Abstract
Extensive studies on floral transition in model species have revealed a network of regulatory interactions between proteins that transduce and integrate developmental and environmental signals to promote or inhibit the transition to flowering. Previous studies indicated FLOWERING PROMOTING FACTOR 1 (FPF1) gene was involved in the promotion of flowering, but the molecular mechanism was still unclear. Here, FPF1 homologous sequences were screened from diploid Gossypium raimondii L. (D-genome, n = 13) and Gossypium arboreum L. genome (A-genome, n = 13) databases. Orthologous genes from the two species were compared, suggesting that distinctions at nucleic acid and amino acid levels were not equivalent because of codon degeneracy. Six FPF1 homologous genes were identified from the cultivated allotetraploid Gossypium hirsutum L. (AD-genome, n = 26). Analysis of relative transcripts of the six genes in different tissues revealed that this gene family displayed strong tissue-specific expression. GhFPF1, encoding a 12.0-kDa protein (Accession No: KC832319) exerted more transcripts in floral apices of short-season cotton, hinting that it could be involved in floral regulation. Significantly activated APETALA 1 and suppressed FLOWERING LOCUS C expression were induced by over-expression of GhFPF1 in the Arabidopsis Columbia-0 ecotype. In addition, transgenic Arabidopsis displayed a constitutive shade-avoiding phenotype that is characterized by long hypocotyls and petioles, reduced chlorophyll content, and early flowering. We propose that GhFPF1 may be involved in flowering time control and shade-avoidance responses.
Introduction
Cotton (Gossypium spp.) is one of the most important natural fiber crops in the world. In addition to its economic importance, cotton has attracted considerable scientific interest from plant breeders, taxonomists, developmental geneticists, and evolutionary biologists because of its unique reproductive developmental aspects and speciation history [1]–[3]. The genus Gossypium L. contains more than fifty species, which are cytogenetically differentiated into eight genomic groups (A–G, and K). Most of the species are diploid (n = 13), but five are allopolyploids (n = 26), originating from an interspecific hybridization event between A- and D-genome diploid species. Not only two diploids of A-genome, Gossypium herbaceum L. and G. arboreum L., but also two allopolyploids of AD-genome, G. hirsutum L. and G. barbadense L. were domesticated by humans for their fiber demands [3]. Because of environmental pressures, such as land use and climatic change, the earliness of cotton has become a vital subject for plant breeders. Several traits co-operate to influence the early ripeness of upland cotton (G. hirsutum L.), with flowering time being especially important. In the seed crop, floral transition is a key developmental switch in the life cycle of cotton as it contributes to the production of dry matter. Furthermore, shifting of the seasonal timing of reproduction is a major goal of plant breeding research as it will produce novel varieties better adapted to local environments and effects of climate change [4].
Plant growth originates from a small number of undifferentiated cells called meristems. The apical meristems being indeterminate or determinate contribute to the fate of shoot architecture. Indeterminate apical meristems retain a population of vegetative stem cells indefinitely which perform tissue and organ differentiation below and on the flanks of the main-stem. Meanwhile the determinate apical meristems undergo terminal differentiation, commonly in a flower or inflorescence. In annual plants, the floral induction process occurs when vegetative shoot meristems develop into inflorescence meristems, and give rise to flowers [5]. The use of the model species Arabidopsis thaliana and Antirrhinum majus has led to significant progress in the understanding of the floral transition. Numerous genes involved in the control of flowering time have been identified, and the roles they played in molecular and genetic pathways were also characterized. Previous studies uncovered that four main floral pathways, namely vernalization, photoperiod, gibberellin, and autonomous pathways converged to regulate floral integrator genes such as LEAFY (LFY), APETALA 1 (AP1), FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), which in turn could activate genes required for reproductive development [6]. Another transcription factor FLOWERING LOCUS C (FLC) containing MADS domain was an inhibitor of flowering, acting as an important convergence point for the autonomous and vernalization pathways in Arabidopsis thaliana [7]–[9].
FLOWERING PROMOTING FACTOR 1 (FPF1) gene was originally understood on account of its role in flowering. Over-expression of FPF1 (Y11988) led to early flowering in Arabidopsis [10]. FPF1 is expressed in apical meristems immediately after the photoperiodic induction of flowering in long-day plants that could flower in response to long days [10]. Previous studies indicated that FPF1 might play an important role in modulating the competence of apical meristems to rapidly respond to the floral meristem identity genes AP1and LFY. During the transition to flowering in Arabidopsis, FPF1 is normally activated at a similar time as LFY, and earlier than AP1. Over-expression of AtAP1 and AtFPF1 shows a synergistic effect in the shortening of the time to flowering both under long-day and short-day conditions [11]. Two closely related genes, FLP1 and FLP2 (FPF1-Like genes), have been identified in Arabidopsis. Constitutive over-expression of each gene causes earlier flowering under both long and short day conditions [12]. Up till now, homologous genes of FPF1 have also been characterized in rice (Oryza sativa) and tobacco (Nicotiana tabacum) [13], [14]. OsRAA1, a homolog of FPF1 in rice, shares similarity of 58% with AtFPF1 at amino acids level. Evidence revealed that over-expression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf shape, heading time and root development [13].
In summarize, AtFPF1/OsRAA1 gene family takes parts in several aspects of plant development. Currently, little is known about the underlying mechanism of these, or of any crossover between this gene family and other floral molecules. This paper described our work to dissect these mechanisms. In our previous study, a high-quality, normalized, full-length cDNA library with a total of 14,373 unique ESTs was generated to provide sequence information for gene discovery related to flower development in upland cotton [15]. The publication of G. raimondii L. genome sequence by the Cotton Genome Project in 2012 facilitates gene excavation vital for the genetic improvement of cotton quality and productivity, as well as serving as a reference for the assembly of the tetraploid G. hirsutum L. genome [16]. G. raimondii L. genome (http://cgp.genomics.org.cn/) and G. arboreum L. genome databases (unpublished) were screened to identify and compare homologs of AtFPF1 from the two diploid cotton species. The genes were cloned and characterized from G. hirsutum L. as one of the major cultivated species. In addition, their expression pattern in cotton, and ectopic expression in Arabidopsis were analyzed.
Materials and Methods
Plant Material and Growth Conditions
A genetic standard line TM-1 (G. hirsutum L.) and a short-season cotton variety CCRI 36 (G. hirsutum L.) were grown in a greenhouse (Anyang, China) with an optimal temperature of 28°C. In order to minimize experimental error, two varieties were planted in the soil possessing equal fertility and managed using standard agricultural practices. Shoot apices from about three hundred plants of the two varieties were harvested when three true leaves came out. Roots, stems, young leaves, mature leaves, flowers, and fibers from mature CCRI 36 and TM-1 plants were collected and mixed together.
Seeds of Arabidopsis thaliana (Columbia-0 ecotype: wild-type or GhFPF1 over-expressing transgenic plants) were sterilized in 75% (v/v) absolute alcohol for 30 s and 10% H2O2 (v/v) for 10 min. After four washes in sterilized double-distilled water (ddH2O), seeds were sown onto sterilized 1/2 MS solid medium (PH 5.8) containing 1.5% sucrose, 0.7% agar (and 50 mg/L kanamycin for transgenic lines). 10-cm petri plates containing medium and seeds were chilled for 48–72 h at 4°C in the dark, and then transferred into an illuminating incubator with 100 µmol·m−2 s−1 fluorescent light at 22°C. Two weeks later, normal seedlings were transplanted into the soil in a growth chamber which provided the plants with continuous 150 µmol·m−2 s−1 fluorescent light at room temperature 22°C. As for the length of hypocotyl and petiole, chlorophyll content and flowering time assay, seeds were directly sowed into the soil in the growth chamber. Ten days later, seedlings were transplanted into flowerpots grown under the same conditions. All Arabidopsis plants were grown under long-day conditions with high red to far red (R/FR ratio: 4.5) provided by fluorescent lamps.
Gene Screening and Sequence Comparative Analysis
Candidate sequences were obtained by performing Blast searches of genome databases of G. raimondii L. (http://cgp.genomics.org.cn/) and G. arboreum L. (unpublished) using Arabidopsis thaliana FPF1 gene as a query sequence. Multiple sequence alignment of the deduced amino acids was performed using the software ClustalX2 (http://www.ebi.ac.uk). The phylogenetic tree was constructed by Molecular Evolutionary Genetics Analysis (MEGA) software 4.1 (http://www.megasoftware.net). Sequence of promoter was analyzed in the database of plant cis-acting regulatory DNA elements (PlantCARE) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Gene Isolation and RNA Analysis
Different organs from upland cotton and Arabidopsis were harvested, and immediately plunged into liquid nitrogen. Total RNA was isolated from cotton and Arabidopsis using the EASY spin plus RNA reagent kit RN38 (AIDLAB, Beijing, China) following manufacturer’s instructions. Poly (dT) cDNA was prepared from the total RNA using the Superscript III First-Strand Synthesis System (Invitrogen, USA) according to the manufacturer’s protocol. Genomic DNA was extracted as described [17].
Open reading frames (ORFs) of GhFPF1and homologous genes were amplified from G. hirsutum L. CCRI 36 using the primers described in Table S1. The running conditions were as follows: denaturation at 94°C for 2 min, followed by 33 cycles at 94°C for 30 s, annealing at 58°C for 1 min and extension at 72°C for 30 s. To obtain the 5′-terminal and 3′-terminal sequences of GhFPF1, 5′- and 3′-RACE were performed using the SMART RACE cDNA Amplification Kit (Clontech, USA) according to the Kit’s protocol in conjunction with primers GPS1 and GPS2 (Table S1).
Quantitative polymerase chain reactions (QRT -PCRs) were performed to measure relative expression of homologous genes using specific primers (Table S2), with Histone 3 (AF024716) to be an internal control [18]. For Arabidopsis, AtUBQ5 (AT3G62250) was used as the control. Transcriptional changes of flowering related genes AtFPF1, AtLFY, AtAP1, AtCO, AtFT, AtSOC1, AtFLC and AtPHYB were examined in the wild as well as transgenic plants. Each qRT-PCR experimental condition was independently repeated three times and in each of these three biological repetitions, three technical replicates were made. All of the amplification of interested genes was analyzed on ABI 7500 system (Applied Biosystems, USA) with SYBR Green I (with Rox) reagents to detect the target sequences. The running conditions were as follows: holding stage at 50°C for 2 min, 94°C for 10 min, followed by 40 cycles at 95°C for 15 s, 60°C for 1 min. Then a melting curve was performed from 65 to 95°C to verify the specificity of the amplified product. QRT-PCR data was processed to measure the relative expression of genes in accordance with the 2−ΔΔCT method [19].
Jasmonic Acid and Salicylic Acid Treatment
Jasmonic acid (JA; Sigma, USA) was dissolved in a small quantity of ethanol, and further diluted with ddH2O to a concentration of 200 µmol·L−1. Salicylic acid (SA; Sigma, USA) was directly dissolved into heated ddH2O water to a final concentration of 400 µmol·L−1. Upland cotton cultivar CCRI 36 was planted in the greenhouse with an optimal temperature of 28°C. One-month-old plants were sprayed with JA and SA solutions when four true leaves had developed [20], [21], [36]. The top two leaves were removed at 0, 0.5, 6, 12, 24, and 48 h time-point. Harvested leaves were immediately plunged into liquid nitrogen, and stored at –80°C for later RNA extraction and relative transcript analysis. Nonexpressor of Pathogenesis-Related Genes 1 (GhNPR1, DQ409173), a SA- and JA-inducible gene in upland cotton was used as a positive control gene here [21].
Vector Construction and Genetic Transformation
The open reading frames of GhFPF1 (GenBank Accession No. KC832319) was cloned into the binary vector pBI121 using an In-Fusions ™ Advantage PCR Cloning Kit (Clontech, USA) via BamHI and SacI sites (New England BioLabs, USA). The recombinant plasmid containing CaMV35S::GhFPF1 was introduced into Agrobacterium tumefaciens (strain LBA4404) and then transformed into Arabidopsis (Columbia-0 ecotype) according to the method of floral dip [22]. Transformants were selected on 1/2 MS medium containing 50 mg l−1 kanamycin, and further confirmed at both the genomic DNA and transcriptional mRNA level. To confirm that GhFPF1had been integrated into the Arabidopsis genome, RT-PCR was performed with the 35S promoter primer (forward) and the GhFPF1-specific primer (reverse). To detect the mRNA expression of GhFPF1in T3 transgenic lines COL-3, COL-4 and COL-7, 18-day-old intact plants were sampled for quantitative RT-PCR.
Flowering Time Calculation and Chlorophyll Content Determination
At least twenty individual plants of the homozygous T3 transgenic lines and wild-type, were governed under the long-day conditions (16 h light/8 h dark). Flowering time was measured by counting the numbers of rosette and cauline leaves and days of the appearance of the first flower. Statistical significance analysis was conducted using the software Sigma Stat 3.5 (Systat Software, San Jose, CA, USA) by One-Way ANOVA analysis. Leaves to determine the content of chlorophyll were harvested from 24-day-old Arabidopsis. Chlorophyll was extracted through 95% acetone-alcohol according to the method as described previously [23] and monitored in a 96-well microplate reader (BioTek, USA).
Ethics Statement
We did not make use of human or vertebrate animal subjects and/or tissue in our research.
Results
Gene Screening and Sequence Comparative Analysis
To investigate the homologs of FLOWERING PROMOTING FACTOR 1 (FPF1) in cotton, the genome databases of G. raimondii L. and G. arboreum L. were screened with the coding region of Arabidopsis FPF1 gene as the reference sequence. Six sequences, which covered complete ORFs, were selected from each database. The twelve ORFs were predicted to encode small proteins of 99 to 113 amino acids in length. Pair-wise alignment of these predicted proteins suggested that they shared 50–68% similarity to the AtFPF1 protein. In addition, these proteins were rich in the three amino acids Ser, Val, and Leu.
Further, pair-wise alignment of the amino and nucleic acids was conducted to explore the differences between G. raimondii L. and G. arboreum L. orthologous genes. Nucleic acid alignment revealed that five of the six orthologous pairs exhibited 95–99% similarity to each other, and the other pair DF10009151- Garb22630, had 84% identity (Table 1). Two pairs among them, DF10021325-Garb08271 and DF10039615-Garb17683, owned the same deduced amino acids sequence because of codon degeneracy. One or two amino acids were different between the three pairs of orthologous DF10000980-Garb07734, DF10029023-Garb34130 and DF10007455-Garb19766. The remaining pair, DF10009151-Garb22630, showed greater divergences, with just 86% homology at amino acid level. Comparative analysis revealed that a short length of nucleic acids was missing from Garb22630 (Figure S1).
Table 1. Pair-wise alignment between G. raimondii L. and G. arboreum L. orthologous sequences on amino acid and nucleic acid levels.
| G. raimondii L. | DF10021325 | DF10039615 | DF10007455 | DF10000980 | DF10029023 | DF10009151 |
| G. arboreum L. | Garb08271 | Garb17683 | Garb19766 | Garb07734 | Garb34130 | Garb22630 |
| Similarities of nucleic acid/amino acids (Identity %) | 97/100 | 99/100 | 95/98 | 99/99 | 97/99 | 84/86 |
Isolation of FPF1 Homologous Genes in G. hirsutum L. and Sequence Analysis
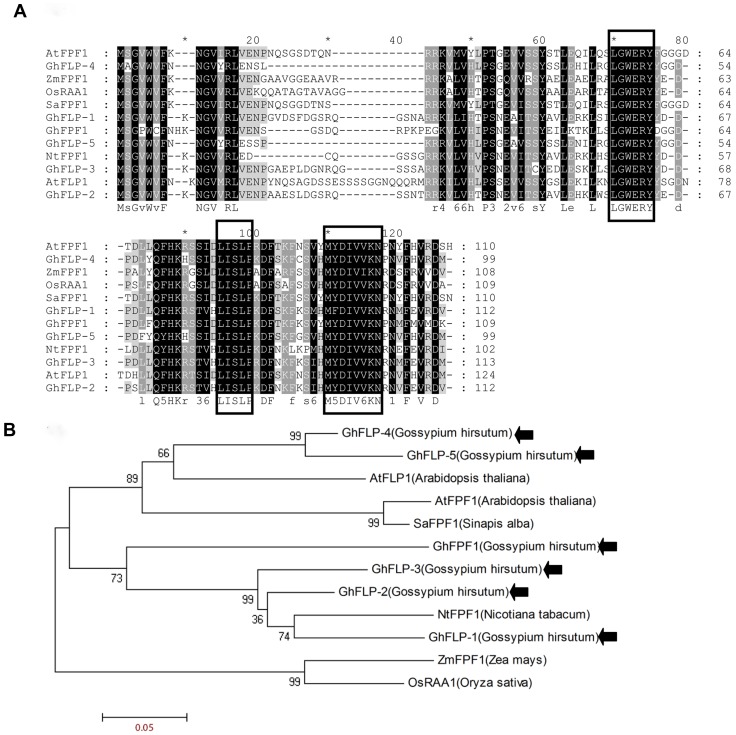
Six genes were identified from CCRI36 and named as GhFPF1, GhFLP-1, GhFLP-2, GhFLP-3, GhFLP-4, and GhFLP-5 corresponding to the sequences of DF10009151, DF10007455, DF10029023, DF10021325, DF10000980, and DF10039615 in G. raimondii L., respectively. The small FPF1 gene family has been characterized in several species including Arabidopsis thaliana, white mustard (Sinapis alba), rice (Oryza sativa), tobacco (Nicotiana tabacum) and maize (Zea mays). Multiple alignment (Figure 1A) of amino acid sequences revealed that distinctions between the homologous proteins occurred in the N-terminus relative to C-terminus. Results suggested that there were three conserved domains present in the protein family. The first motif, -LGWERY- was located in the middle section of the protein, while the second and third conserved motifs, -D/HLISLP- and -MY/FDIVVKN-, were found closer to the C-terminus. Phylogenetic analysis of the FPF1 gene family indicated that these proteins could be divided into three clades, represented by NtFPF1, ZmFPF1/OsRAA1 and AtFPF1. GhFPF1, GhFLP-1, GhFLP-2, and GhFLP-3 were placed into NtFPF1 clade; GhFLP-4 and GhFLP-5 were in the same clade with AtFPF1. None of them were placed into the ZmFPF1/OsRAA1 branch (Figure 1B).
Figure 1. Multiple sequence alignment of FPF1 protein family from G. hirsutum L., and other species.
A. Multiple alignment of FPF1 protein sequences in several species. AtFPF1 (Y11988) and AtFLP1 (AL353995) are Arabidopsis thaliana genes; SaFPF1 (Y11987), NtFPF1 (AY496934), ZmFPF1 (ACG44143) and OsRAA1 (AY659938) are from Sinapis alba, Nicotiana tabacum, Zea mays and Oryza sativa. B. Phylogenetic tree of the FPF1 proteins in the above plants as determined by the MEGA 4.1 software package.
GhFPF1 had Higher Transcriptional Levels in the Floral Apices of CCRI 36
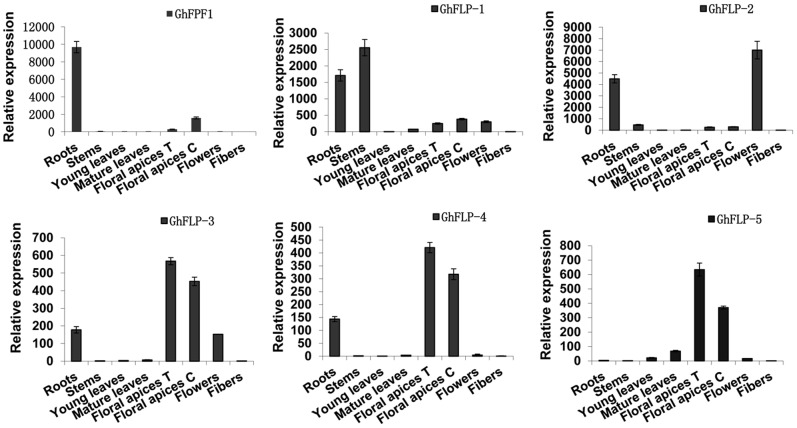
QRT-PCR was used to profile relative expression of above six genes in different tissues of upland cotton. Roots, stems, leaves, flowers and fibers are mixed samples from CCRI 36 and TM-1 while floral apices T and C represent floral apices from CCRI 36 and TM-1, respectively (Figure 2). GhFPF1 gene family displayed tissue-specific expression because abundant transcripts of the six genes were found in roots, floral apices, flowers, and stems, but were barely detectable in leaves or fibers. GhFLP-3, GhFLP-4 and GhFLP-5, in particular, had only some but not much expression in floral apices relative to other tissues. More importantly, we focused on the contrastive analysis of gene expression in floral apices of CCRI 36 (a short-season cotton variety) and TM-1 (a genetic standard line). Results uncovered that GhFPF1 had more than four-fold transcript levels in CCRI 36 than in TM-1. Higher expression of GhFPF1 in the short-season cotton suggested that it was the most possible FPF1 orthologous gene as AtFPF1 involved in the promotion of flowering.
Figure 2. Expression patterns of GhFPF1, GhFLP-1, GhFLP-2, GhFLP-3, GhFLP-4, and GhFLP-5 in G. hirsutum L.
Relative expression of GhFPF1 and its homologs were measured in different tissues of upland cotton CCRI 36 (a short-season cotton variety) and TM-1 (a genetic standard line) using qRT-PCR. Roots, stems, leaves, flowers and fibers stand for mixed samples from CCRI 36 and TM-1. Floral apices from CCRI 36 and TM-1 were harvested and named as floral apices T and C respectively. Error bars represent standard deviation (SD).
Gene Structure Analysis of GhFPF1 and its Response to JA and SA
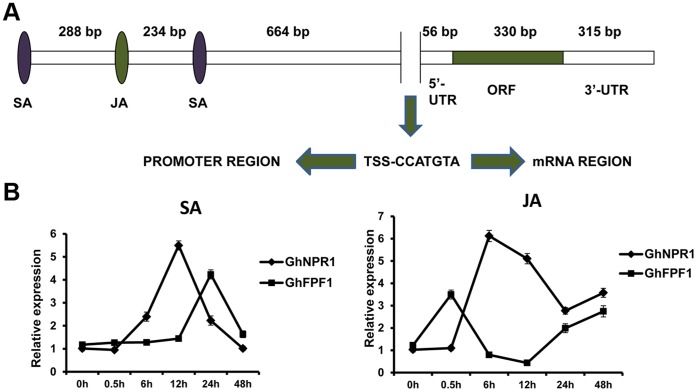
A 5′- and 3′- RACE strategy was performed to gain transcription initiation and termination sites of GhFPF1. A full-length cDNA of 701 bp composed of 56 bp 5′-UTR, 315 bp 3′-UTR and 330 bp ORF was isolated from the cDNA pool of one-week-old seedlings (Figure 3A). Comparison of genomic and cDNA sequence revealed that there was no intron. Sequences of GhFPF1 and GhFPF1-like genes were submitted to NCBI (Accession number: KC832319, KF830866-KF830870).
Figure 3. Gene structure of GhFPF1, and its response to plant hormones.
A. The gene structure of GhFPF1. JA and SA represent response elements of JA and SA; TSS, transcriptional start site. B. GhFPF1 and GhNPR1 expression profiles in the first forty-eight hours after treatment with SA and JA. GhNPR1 (DQ409173), a known JA- and SA-inducible gene, was used as a positive control here. Error bars represent SD.
A 2076 bp promoter region of GhFPF1 was identified from the genomic DNA of upland cotton, containing one and three response elements of methyl jasmonate and salicylic acid respectively. Treatment assay and the following qRT-PCR were performed to evaluate whether GhFPF1 could be regulated by JA and SA plant hormones (Figure 3B). NPR1 (nonexpressor of pathogenesis-related genes 1), a pathogen-related gene regulating SA-dependent defense response, systemic acquired resistance, and mediating crosstalk between SA and JA [24], [25], was used as a SA- and JA-inducible reference gene. According to figure 3B, GhFPF1 and GhNPR1 exhibited the similar expression trend when plants were treated with SA and JA, respectively. After 12h treating with SA, the expression of GhNPR1 reached its peak but peak expression of GhFPF1 occurred 12h later. However, GhFPF1 was responsive to JA earlier than GhNPR1. GhFPF1 transcripts were decreased 0.5h later and began to rise till 12h after the JA treatment, but the transition points of expression trend of GhNPR1 was lagging. GhFPF1 showed different expression fluctuations exposed to exogenous SA and JA but it was positively regulated by the two phytohormones over most time of 48h, suggesting that GhFPF1 could be responsive to both JA and SA, the latter later.
Over-expression of GhFPF1 Promoted Flowering in Arabidopsis
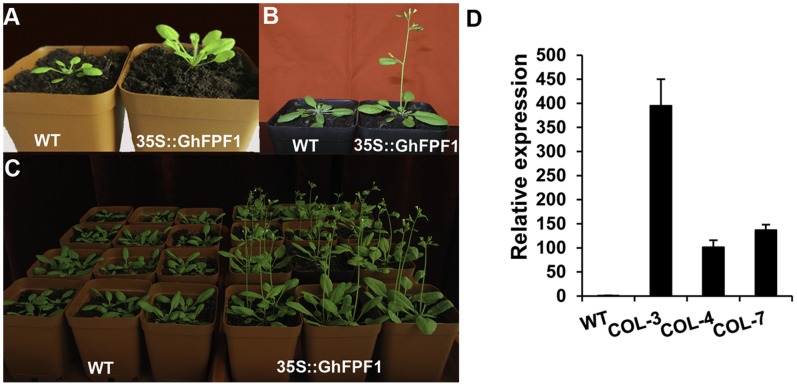
To discuss whether GhFPF1 could modulate flowering time in plants, the open reading frame of GhFPF1was transformed into the Arabidopsis Columbia-0 ecotype under the control of the cauliflower mosaic virus 35S promoter. Seven independent transgenic lines were obtained, and the GhFPF1 transcript was analyzed in three transgenic lines COL-3, COL-4, and COL-7 (Figure 4D). Homozygous T3 lines and the wild-type were grown under long-day conditions of 16 h light/8 h dark. Twenty-four days after sowing, most plants of the transgenic lines had started bolting, while wild-type plants remained in vegetative growth with six rosette leaves developed (Figure 4A). Six days later, the transgenic plants flowered in succession, whereas the wild-type plants had just completed their basal rosette development at this point (Figure 4B and C). To determine flowering time, the number of rosette and cauline leaves and days of the first flower opening up were counted (Table 2). Results turned out that transgenic plant produced flower buds six days earlier than 36.8 days to flowering in the wild-type on average. This corresponds to a reduction in the number of leaves from 15.4 in wild-type to the fewest 11.5 in the transgenic line. Data demonstrated that over-expression of GhFPF1 in Arabidopsis promoted flowering under inductive photoperiods.
Figure 4. over-expression of GhFPF1 promoted flowering in Arabidopsis.
A. 24-day-old plants grown under long-day conditions. The wild-type control developed six small rosette leaves (left) by the time that the GhFPF1 transgenic plant had started bolting on (right). B and C. One-month-old plants grown under long-day conditions. The wild-type control was still at the vegetative stage (left), whereas the transgenic plant(s) had flowered (right). D. Relative transcriptional analysis of GhFPF1 in 18-day-old transgenic Arabidopsis lines COL-3, COL-4 and COL-7.
Table 2. Flowering time under long-day conditions, as measured by days to flowering and number of basal rosette and cauline leaves.
| Line NO | Flowering time (days after sowing) | P value | Rosette leaves + cauline leaves | P value |
| WT | 36.8±0.47 | 15.4±0.55 | ||
| COL1 | 30.0±0.44* | <0.05 | 11.5±0.57* | <0.05 |
| COL2 | 30.9±0.34* | <0.05 | 12.4±0.42* | <0.05 |
| COL3 | 30.3±0.53* | <0.05 | 11.9±0.42* | <0.05 |
| COL4 | 33.3±0.74* | <0.05 | 12.7±0.41* | <0.05 |
| COL5 | 32.2±0.72* | <0.05 | 13.3±0.57* | <0.05 |
| COL6 | 32.5±0.67* | <0.05 | 13.3±0.56* | <0.05 |
| COL7 | 30.7±0.66* | <0.05 | 12.2±0.32* | <0.05 |
Asterisks indicate significant variation differences between the wild-type and each 35S::GhFPF1 transgenic population line as determined by one-way ANOVA analysis. Means with SD from twenty plants of every line were shown.
To gain further insight into how GhFPF1 regulates the floral transition, we examined the expression levels of six genes associated with the promotion of flowering of Arabidopsis: FPF1, LFY, AP1, CONSTANS (CO), FT, SOC1, and the flowering repressor, FLC both in wild-type and transgenic plants of vegetative (18-day-old) and bolting on (24-day-old). Relative qRT-PCR indicated that the level of AtFLC transcripts in transgenic plants of 18-day-old was one-eighth of that in wild-type plants under long-day conditions. Meanwhile, AtSOC1 and AtAP1 transcripts were up-regulated by two to three-fold relative to the wild-type. There was no obvious change in the relative transcript amount of AtFPF1, AtLFY, AtFT, or AtCO (Figure 5A). In 24-day-old transgenic plants collected at the same time of 11 am (3 hours exposure to light), the transcripts of AtFPF1, AtLFY, AtFT, AtCO were up-regulated by four times more or less, but AtSOC1 transcripts remained at the same levels as in 18 days (Figure 5A and B). Remarkably, AtAP1 was activated highly with an increase of 18.5-fold compared to the wild-type; meanwhile, the expression of the flowering repressor AtFLC was almost completely suppressed due to the ectopic expression of GhFPF1. Combining these, it could be hypothesized that early flowering conferred by GhFPF1 over-expression in Arabidopsis might be mediated through AtAP1 and AtFLC.
Figure 5. Relative qRT-PCR analysis of genes involved in floral regulation in Arabidopsis.
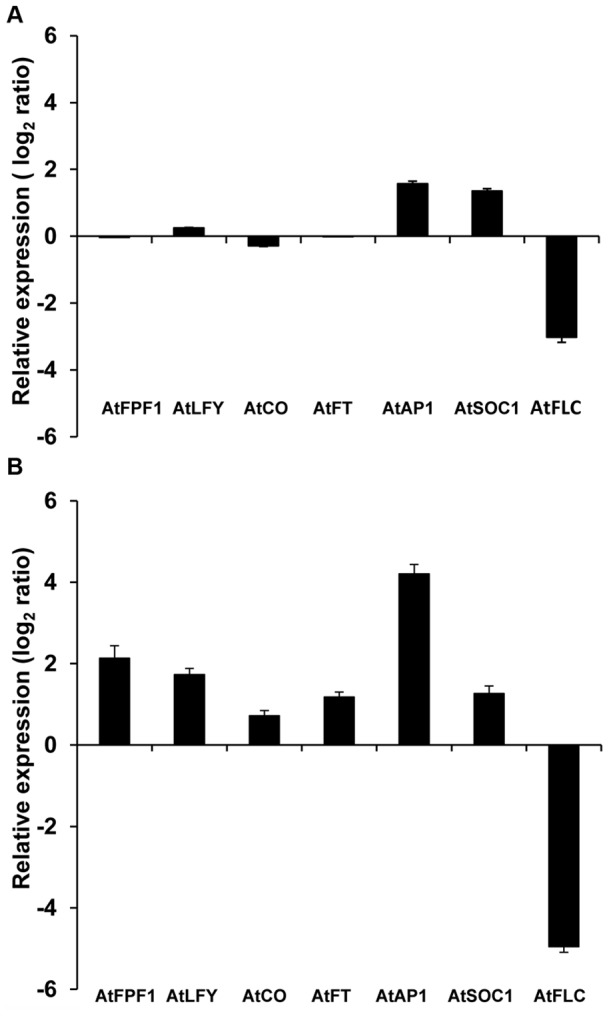
The AtUBQ5 gene was used for calibration and the histogram was drawn based on the log2 scale of the ratio of gene expression in transgenic plants relative to wild-type. Entire 18-day-old plants of vegetative (A) and the above-ground parts of 24-day-old plants of starting bolting on (B) were analyzed. Interested genes were AtFPF1 (AT5G24860), AtLFY (AT5G61850), AtCO (AT5G15840), AtFT (AT1G65480), AtAP1 (AT1G69120), AtSOC1 (AT2G45660), and AtFLC (AT5G10140). All plants were grown under long-day conditions of 16 h light/8 h dark. Data in graph were mean values with standard deviation (error bar) from three replicates.
Over-expression of GhFPF1 Triggered Shade Avoidance Syndrome (SAS) in Arabidopsis
Except accelerated flowering time, compared with wild-type, transgenic Arabidopsis generated longer hypocotyls and petioles (Table 3), as well as the upward of movement of leaves. Also chlorophyll content was found to be reduced in transgenic plants (Figure 6). Taken together, transgenic plants were recognized as so-called shadow avoidance syndrome (SAS). The elongated appearance and early flowering response were similar to phenotype of phyB mutants. PHYB, acting as the major phytochrome in light-grown plants played a predominant role in shade avoidance syndrome [48]. Since the plants were provided sufficient fluorescent light with red to far-red ratio 4.5, transcripts of PHYB were measured using qPCR in transgenic and wild Arabidopsis further. The result revealed that expression of PHYB in transgenic plants was decreased by more than fifty percent (Figure 6E).
Table 3. Statistics of hypocotyl and petiole lengths of wild and transgenic plants grown under long-day conditions with fluorescent lamps.
| Line NO | Hypocotyl length (mm) | P value | Petiole length (mm) | P value |
| WT | 3.8±0.04 | 7.0±0.18 | ||
| 35S:: GhFPF1 | 6.5±0.07** | <0.01 | 11.2±0.21** | <0.01 |
Hypocotyl lengths of wild and transgenic lines were quantified from thirty ten-day-old plants. Petiole lengths of true leaves of the basal rosette were measured and the data were gathered from thirty wild type and transgenic 24-day-old plants, respectively. Mean values (±SD) were shown and statistical analysis was evaluated by the same way as table 2.
Figure 6. Over-expression of GhFPF1 led to shade-avoidance responses in transgenic plants.
A. Phenotype of hypocotyl of ten-day-old wild type (left) and transgenic plants (right) grown under long-day conditions. B. The seventh rosette leaves of wild type (left) and transgenic (right) 24-day-old plants grown under the same conditions as (A). C. Shade-avoidance responses in 24-day-old transgenic plants (right) which grew fast with upward leaves. All Arabidopsis plants were grown under long-day conditions with high red/far red (R/FR ratio: 4.5) light provided by fluorescent lamps. D. Chlorophyll content of leaves in transgenic plants was lower than that in wild-type and the difference was very significant (P<0.01) assessed by T-test. E. The transcript levels of AtPHYB (AT2G18790) in wild-type and GhFPF1 over-expression transgenic plants. The AtUBQ5 gene was used as calibrator.
Discussion
GhFPF1 gene belongs to a novel gene family that seems to be conserved in both higher and lower plants. Until now, as were characterized in several species, members of this gene family were short in length as well as lacking in intron of their genomic sequences [10]–[12]. Twelve FPF1 homologs were identified from the diploid cotton genomic databases of G. raimondii L. and G. arboretum L. Orthologous sequences from the two cotton species were compared with each other, suggesting that nucleic acid sequences of the six pairs of orthologs were distinct, though two pairs of orthologous genes possessed the same deduced protein sequence as a result of codon degeneracy. High constraints of genetic divergence might occur during speciation for five genes had higher synonymous changes between the two species.
Previous studies have indicated that for each gene studied, allotetraploid species of cotton should have two homologs which represent descendants from the A-genome, and D-genome donors, evolving independently at the time of polyploidy formation [26]. Meanwhile, on allopolyploidy, genomic changes will take place, including chromosomal rearrangement and changes in gene expression. During the cloning and identification of the FPF1 homologous genes in G. hirsutum L., at least eight single clones were sequenced for every gene. It was discovered that five genes contained cDNA sequences identical to D-genome sequences from G. raimondii L., whereas only one gene GhFLP3 was found to have the same sequence as the A-genome sequence from G. arboreum L. The finding revealed that this gene family exhibited subgenome-specific expression bias to D-subgenome.
When plants undergo the transition to flowering, the vegetative shoot apical meristem will be transformed into an inflorescence meristem. Inflorescence meristems can respond to both environmental and endogenous flowering signals to give rise to floral meristems, which go on to produce the various floral organs in succession [27]. Axillary buds of upland cotton can be either vegetative or floral buds which develop into vegetative shoots or reproductive shoots, respectively. Two types of axillary buds are distinct from each other for apical meristems of the vegetative buds are small and either domed or tapered in shape, with one or two layers of tunica cells but the apical meristems of the floral buds are large and columnar, with two or three layers of tunica cells in floral buds with flat surfaces [28]. It was reported that the short-season cotton varieties of CCRI16 and CCRI36 initiated morphological differentiation of the floral bud at the developmental stage of two true leaves flattened, whereas the late-maturing CCRI12 activated this process when there were three true leaves expanded [28], [29]. TM-1, a genetic standard line as well as a late-maturing variety was also found to begin morphological differentiation of the floral bud when three true leaves were expanded (Figure S2). To select specific genes that are involved in the floral development of short-season cotton, the relative expression of GhFPF1 and GhFPF1-like genes in the floral apices of CCRI 36 and TM-1 were analyzed and compared at the developmental stage of three true leaves expanded. Transcripts of GhFPF1 were discovered to be more abundant in the floral apices of CCRI 36, suggesting that GhFPF1 might be involved in the floral regulation of short-season cotton.
In the study we found that GhFPF1 was predominantly expressed in roots and floral apices, the former more. In Arabidopsis thaliana, a number of genetic pathways controlling flowering time have been identified. In fact, most of the genes involved were preferentially expressed in the shoot apical meristem (SAM) and the root tip, such as PHYA, CRY2, FLC and so on, but, surprisingly, only a few were expressed preferentially or exclusively in leaves for example FT [7], [30], [31]. Day length and light quality are essentially perceived by photosynthetic organs, expanded leaves and stem, whereas water and mineral availability are perceived by the roots. The root system is presumably capable of reacting to the critical environmental changes and, as a result, influences the flowering process to some extent. In Sinapis, analyses of changes in the contents of phloem and xylem saps during the floral transition have disclosed a complex shoot-to-root-to-shoot signalling loop involving both nutrients and hormones [32], [33]. Higher transcripts of GhFPF1 in roots implicates that it could be involved in multiple functions in Gossypium hirsutum L. Moreover, members of GhFPF1 gene family displayed tissue specific expression, which is not rare. Most genes controlling flowering time were expressed across a wide range of organs and tissues, but a survey of available data on their spatial expression patterns revealed that many genes showed preferential expression in more limited areas such as SAM and RAM [34].
Jasmonic and salicylic acid response elements were found in the promoter region of GhFPF1, which suggested that GhFPF1 may be regulated by the plant hormones JA and SA. Response of GhFPF1 to JA (wound signal) and SA treatments could hint involvement of GhFPF1 in plant defense responses. Previous studies implicated both positive and negative roles of SA in affecting flowering time via influencing the expression of flowering regulatory genes FLC and FT [35]–[38]. Also SA can links flowering time with some stress signalling coming from defense responses or poor-nutrition [36], [38]. Recent studies provided new insights into the mechanisms of salicylic acid (SA) perception and NPR1 was proposed to be with NPR3/NPR4, resembling the multi-receptor of SA in diverse immune responses such as basal defense, systemic acquired resistance establishment, and effector-triggered immunity (ETI) [39]. GhFPF1 and GhNPR1 shared the similar expression trend to exogenous SA but the association between FPF1, SA, plant defense responses and flowering time requires further investigation. Little has been reported about the relationship between JA and the regulation of flowering time. Thus, it seems that JA may not have a direct effect upon the transition to flowering. Because of domestication, upland cotton has become a compact day-neutral and an annual row-crop from a lanky photoperiodic and perennial plant. Moreover some growth characteristics of cotton are different from other species as vegetative growth will proceed after the initiation of reproductive growth. Also flowering and fruit set are not synchronized but continue through the growing season. These competing sinks in upland cotton may give rise to flowering mechanism different from Arabidopsis.
Ectopic expression of GhFPF1 in Arabidopsis led to earlier flowering, and a decreased number of rosette and cauline leaves. When compared to the wild-type, transgenic plants had an increased expression of AtAP1, and suppressed expression of AtFLC. The MADS-domain transcription factor AP1, acts as a floral meristem identity gene that controls the onset of Arabidopsis flower development. AP1 expression is first observed throughout the emerging floral primordia, and is later confined to the outer whorls of floral buds, where it is also involved in the specification of sepals and petals [40], [41]. Another MADS-box transcription factor, FLC is a major repressor of flowering in Arabidopsis. It binds to the first intron of FT, and the promoter of SOC1, in each case inhibiting transcriptional activity. The FT protein interacts with FD to stimulate the activity of AP1. SOC1 can bind to the promoter of LFY to activate its transcription. The actions of AP1 and LFY promote the development of the inflorescence meristem, which leads to the production of flowers [42]–[44]. Ectopic expression of GhFPF1 in 18-day-old Arabidopsis caused slight increases in AtAP1 and AtSOC1expression levels. AtAP1 transcript was induced to a much higher level six days later, when the transgenic Arabidopsis had started bolting. AtSOC1expression remained at the same levels as before. AtFLC expression was suppressed at an extremely low level at both time points. This reveals that the promotion flowering of over-expression of GhFPF1 in Arabidopsis is possibly dependent on AtAP1 and AtFLC. Previous studies had confirmed that the FPF1 gene family took a positive effect on flowering time regulation [10], [13], [14]. Until now, little was known about the molecular mechanism of FPF1 in floral regulation pathways. It is expected that the role of FPF1 in the transition to flowering will be the focus of future research.
To grow and develop optimally, all organisms need to perceive and process information from their environment. As sessile organisms, plants need to sense and respond to external stimuli more than most organisms. Therefore, plants have to adapt their developmental pattern to the environmental changes to ensure survival and reproduction. Light influences every developmental transition from seed germination and seedling emergence to flowering. For shade-intolerant plants, such as Arabidopsis thaliana, a reduction in the red to far-red (R: FR) ratio of incoming radiation, which is caused by absorption of red light and reflection of far-red radiation by canopy leaves, signals the proximity of neighboring plants and triggers the shade avoidance syndrome (SAS) [45]. A common phenotype of the SAS is re-allocation of energy resources from storage organs to stems and petioles so that the plant outgrows its competitors. Other responses induced by reduction in R: FR ratio include increased leaf angle, accelerated leaf senescence and reduced deposition of flxed carbon to storage organs [46]. Shade avoidance in higher plants is regulated by the action of multiple phytochrome (phy) species that detect changes in the red to far-red ratio (R: FR) of incident light to initiate a redirection of growth and an acceleration of flowering [47]. Phytochrome B (phyB) is clearly the most important photoreceptor in the vast majority of responses to shade, in some cases redundantly with other members of its clade. In Arabidopsis, phyB mutants display a constitutive shade-avoiding phenotype that is characterized by long hypocotyls and petioles, reduced chlorophyll content, early flowering [48]. In the previous study, transgenic Arabidopsis over-expressing AtFPF1 was also deemed to share the similar phenotype to phyB mutant, and the authors deduced that the lack of phytochrome B leads to an enhanced responsiveness to GA [10]. Also they found that constitutively FPF1-expression plants contained slightly higher amounts of GA4 and GA20 than wild-type plants did. Here the transcripts of key genes in gibberellin biosynthesis GA3OX1 and GA20OX1 were analyzed and proved to show little difference between wild and transgenic plants. The expression of PHYB was checked to have only 44% amount of that in wild-type. In addition, the transgenic Arabidopsis presented typical shade avoidance responses such as early flowering, longer hypocotyls and petioles, reduced chlorophyll content under high R: FR ratio light conditions. Though functional complementation assay should be developed further, suppressed expression of PHYB in transgenic Arabidopsis of over-expressing of GhFPF1 may be the reason why shade avoidance syndrome (SAS) was induced presumably.
In summary, identification of FPF1 homologous genes in G. raimondii L. (D- genome) and G. arboreum L. (A- genome), also further comparing the sequences provided the groundwork necessary for understanding the distinctions and similarities between the sequences in the same genus of the different species. Strongly activated AP1 and suppressed FLC expression in transgenic plants suggested some points for understanding the mechanism of GhFPF1 to promote flowering. It seemed that transgenic Arabidopsis exhibited typical shade avoidance responses which might be caused by reduced expression of PHYB. Nevertheless, other proofs should be complemented in the future.
Supporting Information
Pair-wise alignment of FPF1 homologs DF10009151 ( G. raimondii L.) and Garb22630 ( G. arboreum L.).
(TIF)
Paraffin section analysis of flower bud differentiation in G. hirsutum L. TM-1. A, B, C and D corespond to four developmental stages in shoot apices when there were two cotyledons, one, two, and three true leaves flattened. Images of representative paraffin sections were shown here. Yellow arrows pointed out the shoot apical meristem (SAM) of every stage. White and red arrows indicated vegetative bud primordium (VP) and floral bud primordium (FP) respectively.
(TIF)
Cloning primers used for amplification of GhFPF1 and homologous genes.
(DOCX)
Quantitative PCR primers of target genes used in the study.
(DOCX)
Acknowledgments
We would like to thank doctoral candidate Xu DB for his help with the application of the laser confocal microscope. We wish to express our gratitude to teacher Suo TP for his guidance of the Scanning Electron Microscopy. Thanks for the seeds of CCRI 36 and TM-1 provided by teacher Wang L.
Funding Statement
This research was supported by the Earmarked Fund for China Agriculture Research System (CARS-18) and funded by the National High-tech Research and Development Projects of China (2011AA10A102). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lightfoot DJ, Malone KM, Timmis JN, Orford SJ (2008) Evidence for alternative splicing of MADS-box transcripts in developing cotton fibre cells. Mol Genet Genomics 279: 75–85. [DOI] [PubMed] [Google Scholar]
- 3. Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186. [Google Scholar]
- 4. Jung C, Muller AE (2009) Flowering time control and applications in plant breeding. Trends Plant Sci 14: 563–573. [DOI] [PubMed] [Google Scholar]
- 5. Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, et al. (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana, Nat Genet. 40: 1489–1492. [DOI] [PubMed] [Google Scholar]
- 6. Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289. [DOI] [PubMed] [Google Scholar]
- 7. Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci 97: 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kania T, Russenberger D, Peng S, Apel K, Melzer S (1997) FPF1 promotes flowering in Arabidopsis. Plant Cell 9: 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Melzer S, Kampmann G, Chandler J, Apel K (1999) FPF1 modulates the competence to flowering in Arabidopsis. Plant J 18: 395–405. [DOI] [PubMed] [Google Scholar]
- 12.Roland Borner, Grit Kampmann, Klaus Apel, Siegbert Melzer (2000) The FPF1 gene family and flowering time control in Arabidopsis. 11th International Conference on Arabidopsis Research: June 24–28, Madison, Wisconsin, USA. Abstract can be downloaded from http://arabidopsis.org/servlets/TairObject?type=publication&id=1546989.
- 13. Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, et al. (2004) Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol 135: 1502–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smykal P, Gleissner R, Corbesier L, Apel K, Melzer S (2004) Modulation of flowering responses in different Nicotiana varieties. Plant Mol Biol 55: 253–262. [DOI] [PubMed] [Google Scholar]
- 15. Lai D, Li H, Fan S, Song M, Pang C, et al. (2011) Generation of ESTs for flowering gene discovery and SSR marker development in upland cotton. PLoS ONE 6: e28676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang K, Wang Z, Li F, Ye W, Wang J, et al. (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44: 1098–1103. [DOI] [PubMed] [Google Scholar]
- 17. Attitalla IH (2011) Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J Biol Sci 14: 998–999. [PubMed] [Google Scholar]
- 18. Guan XY, Li QJ, Shan CM, Wang S, Mao YB, et al. (2008) The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol Plant 134: 174–182. [DOI] [PubMed] [Google Scholar]
- 19. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 20. Chen LG, Zhang LP, Yu DQ (2010) Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol Plant Microbe In 5: 558–565. [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y, Wang X, Cheng C, Gao QQ, Liu JY, et al. (2008) Molecular cloning and characterization of GhNPR1, a gene implicated in pathogen responses from cotton (Gossypium hirsutum L.). Biosci Rep 28: 7–14. [DOI] [PubMed] [Google Scholar]
- 22. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 23. Yang MW (2002) Study on Rapid Determination of Chlorophyll Content of Leaves. Chinese Journal of Spectroscopy Laboratory 19: 479–481. [Google Scholar]
- 24. Mukhtar MS, Nishimura MT, Dang J (2009) NPR1 in plant defense: it’s not over ‘til it’s turned over. Cell 137: 804–806. [DOI] [PubMed] [Google Scholar]
- 25. Spoel SH, Dong X (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351. [DOI] [PubMed] [Google Scholar]
- 26. Cronn RC, Small RL, Wendel JF (1999) Duplicated genes evolve independently after polyploid formation in cotton. Proc Natl Acad Sci 96: 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ren G, Chen Y, Dong H, Chen S (2000) Studies on flower bud differentiation and changes of endogenous hormones of Gossypium hirsutum. Acta Botanica Boreali-Occidentalia Sinica 20: 847–851. [Google Scholar]
- 29. Li J, Fan SL, Song MZ, Pang CY, Wei HL, et al. (2013) Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L. Plant Cell Rep. 32: 1675–1686. [DOI] [PubMed] [Google Scholar]
- 30. Tóth R, Kevei E, Hall A, Millar AJ, Nagy F, et al (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takada S, Goto K (2003) TERMINAL FLOWER 2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Havelange A, Lejeune P, Bernier G (2000) Sucrose/cytokinin interaction in Sinapis alba at floral induction: a shoot-to-root-to-shoot physiological loop. Physiol Plant 109: 343–350. [Google Scholar]
- 33. Bernier G, Corbesier L, Périlleux C (2002) The flowering process: on the track of controlling factors in Sinapis alba. Russ. J. Plant Physiol 49: 445–450. [Google Scholar]
- 34. Bernier G, Périlleux C (2005) A physiological overview of the genetics of flowering time control. Plant Biotechnol J 3: 3–16. [DOI] [PubMed] [Google Scholar]
- 35. Korves TM, Bergelson J (2003) A developmental response to pathogen infection in Arabidopsis. Plant Physiol 133: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martinez C, Pons E, Prats G, Leon J (2004) Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J 37: 209–217. [DOI] [PubMed] [Google Scholar]
- 37. Wada KC, Yamada M, Shiraya T, Takeno K (2010) Salicylic acid and the flowering gene FLOWERING LOCUS T homolog are involved in poor-nutrition stress-induced flowering of Pharbitis nil. J Plant Physiol 167: 447–452. [DOI] [PubMed] [Google Scholar]
- 38. Wang GF, Seabolt S, Hamdoun S, Ng G, Park J, et al. (2011) Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol 156: 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pajerowska-Mukhtar KM, Emerine DK, Mukhtar MS (2013) Tell me more: roles of NPRs in plant immunity. Trends in Plant Sci 18: 402–411. [DOI] [PubMed] [Google Scholar]
- 40. Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734. [DOI] [PubMed] [Google Scholar]
- 41. Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, et al. (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- 42. Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- 43. Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192. [DOI] [PubMed] [Google Scholar]
- 44. Searle I, He Y, Turck F, Vincent C, Fornara F, et al. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ballar CL, Sanchez RA, Scopel AL, Casal JJ, Ghersa CM (1987) Early detection of neighbour plants by phytochrome perception of spectral changes in reflected sunlight. Plant Cell Environ 10: 551–57. [Google Scholar]
- 46. Ballare CL (1999) Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci 4: 97–102. [DOI] [PubMed] [Google Scholar]
- 47. Franklin KA (2005) Phytochromes and Shade-avoidance Responses in Plants. Annals of Botany 96: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pair-wise alignment of FPF1 homologs DF10009151 ( G. raimondii L.) and Garb22630 ( G. arboreum L.).
(TIF)
Paraffin section analysis of flower bud differentiation in G. hirsutum L. TM-1. A, B, C and D corespond to four developmental stages in shoot apices when there were two cotyledons, one, two, and three true leaves flattened. Images of representative paraffin sections were shown here. Yellow arrows pointed out the shoot apical meristem (SAM) of every stage. White and red arrows indicated vegetative bud primordium (VP) and floral bud primordium (FP) respectively.
(TIF)
Cloning primers used for amplification of GhFPF1 and homologous genes.
(DOCX)
Quantitative PCR primers of target genes used in the study.
(DOCX)