 0.5* |
| diacylglycerols (DAG) | ||
| C32:0 DAG | 0.7** | 0.9 |
| C34:2 DAG | 0.4** | 0.8 |
| C34:1 DAG | 0.2** | 0.9 |
| C36:4 DAG (C16:0/C20:4 DAG) | 0.3** | 0.5** |
| C38:5 DAG (C18:0/C20:4 DAG) | 0.6** | 0.6* |
| triacylglycerols (TAG) | ||
| C48:2 TAG | 0.4** | 1.0 |
| C50:2 TAG | 0.4** | 1.2 |
| C52:4 TAG | 0.7* | 1.7 |
| C52:3 TAG | 0.7 | 1.2 |
| C52:1 TAG | 0.6* | 1.1 |
| C54:5 TAG | 0.6** | 1.0 |
| C52:4 TAG | 0.5** | 0.9 |
| C54:3 TAG | 0.5** | 0.9 |
| C56:6 TAG | 0.7 | 0.9 |
| lysophosphatidyl choline (LPC) | ||
| C16:0 LPC | 0.9 | 1.2 |
| phosphatidylcholine (PC) | ||
| C34:1 PC | 0.8 | 1.2 |
| C36:1 PC | 0.9 | 1.1 |
| acyl carnitine | ||
| C16:0 acylcarnitine | 1.2 | 0.9 |
p<0.05
p<0.01 with >1.4-fold or <0.7-fold of control
Human Ces3 (CES1) is more active in obesity and diabetes
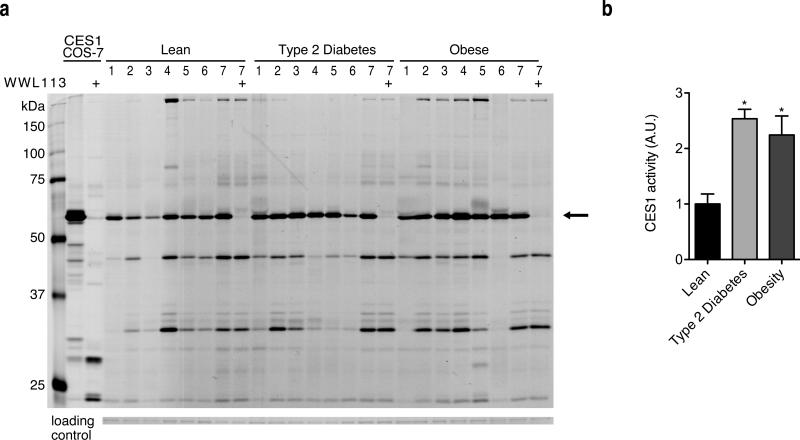
To explore the extent to which our findings in rodents may translate to humans, the activity of human CES1 (the orthologue of mouse Ces3) was measured by gel-based ABBP in adipose tissue biopsies of lean (BMI < 25), obese (BMI 31.8-55.8, mean 37.2; mean Hb a1c 4.9), and type 2 diabetic (BMI 32.7-61, mean 41.7; Hb a1c 6.7-11.2, mean 8.7) individuals. Similar to what we observed with Ces3 in mice, we found hCES1 was a highly active serine hydrolase in white fat. Though there was heterogeneity among human samples, the levels of hCES1 activity were increased roughly two-fold in obese individuals and patients with type 2 diabetes compared to lean subjects (Fig. 5). Enhanced hCES1 activity, and the consequent release of greater numbers of fatty acids into the circulation, may be a heretofore unrecognized feature of the pathogenesis of obesity-diabetes. Notably, WWL113 also inhibited hCES1, as assessed using gel-based ABPP (Fig. 5a) and a substrate hydrolysis assay (Supplementary Fig. 12; IC50 ~ 50 nM).
Figure 5. Human Ces3 (CES1) is more active in adipose tissue of obese and type 2 diabetic individuals.
(a) ABPP profiles of WAT samples from 7 lean, obese, and type 2 diabetic individuals. Note that hCES1 (black arrow) is the most active serine hydrolase present, and that WWL113 (10 μM) inhibits the human homologue of Ces3. Left-most two lanes are lysates of COS-7 cells ectopically expressing hCES1. (b) Bar graph shows quantification of hCES1 activity relative to a Comassie-stained band that is similarly abundant in all samples (loading control). Error bars represent s.e.m. and *p < 0.05 vs. lean controls.
DISCUSSION
Renewed interest in phenotypic screens for drug discovery is being driven by advances in chemical proteomics that are enabling identification of the molecular targets of bioactive compounds. Common chemical proteomic strategies for target identification involve generating affinity matrices or photoreactive derivatives of the bioactive small-molecule.4 MS-based proteomics of enriched proteins then provides a list of likely targets, and molecular biology (e.g., RNAi) or other means are used to confirm the target(s) responsible for the observed biological effect. While productive, these approaches require derivatization of screening hits, which can impair target interactions. Even in cases where target identification is successful, considerable medicinal chemistry is often required to convert screening hits into probes that can be used for biological studies.
Here, we have employed an alternative approach based on phenotypic screening of a focused small-molecule library of carbamates designed to inhibit a specific enzyme class (serine hydrolases), followed by competitive ABPP to identify the protein targets of bioactive hits. The special features of carbamates, which include drug-like structures and an irreversible mechanism of inhibition, facilitate swift in vivo validation of new targets without requiring extensive medicinal chemistry optimization. Our phenotypic assay selected molecules that promote adipocyte differentiation and lipid storage in fat cells. The notion that molecules that induce fat cell formation would be beneficial in diabetes may seem counterintuitive given the association between obesity and the development of insulin resistance and diabetes. However, obesity-linked insulin resistance is associated with increased levels of circulating free fatty acids and ectopic lipid deposition.14 In obesity, the hypertrophied adipocyte is not able to properly store excess fatty acids and, as a result, these lipids deposit in other tissues where they hinder insulin action.34 Compounds that can restore normal lipid partitioning amongst tissues and potentiate adipocyte function (e.g., TZDs) may thus enhance systemic insulin action. Several adipogenic carbamates from our screen inhibited the same target, a ~60 kDa serine hydrolase that was identified by competitive ABPP-MudPIT as Ces3 (also known as Ces1d or TGH).
Ces3 has been shown through genetic methods to be involved in lipolysis,27 the process whereby the adipocyte hydrolyzes stored triglycerides into fatty acids to be used as fuel by other tissues in times of need. Hormonally-induced lipolysis is controlled by catecholamines and insulin and is mediated by other serine hydrolases (ATGL/desnutrin, HSL, MGLL).24 In contrast, Ces3 has been reported to be important for basal lipolysis,27 in agreement with our observation that WWL113 and WWL229 block this process. Increased adipocyte lipolysis is a central feature of insulin resistance and type 2 diabetes:32,34 it generates surplus fatty acids that deposit as ectopically in tissues, and excess glycerol that the liver uses to boost glucose production. Compounds that inhibit lipolysis can improve insulin sensitivity in type 2 diabetes patients.35,36 Strategies to target obesity-linked increases in lipolysis have focused on inhibition of hormone-induced lipolysis, a process that is part of the normal response to fasting and feeding. Animal models have questioned the wisdom of systemic blockade of hormone-induced lipolysis,37 and these drug discovery efforts have yet to translate into clinical agents. Here, we have shown for the first time that pharmacologic restrain of basal lipolysis (via Ces3 inhibition) is sufficient to elicit therapeutic benefits in vivo. Because Ces3/hCES1 inhibitors will block primarily basal lipolysis, they may prove safer than molecules that hamper hormone-induced lipolysis.
Treatment with WWL113 ameliorated multiple features of metabolic syndrome in models of genetic and diet-induced obesity and insulin resistance/diabetes. The compound acts primarily at the two tissues of greatest Ces3 activity, adipose and liver. In fat, Ces3 inhibition blocks basal lipolysis and enhances adipocyte function. In liver, WWL113 treatment decreases expression of lipogenic genes while simultaneously increasing expression of fatty acid oxidation and ketogenesis enzymes. These changes, likely coupled to reduced fatty acid and glycerol efflux from adipocytes, prevent steatosis and enhance liver insulin sensitivity. Ces3 has been shown to be important for hydrolysis of hepatic TG stores prior to re-esterification and assembly into apoB-containing VLDL-TG particles.31 Inhibition of Ces3 in rat primary hepatocytes decreases VLDL secretion, which may appear at odds with the lack of lipid accumulation in the liver of WWL113-treated mice.38 However, Ces3 null primary hepatocytes show increased fatty acid oxidation.39 Thus, the phenotype we observe in treated mice likely reflects the balance of decreased synthesis and increased oxidation of hepatic fatty acids, and reduced influx of adipocyte-derived fatty acids.
Contrary to the profile of WWL113 in cultured adipocytes, competitive ABPP-MudPIT of tissues revealed that this compound also inhibits the closely related carboxylesterases Ces1f, Ces1 (Ces1g), Ces1c in vivo. While we cannot exclude the possibility that these poorly charaterized enzymes might contribute to the effects of WWL113, we should mention that all the major effects of WWL113 treatment are recapitulated by genetic ablation of Ces3.39 Mice lacking Ces3 fed a standard diet show decreased plasma triglycerides and free fatty acids, lack of liver steatosis, enhanced insulin sensitivity and glucose tolerance, augmented hepatic fatty acid oxidation, and increased energy expenditure (which may explain the resistance to weight gain in WWL113-treated mice, though we have no direct evidence of this). In contrast, Ces1 null mice develop obesity, hyperlipidemia, fatty liver, hyperinsulinemia, and insulin resistance on a normal diet.40 Moreover, consistent with the effect of WWL113 in cultured adipocytes, RNAi studies have shown that Ces3 knockdown blocks basal lipolysis without a major effect on hormone-induced lipolysis.27 Finally, we have shown that a second structurally-unrelated carbamate that shows complete Ces3 selectivity (WWL229) recapitulates all the effects seen with WWL113 in adipocytes. Thus, we believe that our set of Ces3-directed chemical probes has provided the first pharmacological evidence supporting the utility of Ces3 inhibition in models of obesity-diabetes. It is interesting to note that recently described liver-specific Ces3 null mice have decreased cholesterol and triglycerides in plasma, but moderately increased levels in liver.41 More importantly, these mice do not show enhanced glucose tolerance, and females display decreased insulin sensitivity. These findings suggest that adipose tissue may be a primary determinant of the benefits we have observed upon pharmacologic inhibition of Ces3.
The carboxylesterase family is considerably more complex in mice than humans, and many mouse enzymes lack human orthologues (e.g., Ces1f, Ces1c).42 In humans, hCES1 (the orthologue of mouse Ces3) has been studied primarily as a liver detoxification enzyme.43 However, increased hCES1 mRNA in adipose tissue has been associated with obesity and metabolic impairments in obesity-discordant sibling studies,44 and in several other human populations.45-47 Our data indicates that hCES1 activity is increased in WAT of obese and type 2 diabetics. Hence, CES1 inhibition in humans may produce similar metabolic benefits as are seen in mice treated with a Ces3 inhibitor.
We have outlined an approach integrating phenotypic screening with ABPP that can simplify the process of target identification and in vivo validation of screening results. Broad-spectrum activity-based probes have been developed for multiple enzyme classes beyond serine hydrolases.48-50 In principle, our approach should be applicable to evaluate the function of other enzyme classes in any desired biological output that can be reduced to a cell-based assay. Our work has also uncovered multiple poorly-annotated serine hydrolases active during adipogenesis, underscoring the potential of ABPP to contribute to the discovery of biochemical pathways that support complex cellular processes.
METHODS
Methods and associated references are available in the online version of the paper.
Supplementary Material
Acknowledgements
We thank Natasha Kralli and Peter Tontonoz for valuable discussions. E.S. is supported by a Career Development Award from the American Diabetes Association, The McDonald's Center for Type 2 Diabetes and Obesity, and NIH grant DK081003. B.F.C is supported by NIH grant CA087660. M.J.W. is supported by grants and a Senior Research Fellowship from the National Health and Medical Research Council of Australia. A.P.R. is supported by a Career Development Fellowship from the National Health and Medical Research Council of Australia. E.D. was supported by an Anxeles Alvariño fellowship from the Xunta de Galicia, Spain.
Footnotes
Author Contributions. E.S., E.D., and B.F.C. designed experiments. E.S., E.D., J.P., C.G. and A.G. performed cell-based, biochemical, and in vivo experiments. W.L and J.W.C. synthesized compounds. J.T. and D.K.N. performed lipidomic analysis. A.G., E.D. and K.-L. H. performed proteomic experiments. D.P. provided technical help. A.P.R., M.J.W., and P.E.O. provided human samples. E.D., J.P., C.G., A.G., J.T., D.K.N., K.-L. H., S.N., and B.F.C. and E.S. analyzed data. E.D., B.F.C. and E.S. wrote the manuscript.
Competing Financial Interests Statement
B.F.C. is a cofounder and advisor for a biotechnology company interested in developing inhibitors for serine hydrolases as therapeutic targets.
REFERENCES
- 1.Kodadek T. Rethinking screening. Nat Chem Biol. 2010;6:162–165. doi: 10.1038/nchembio.303. [DOI] [PubMed] [Google Scholar]
- 2.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–19. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 3.Hahn CK, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16:281–94. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–24. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 5.Moellering RE, Cravatt BF. How chemoproteomics can enable drug discovery and development. Chem Biol. 2012;19:11–22. doi: 10.1016/j.chembiol.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laggner C, et al. Chemical informatics and target identification in a zebrafish phenotypic screen. Nat Chem Biol. 2012;8:144–6. doi: 10.1038/nchembio.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wacker SA, Houghtaling BR, Elemento O, Kapoor TM. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat Chem Biol. 2012;8:235–7. doi: 10.1038/nchembio.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ong SE, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–22. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj L, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–4. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 11.Heal WP, Dang TH, Tate EW. Activity-based probes: discovering new biology and new drug targets. Chem Soc Rev. 2011;40:246–57. doi: 10.1039/c0cs00004c. [DOI] [PubMed] [Google Scholar]
- 12.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–91. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 13.Waki H, et al. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab. 2007;5:357–70. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 14.McGarry JD. Banting lecture 2001: dysregulation of fatty acid metabolism in the etiology of type 2 diabetes. Diabetes. 2002;51:7–18. doi: 10.2337/diabetes.51.1.7. [DOI] [PubMed] [Google Scholar]
- 15.Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–63. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wani JH, John-Kalarickal J, Fonseca VA. Dipeptidyl peptidase-4 as a new target of action for type 2 diabetes mellitus: a systematic review. Cardiol Clin. 2008;26:639–48. doi: 10.1016/j.ccl.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Morak M, et al. Differential activity-based gel electrophoresis for comparative analysis of lipolytic and esterolytic activities. J Lipid Res. 2009;50:1281–92. doi: 10.1194/jlr.M800566-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schicher M, et al. Functional proteomic analysis of lipases and esterases in cultured human adipocytes. J Proteome Res. 2010;9:6334–44. doi: 10.1021/pr1005795. [DOI] [PubMed] [Google Scholar]
- 20.Birner-Gruenberger R, et al. Functional fat body proteomics and gene targeting reveal in vivo functions of Drosophila melanogaster alpha-Esterase-7. Insect Biochem Mol Biol. 2012;42:220–9. doi: 10.1016/j.ibmb.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Bachovchin DA, et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci U S A. 2010;107:20941–6. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–7. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J Am Chem Soc. 2007;129:9594–5. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 24.Zechner R, et al. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–91. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehner R, Vance DE. Cloning and expression of a cDNA encoding a hepatic microsomal lipase that mobilizes stored triacylglycerol. Biochem J. 1999;343(Pt 1):1–10. [PMC free article] [PubMed] [Google Scholar]
- 26.Soni KG, et al. Carboxylesterase 3 (EC 3.1.1.1) is a major adipocyte lipase. J Biol Chem. 2004;279:40683–9. doi: 10.1074/jbc.M400541200. [DOI] [PubMed] [Google Scholar]
- 27.Wei E, Gao W, Lehner R. Attenuation of adipocyte triacylglycerol hydrolase activity decreases basal fatty acid efflux. J Biol Chem. 2007;282:8027–35. doi: 10.1074/jbc.M605789200. [DOI] [PubMed] [Google Scholar]
- 28.Okazaki H, et al. Identification of a novel member of the carboxylesterase family that hydrolyzes triacylglycerol: a potential role in adipocyte lipolysis. Diabetes. 2006;55:2091–7. doi: 10.2337/db05-0585. [DOI] [PubMed] [Google Scholar]
- 29.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:4686–7. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 30.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–9. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Quiroga AD, Lehner R. Role of endoplasmic reticulum neutral lipid hydrolases. Trends Endocrinol Metab. 2011;22:218–25. doi: 10.1016/j.tem.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–71. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boelsterli UA, Bedoucha M. Toxicological consequences of altered peroxisome proliferator-activated receptor gamma (PPARgamma) expression in the liver: insights from models of obesity and type 2 diabetes. Biochem Pharmacol. 2002;63:1–10. doi: 10.1016/s0006-2952(01)00817-6. [DOI] [PubMed] [Google Scholar]
- 34.Cusi K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr Diab Rep. 2010;10:306–15. doi: 10.1007/s11892-010-0122-6. [DOI] [PubMed] [Google Scholar]
- 35.Bajaj M, et al. Sustained reduction in plasma free fatty acid concentration improves insulin action without altering plasma adipocytokine levels in subjects with strong family history of type 2 diabetes. J Clin Endocrinol Metab. 2004;89:4649–55. doi: 10.1210/jc.2004-0224. [DOI] [PubMed] [Google Scholar]
- 36.Fulcher GR, Walker M, Catalano C, Agius L, Alberti KG. Metabolic effects of suppression of nonesterified fatty acid levels with acipimox in obese NIDDM subjects. Diabetes. 1992;41:1400–8. doi: 10.2337/diab.41.11.1400. [DOI] [PubMed] [Google Scholar]
- 37.Girousse A, Langin D. Adipocyte lipases and lipid droplet-associated proteins: insight from transgenic mouse models. Int J Obes (Lond) 2012;36:581–94. doi: 10.1038/ijo.2011.113. [DOI] [PubMed] [Google Scholar]
- 38.Gilham D, et al. Inhibitors of hepatic microsomal triacylglycerol hydrolase decrease very low density lipoprotein secretion. FASEB J. 2003;17:1685–7. doi: 10.1096/fj.02-0728fje. [DOI] [PubMed] [Google Scholar]
- 39.Wei E, et al. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 2010;11:183–93. doi: 10.1016/j.cmet.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Quiroga AD, et al. Deficiency of carboxylesterase 1/esterase-x results in obesity, hepatic steatosis, and hyperlipidemia. Hepatology. 2012;56:2188–98. doi: 10.1002/hep.25961. [DOI] [PubMed] [Google Scholar]
- 41.Lian J, et al. Liver specific inactivation of carboxylesterase 3/triacylglycerol hydrolase decreases blood lipids without causing severe steatosis in mice. Hepatology. 2012;56:2154–62. doi: 10.1002/hep.25881. [DOI] [PubMed] [Google Scholar]
- 42.Holmes RS, et al. Recommended nomenclature for five mammalian carboxylesterase gene families: human, mouse, and rat genes and proteins. Mamm Genome. 2010;21:427–41. doi: 10.1007/s00335-010-9284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu Rev Pharmacol Toxicol. 1998;38:257–88. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 44.Jernas M, et al. Regulation of carboxylesterase 1 (CES1) in human adipose tissue. Biochem Biophys Res Commun. 2009;383:63–7. doi: 10.1016/j.bbrc.2009.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinberg GR, Kemp BE, Watt MJ. Adipocyte triglyceride lipase expression in human obesity. Am J Physiol Endocrinol Metab. 2007;293:E958–64. doi: 10.1152/ajpendo.00235.2007. [DOI] [PubMed] [Google Scholar]
- 46.Nagashima S, et al. Depot-specific expression of lipolytic genes in human adipose tissues-- association among CES1 expression, triglyceride lipase activity and adiposity. J Atheroscler Thromb. 2011;18:190–9. doi: 10.5551/jat.6478. [DOI] [PubMed] [Google Scholar]
- 47.Marrades MP, Gonzalez-Muniesa P, Martinez JA, Moreno-Aliaga MJ. A dysregulation in CES1, APOE and other lipid metabolism-related genes is associated to cardiovascular risk factors linked to obesity. Obes Facts. 2010;3:312–8. doi: 10.1159/000321451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 49.Schmidinger H, Hermetter A, Birner-Gruenberger R. Activity-based proteomics: enzymatic activity profiling in complex proteomes. Amino Acids. 2006;30:333–50. doi: 10.1007/s00726-006-0305-2. [DOI] [PubMed] [Google Scholar]
- 50.Edgington LE, Verdoes M, Bogyo M. Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes. Curr Opin Chem Biol. 2011;15:798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1:1067–71. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–15. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 54.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 55.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer invasiveness. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cociorva D, D, L.T., Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinformatics. 2007;16:13.4.1–13.4.14. doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- 57.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 59.Niphakis MJ, Johnson DS, Ballard TE, Stiff C, Cravatt BF. O-hydroxyacetamide carbamates as a highly potent and selective class of endocannabinoid hydrolase inhibitors. ACS Chem Neurosci. 2012;3:418–26. doi: 10.1021/cn200089j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nomura DK, et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011;334:809–13. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.