Abstract
Defects in mtDNA replication are the principle cause of severe, heritable metabolic disorders classified as mitochondrial diseases. In vitro analysis of the biochemical mechanisms of mtDNA replication has proven to be a powerful tool for understanding the origins of mitochondrial disease. Mitochondrial single-stranded DNA binding protein (mtSSB) is an essential component of the mtDNA replication machinery. To facilitate ongoing biochemical studies, a recombinant source of mtSSB is needed to avoid the time and expense of human tissue culture. This chapter focuses on the sub-cloning, purification, and initial functional validation of recombinant human mitochondrial single-stranded DNA binding protein. The cDNA encoding the mature form of the human mtSSB protein was amplified from a HeLa cDNA library, and recombinant human mtSSB was over-produced in E. coli. A procedure was developed to rapidly purify milligram quantities of homogenous, nuclease-free mtSSB that avoids DNA-cellulose chromatography. We show that, similar to E. coli SSB, human mtSSB assembles into a tetramer and binds single-stranded oligonucleotides in a 4-to-1 protein:oligonucleotide molar ratio.
Keywords: mitochondria, single stranded DNA binding protein, DNA replication, EMSA, tetramer, mtDNA, protein purification
1. Introduction
Human mitochondria possess a circular, double-stranded DNA chromosome (16,569 bp) that is indispensible for the healthy growth of cells. Mitochondrial DNA (mtDNA) encodes 2 rRNAs, 22 tRNAs and 13 mRNAs essential for electron transport and oxidative phosphorylation. Mutation of mtDNA causes a wide spectrum of neuromuscular degenerative diseases affecting many different tissues, and defects in mtDNA replication are the principle cause of metabolic disorders classified as “mitochondrial diseases”. All of the factors required for replication and repair of the mitochondrial genome are known to be encoded by the nucleus, imported into mitochondria as pre-proteins, and proteolytically processed into their mature forms. Efforts to define the genetics and the biochemical mechanisms by which cells maintain the integrity of their mtDNA are essential to understanding the origins of mitochondrial diseases.
Mitochondrial single-stranded DNA binding protein (mtSSB) was discovered in an analysis of protein-mtDNA complexes derived from rat liver mitochondria that had been lysed with SDS, which revealed nucleoprotein fibrils within the single-stranded portions of both stable and expanding D-loops in replicative intermediates of rat liver mtDNA (1). Current models of mtDNA replication predict replication intermediates that contain large regions of single-stranded DNA, and the abundant presence of 16 kDa mtSSB in these nucleoprotein fibrils strongly suggests that the mtSSB protein is an essential component of the mtDNA replication machinery. Also, native mtSSBs isolated from Drosophila, Xenopus, and HeLa have been reported to stimulate DNA polymerase γ from these organisms, dependent on the substrate utilized in vitro (2–4). In addition, mtSSB has been shown in vitro to relieve replication stalling by Xenopus laevis pol γ within dT-rich template DNA sequences (5). More recently, human mtSSB has been shown to stimulate activity of the mtDNA helicase, and in vitro mtSSB is part of a minimal replisome complex along with the helicase, DNA polymerase γ and its accessory protein (6,7). Mutations in the nuclear genes encoding mtDNA replication components, specifically the DNA polymerase γ (POLG and POLG2) and the helicase (PEO1), have been clearly linked with mitochondrial disease (8,9). The gene for human mtSSB has been cloned, and the protein has been shown to be homologous to E. coli SSB (10). Although mtSSB has not yet been identified as a disease locus in humans, it is essential in yeast (11), and participation in mtDNA replication makes mtSSB an obvious candidate locus for mitochondrial disease in humans.
Existing procedures for the purification of native mitochondrial or E. coli SSBs consistently utilize single-stranded DNA-cellulose chromatography as a primary purification step (2,12,13). In our hands, recombinant mtSSB prepared from E. coli BL21(DE3) lysates by such protocols contained unacceptably high levels of nuclease activity that could not be resolved by additional chromatography on Affi-Gel Blue (14) or hydroxylapatite. Previous reports describing the purification of recombinant human mtSSB rely on animal cell culture as the source of protein and/or affinity tags to aid purification (6,15,16). Here, we describe over-expression in E. coli of the mature form of human mtSSB without an affinity tag and without the mitochondrial targeting sequence, and a procedure for rapid purification of milligram quantities of homogenous, nuclease-free mtSSB that does not use DNA-cellulose chromatography is presented.
2. Materials
Utilizing oligonucleotide primers designed from the published nucleotide sequence of the human gene (10), the cDNA for the human mtSSB was amplified from a HeLa cDNA library. Expression of the full-length human cDNA (aa 1–148, MW 17249) in E. coli (JM105) from a pQE9 expression vector (Qiagen) generated recombinant protein that was almost entirely insoluble (data not shown). Similarly, Li and Williams demonstrated that alteration of the N-terminal amino acid sequence of murine mtSSB, such as by insertion of an affinity-tag to aid protein purification or even by the simple retention of the mitochondrial targeting presequence, adversely affected DNA binding and/or tetramerization of recombinant murine mtSSB (15). Accordingly, we deleted the mitochondrial targeting sequence identified by Tiranti (10) and transferred the cDNA encoding the mature form (aa 17–148, MW 15316) into the pET21a expression vector (Novagen). The resulting plasmid, pET21aHmtSSB, was used to transform E. coli JM105(DE3) to ampicillin resistance. (see Notes 1 and 2). Ammonium sulfate was from Invitrogen, Carlsbad, CA. Ampicillin, IPTG, myo-inositol, CHES were obtained from Sigma. E. coli single-stranded DNA binding protein was purchased from United States Biochemical. Synthetic oligonucleotides were from Oligos Etc. (Wilsonville, OR).
2.1 Growth and Induction
2X YT bacterial growth media containing 10 g yeast extract, 16 g Bacto-tryptone, 5 g NaCl, sterilized at 121°C in a steam autoclave.
40% (w/v) glucose, sterilized by 0.45 μm filtration.
100 mg/mL ampicillin, sterilized by 0.45 μm filtration.
1 M IPTG is made by dissolving isopropyl-B-D-thiogalactopyranoside in water at 0.238 g/mL, sterilized by 0.45 μm filtration.
Refrigerated centrifuge with a rotor capable of holding 1 liter.
2.2 Cell Lysis
Buffer H contained 30 mM HEPES-KOH (pH 7.6), 1 mM dithiothreitol, 0.25 mM EDTA, 0.25% (w/v) myo-inositol, 0.01% (v/v) NP-40, 0.1 mM PMSF.
Branson Sonifier (model 450), equipped with a flat, 0.5 inch disrupter tip.
Sorvall RC-5B refrigerated centrifuge with an SS34 rotor, or the equivalent.
2.3 Affi-Gel Blue Chromatography
Affi-Gel Blue resin, 100 – 200 mesh (wet) was purchased from Bio-Rad.
Buffer H also containing 50 mM KCl.
Buffer H also containing 800 mM KCl.
Buffer H also containing 0.5 M KSCN.
2.4 Ammonium Sulfate Precipitation
Ammonium sulfate (Invitrogen, Carlsbad, CA).
Ice bath, beaker with stirbar.
Sorvall RC-5B refrigerated centrifuge with an SS34 rotor, or the equivalent.
Dialysis tubing with a 12000 – 14000 MW cutoff.
Dialysis buffer containing 25 mM HEPES-KOH (pH 7.6), 2 mM 2-mercaptoethanol, 0.1 mM EDTA, 10% glycerol, 50 mM KCl.
2.5 MonoS Chromatography
MonoS (HR 5/5) cation exchange column was purchased from GE Healthcare.
Akta FPLC chromatography system was purchased from GE Healthcare.
2.6 MonoQ Chromatography
MonoQ (HR 5/5) anion exchange column was purchased from GE Healthcare.
N-cyclohexyl-2-aminoethanesulfonic acid (CHES) was obtained from Sigma.
Buffer Q contained 50 mM CHES-KOH (pH 9.5), 2 mM 2-mercaptoethanol, 0.1 mM EDTA, 10% glycerol.
Akta FPLC chromatography system was purchased from GE Healthcare.
2.7 Dialysis and Storage
Storage Buffer was Buffer H also containing 0.25 M KCl and 25% glycerol.
Spectrophotometer and quartz cuvette, capable of measuring absorbance at 280 nm.
3. Methods
E. coli JM105(DE3) bearing a pET21a vector encoding the mature form of human mitochondrial SSB was treated with IPTG to induce gene expression and lysed by sonication. Cleared lysates prepared from 1 liter cultures were applied to Affi-Gel Blue resin, extensively washed, and mtSSB was eluted with 0.5 M KSCN before fractional precipitation with 10 – 35% (saturation) ammonium sulfate. Following dialysis, the protein fraction that did not bind MonoS was adjusted to pH 9.3 and applied to a MonoQ FPLC column. Homogenous mtSSB was eluted from MonoQ with a linear salt gradient. Typical yield from a 1 L culture was 4 – 6 mg homogenous mtSSB (see Table 1). Detailed procedures are listed below.
Table 1.
Purification of Recombinant Human Mitochondrial SSB
| Fraction | Volume (mL) | Protein (mg/mL) | Total Protein (mg) | |
|---|---|---|---|---|
| I. | Lysate | 19 | 13 | 250 |
| II. | Affi-Gel Blue | 52 | 0.40 | 21 |
| III. | Ammonium Sulfate | 2.1 | 6.7 | 14 |
| IV. | MonoS | 3.0 | 1.5 | 4.4 |
| V. | MonoQ | 5.8 | 0.74 | 4.3 |
| VI. | Dialysed | 3.6 | 1.2 | 4.3 |
The cDNA for mature mtSSB was expressed in E. coli, and the protein was purified to homogeneity as described in Subheading 3.1 to 3.7.
3.1 Growth and Induction
Grow an overnight culture (30 mL) of JM105(DE3) transformed with the pET21a HmtSSB plasmid at 37°C in 2xYT media supplemented with 0.1 mg/mL ampicillin.
Innoculate 1 L of 2xYT media supplemented with 0.1 mg/mL ampicillin and 0.1% (w/v) glucose with 5 mL of the overnight culture.
Incubate culture at 37°C on a rotary shaker. Periodically monitor growth by measuring light scattering at 600 nm.
When the culture density reaches A600 = 0.5, induce protein expression by adding 1 ml of 1M ITPG to make a final concentration of 1 mM IPTG.
Continue growth at 37°C for 3 hours. Harvest cells by centrifugation for 20 minutes at 4°C at 4700 g, such as in a Beckman J6-M centrifuge with a swinging bucket rotor at 4000 rpm.
Resuspend the cell pellet (~5 g) in 5 mL of Buffer H. Freeze the suspension of induced cells with liquid nitrogen, and store the sample at −80°C.
3.2 Cell Lysis
Thaw suspension of JM105(DE3)pET21aMmtSSB on wet ice. Dilute the suspension with 10 mL of Buffer H also containing 50 mM KCl to hasten thawing. (see Note 3).
Sonicate the sample for 45 seconds on a 50% duty cycle. Allow the sample to cool on ice for 2 minutes. Repeat the sonication and cooling steps two more times. (see Note 4).
Clarify the whole cell lysate by centrifugation for 10 minutes at 17000 g, such as in a Sorvall RC-5B refrigerated centrifuge with an SS34 rotor at 12000 rpm. Hold the supernatant on ice.
3.3 Affi-Gel Blue Chromatography
Pre-equilibrate the Affi-Gel Blue resin in Buffer H also containing 50 mM KCl by gravity settling in a beaker. (see Note 5).
Transfer 14 mL of pre-equilibrated Affi-Gel Blue to a 1.5 cm I.D. chromatography column. Pack and wash the settled resin bed with 5 column volumes of Buffer H containing 50 mM KCl.
Apply the cleared lysate to the Affi-Gel Blue column by gravity flow at a rate not exceeding 0.5 mL per minute.
Flush away unbound material in the lysate by washing the resin with 5 column volumes of Buffer H containing 50 mM KCl at a flow rate of 1.0 – 1.5 mL per minute. (see Note 6).
Elute weakly bound contaminating proteins by washing the resin with 5 column volumes of Buffer H containing 800 mM KCl at a flow rate of 1.0 – 1.5 mL per minute.
Elute recombinant human mtSSB with 5 column volumes of Buffer H containing 0.5 M KSCN at a flow rate of 1.0 – 1.5 mL per minute.
3.4 Ammonium Sulfate Precipitation
Transfer the AffiGel Blue eluate to a glass beaker on an ice bath.
Bring the solution to 10% (saturated) ammonium sulfate by adding solid ammonium sulfate at a ratio of 0.0549 g ammonium sulfate per mL of solution. Ammonium sulfate should be added slowly enough to prevent the accumulation of crystals in the beaker. (see Note 7). After all the ammonium sulfate has dissolved, continue stirring on ice for an additional 15 minutes.
Transfer the slurry to a centrifuge tube, and clarify by centrifugation for 15 minutes at 17000 g, such as in a Sorvall RC-5B refrigerated centrifuge with an SS34 rotor at 12000 rpm.
Transfer the supernatant to a fresh beaker and measure the new volume. Bring the supernatant to 35% (saturated) ammonium sulfate by slowly adding solid ammonium sulfate at a ratio of 0.1489 g ammonium sulfate per mL of solution. After all the ammonium sulfate has dissolved, continue stirring on ice for an additional 15 minutes.
Collect the precipitated protein by centrifugation for 15 minutes at 17000 g, as above.
Resuspend the well-drained protein precipitate in 2.0 mL of cold dialysis buffer, and dialyse for 1 – 2 hours, until the conductivity of the sample approaches that of the dialysis buffer.
Clarify the dialysate by centrifugation for 10 minutes at 17000 g to remove any undissolved material. Hold the supernatant on ice.
3.5 MonoS chromatography
Attach the MonoS (HR 5/5) column to an Akta FPLC chromatography system. (see Note 8).
Equilibrate the MonoS column with at least 5 mL of dialysis buffer, to ensure complete equilibration of the column as judged by stable UV and conductivity baselines for the column effluent.
Inject the clarified dialysate onto the MonoS column, followed by 5 mL of dialysis buffer. Collect the unbound protein fraction that passes directly through the MonoS column. (see Note 9).
3.6 MonoQ chromatography
Measure the volume of the material passing through the MonoS column. Add 0.2 volumes of 0.5 M CHES-KOH (pH 9.5), which will raise the pH from 7.6 to approximately 9.3.
Attach the MonoQ (HR 5/5) column to an Akta FPLC chromatography system. (see Note 8).
Equilibrate the MonoQ column with at least 5 mL of Buffer Q also containing 50 mM KCl. Ensure complete equilibration of the column as judged by stable UV and conductivity baselines for the column effluent.
Apply the near homogenous mtSSB (Fraction IV) to the MonoQ column. Flush unbound material from the column with 4 mL of equilibration buffer.
Elute the column with a 25 mL linear gradient of KCl (50 mM – 500 mM) in Buffer Q.
Identify fractions containing mtSSB as the major peak on the UV-profile. Homogenous mtSSB elutes at approximately 0.24 M KCl.
3.7 Dialysis and Storage
Dialyse the pooled mtSSB peak extensively against Storage Buffer at 4°C.
Determine the protein concentration of homogenous mtSSB with a uv-spectrophotometer, utilizing an extinction coefficient at 280 nm of 76240 M−1cm−1 for tetramers (19060 M−1cm−1 for monomers), as calculated from the primary amino acid sequence (13,17).
Store the dialyzed protein preparation in tightly sealed vials at −20°C.
4 Validation of recombinant mtSSB
Validation experiments were performed both to verify the purity of recombinant, mature human mtSSB as prepared by this method, as well as to confirm the ability of the purified protein to form homotetramers and bind to single-stranded DNA.
4.1 Analysis of Purity
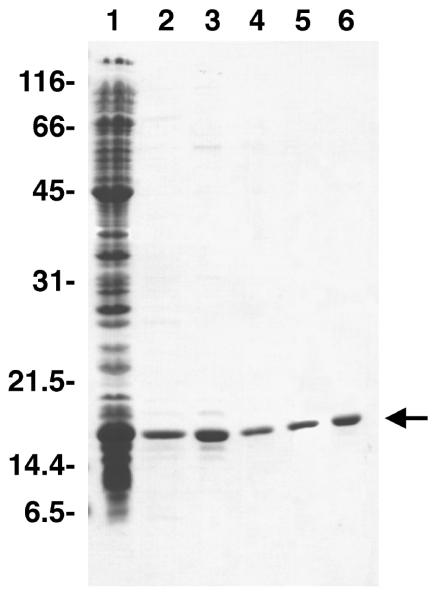
Established procedures for the purification of prokaryotic and mtSSB's often rely upon single-stranded DNA-cellulose chromatography as a terminal purification step. To avoid the possibility of DNA contaminating the mtSSB preparation, as well as to minimize the chances of enriching for other DNA metabolizing enzymes due to their intrinsic affinity to DNA, we sought to develop a rapid protocol for purifying recombinant mtSSB with a high yield that does not utilize DNA affinity chromatography. Samples taken throughout the purification were analyzed by SDS-PAGE (see Fig. 1). The high specificity of Affi-Gel Blue rendered the mtSSB >90% pure after the first chromatographic step. KSCN was removed by ammonium sulfate precipitation, and passage through a MonoS column removed many higher MW contaminants, including a contaminating nuclease activity. Although E. coli SSB and mtSSBs do not readily bind MonoS or MonoQ FPLC columns at neutral pH, MonoQ binds human mtSSB with high capacity at pH 9.5, permitting purification to homogeneity without resorting to DNA-cellulose chromatography.
Fig. 1.
Purification profile of recombinant, human, mitochondrial single-stranded DNA binding protein. Proteins were resolved by 12.5% SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1, cleared lysate; lane 2, Affi-Gel Blue; lane 3, ammonium sulfate; lane 4, MonoS; lane 5, MonoQ; lane 6, dialysis. The positions of molecular weight markers (kDa) are shown, and mtSSB is indicated by the arrow.
4.2 Tetramerization Measured by Analytical Ultracentrifugation
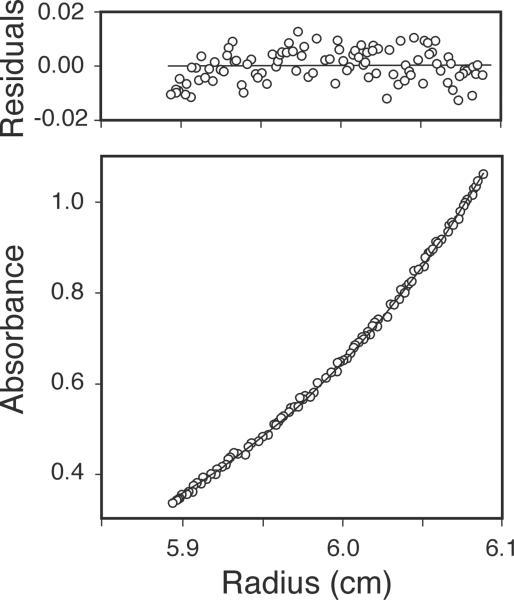
Equilibrium sedimentation analysis was performed to determine the native conformation of purified, recombinant, human mtSSB. In preparation for analytical ultracentrifugation, glycerol was removed from a sample of mtSSB (Fraction V) by dialysis against a buffer containing 30 mM HEPES•KOH (pH 7.6), 2 mM 2-mercaptoethanol, 0.25 mM EDTA, 0.25 M KCl. Protein concentration was determined spectrophotometrically, as in in Subheading 3.7. Samples (100 μL) were adjusted by dilution with dialysis buffer to protein concentrations of 32.1, 16.0, and 10.7 μM mtSSB (monomers), and subjected to equilibrium sedimentation in an Optima XL-A analytical ultracentrifuge (Beckman Instruments) at 11,000 rpm at 4°C prior to obtaining absorbance profiles at 280 nm (see Fig. 2). Profiles were fit with Optima XL-A data analysis software, assuming a partial specific volume of 0.7261 cm3/g and a solvent density of 1.0128 g/cm3. The random distribution of residual absorbance values indicated that all three profiles fit very well to a model of a single species (see Fig. 2) with an average molecular weight of 62,600 ± 1760 Da. Although attempts to model monomer-dimer or monomer-tetramer equilibria were confounded by the inability to detect SSB monomers at these protein concentrations, the subunit dissociation constant is estimated to be less than 10−8 M. Because the recombinant protein has a predicted molecular weight of 15,316 Da, the measured value indicates that recombinant human mtSSB exists as a tetramer in solution, as observed previously for native and recombinant forms of mtSSB from Xenopus laevis (3,18), Drosophila melanogaster (2), and humans (13).
Fig. 2.
Equilibrium analytical ultracentrifugation data. Recombinant human mtSSB (49 μg, fractionV) was subjected to equilibrium sedimentation, and a radial scan of the absorbance profile at 280 nm (circles) was fit to a model of a single ideal species (solid line). The random distribution of small residual values indicates only minor deviation from the ideal model.
4.3 DNA-binding Confirmed by Electrophoretic Mobility Shift Assay
The ability of mtSSB tetramers to bind single-stranded DNA was confirmed by electrophoretic mobility shift assay, as described previously (19). DNA-binding by E. coli SSB, which also exists as a tetramer in solution (20), was assessed as a positive control in side-by-side reactions. Various concentrations of human mtSSB or E. coli SSB were combined in vitro with radiolabeled, single-stranded 34mer oligonucleotide, and mixtures were resolved by native PAGE (see Fig. 3, upper panel). This titration experiment indicates that 40 fmol of oligonucleotide are saturated by approximately four equivalents of either E. coli SSB monomers or mtSSB monomers, confirming the similar DNA-binding strength of each tetrameric species in vitro (see Fig. 3, lower panel).
Fig. 3.
DNA binding properties of mtSSB and E. coli SSB were assessed by electrophoretic mobility shift assay. Binding mixtures (20 μL) contained 15 mM HEPES-KOH (pH 7.6), 0.5 mM EDTA, 1 mM dithiothreitol, 50 μg/mL acetylated BSA, 40 fmol of 32P-labeled 34mer, and the indicated quantities of E. coli SSB or human mtSSB. Bound and unbound oligonucleotides were resolved on 10% native polyacrylamide gels and exposed to a phosphor storage screen. Radioactivity was imaged on a Typhoon 9400 phosphorimager and quantified with NIH Image software.
5. Notes
The E. coli strain JM105 harbors a mutated form of exonuclease I (sbcB15), which eliminates one source of minor exonuclease contamination in some preparations of SSB. Accordingly, JM105(DE3) is the host of choice for production of recombinant mtSSB.
JM105 was modified to contain the gene for T7 RNA polymerase with the Lambda DE3 Lysogenization kit, as described by the manufacturer (Novagen).
All steps in the purification should be performed on ice or at 4°C.
Good lysis is achieved with a Branson Sonifier (model 450) equipped with a flat, 0.5 inch disrupter tip. Operation at an output control setting of 5 delivers approximately 90 – 100 watts of power to the sample. Care should be taken to fully immerse the probe, to avoid foaming that can occur due to the NP-40 in the lysis buffer.
New Affi-Gel Blue resin can leach unbound dye into your biological sample. Prior to first use, new resin should be washed in a solution consisting of 0.1 M acetic acid, 1.4 M NaCl, and 40% 2-propanol until no more dye leaches from the resin, followed by exhaustive washing in water. Then the washed resin can be transferred to the equilibration buffer.
Care should be taken to allow all of one buffer to enter the resin bed before changing to a new solution. This will reduce mixing of the two buffers and ensure a more abrupt transition between washing and elution conditions.
In advance, grind the ammonium sulfate into a powder with a mortar and pestle. Finely ground ammonium sulfate dissolves very readily, which prevents the accumulation of ammonium sulfate crystals in the bottom of the beaker.
All FPLC operations were performed at 4°C, with a flow rate of 1 mL per minute and an upper pressure limit of 5 MPa.
Human mtSSB does not bind to MonoS under these buffer conditions. However, control experiments clearly indicate complete retention of contaminating 3'to 5' exonuclease activity among the proteins that bind the MonoS column (data not shown).
Acknowledgments
The authors would like to thank Dr. Harvey Sage of the Department of Biochemistry at Duke University Medical Center for performing the analytical ultracentrifugation, as well as Drs. Leroy Worth (NIEHS) and Sherine Chan (NIEHS) for thoughtful comments on the manuscript. This work was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences.
References
- 1.Van Tuyle GC, Pavco PA. The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J Cell Biol. 1985;100:251–257. doi: 10.1083/jcb.100.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thommes P, Farr CL, Marton RF, Kaguni LS, Cotterill S. Mitochondrial single-stranded DNA-binding protein from Drosophila embryos. Physical and biochemical characterization. J Biol Chem. 1995;270:21137–21143. doi: 10.1074/jbc.270.36.21137. [DOI] [PubMed] [Google Scholar]
- 3.Mikhailov VS, Bogenhagen DF. Effects of Xenopus laevis mitochondrial single-stranded DNA-binding protein on primer-template binding and 3′-->5′ exonuclease activity of DNA polymerase γ. J Biol Chem. 1996;271:18939–18946. doi: 10.1074/jbc.271.31.18939. [DOI] [PubMed] [Google Scholar]
- 4.Genuario R, Wong TW. Stimulation of DNA polymerase γ by a mitochondrial single-strand DNA binding protein. Cell Mol Biol Res. 1993;39:625–634. [PubMed] [Google Scholar]
- 5.Mikhailov VS, Bogenhagen DF. Termination within oligo(dT) tracts in template DNA by DNA polymerase γ occurs with formation of a DNA triplex structure and is relieved by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 1996;271:30774–30780. doi: 10.1074/jbc.271.48.30774. [DOI] [PubMed] [Google Scholar]
- 6.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 7.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimauro S, Davidzon G. Mitochondrial DNA and disease. Ann Med. 2005;37:222–232. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- 9.Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase γ. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 10.Tiranti V, Rocchi M, DiDonato S, Zeviani M. Cloning of human and rat cDNAs encoding the mitochondrial single-stranded DNA-binding protein (SSB) Gene. 1993;126:219–225. doi: 10.1016/0378-1119(93)90370-i. [DOI] [PubMed] [Google Scholar]
- 11.Van Dyck E, Foury F, Stillman B, Brill SJ. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 13.Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur J Biochem. 1994;221:435–443. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- 14.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 15.Li K, Williams RS. Tetramerization and single-stranded DNA binding properties of native and mutated forms of murine mitochondrial single-stranded DNA-binding proteins. J Biol Chem. 1997;272:8686–8694. doi: 10.1074/jbc.272.13.8686. [DOI] [PubMed] [Google Scholar]
- 16.Li K, Williams RS. Expression, purification, and in vitro assays of mitochondrial single-stranded DNA-binding protein. Methods Mol Biol. 2002;197:295–302. doi: 10.1385/1-59259-284-8:295. [DOI] [PubMed] [Google Scholar]
- 17.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 18.Mahoungou C, Ghrir R, Lecaer JP, Mignotte B, Barat-Gueride M. The amino-terminal sequence of the Xenopus laevis mitochondrial SSB is homologous to that of the Escherichia coli protein. FEBS Lett. 1988;235:267–270. doi: 10.1016/0014-5793(88)81276-6. [DOI] [PubMed] [Google Scholar]
- 19.Chan SSL, Longley MJ, Copeland WC. Modulation of the W748S mutation in DNA polymerase γ by the E1143G polymorphism in mitochondrial disorders. Hum Mol Genet. 2006;15:3473–3483. doi: 10.1093/hmg/ddl424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA- binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]