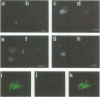
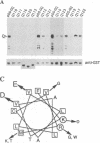
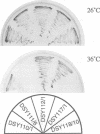
Abstract
NUF1/SPC110, encoding a nuclear filament-related protein which is a component of the yeast spindle pole body (SPB), has been identified in a screen designed to isolate genes encoding targets of yeast calmodulin. Spc110p interacts with calmodulin by two different criteria and the calmodulin interacting region has been localized within the C-terminus of the protein. Point mutations between residues 898 and 917 further define the calmodulin binding site within this region. Mutations in this domain which abolish calmodulin binding in vitro prevent Spc110p function in vivo, demonstrating that calmodulin binding by Spc110p has important functional consequences. In keeping with a role for calmodulin in Spc110p function, we show that calmodulin localizes to the yeast SPB when cells are prepared under appropriate conditions. Non-functional mutant Spc110 proteins which cannot bind calmodulin are present at lowered steady-state levels in the cell; when their level is increased by elevated gene dosage, partial recovery of Spc110p function is seen. Overexpression of calmodulin suppresses the defect(s) associated with the mutant Spc110 proteins, supporting the notion that Spc110p stability is a consequence of its ability to bind calmodulin and pointing to a direct role for calmodulin in Spc110p function.
Full text
PDF

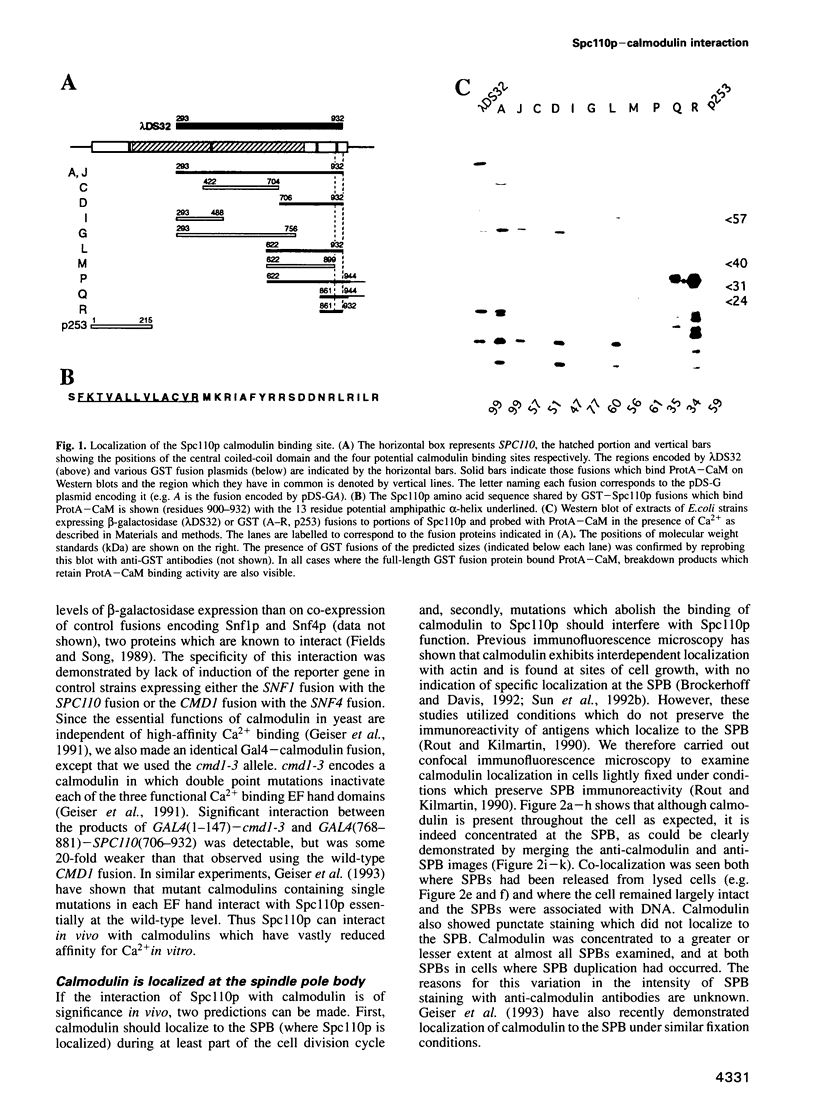
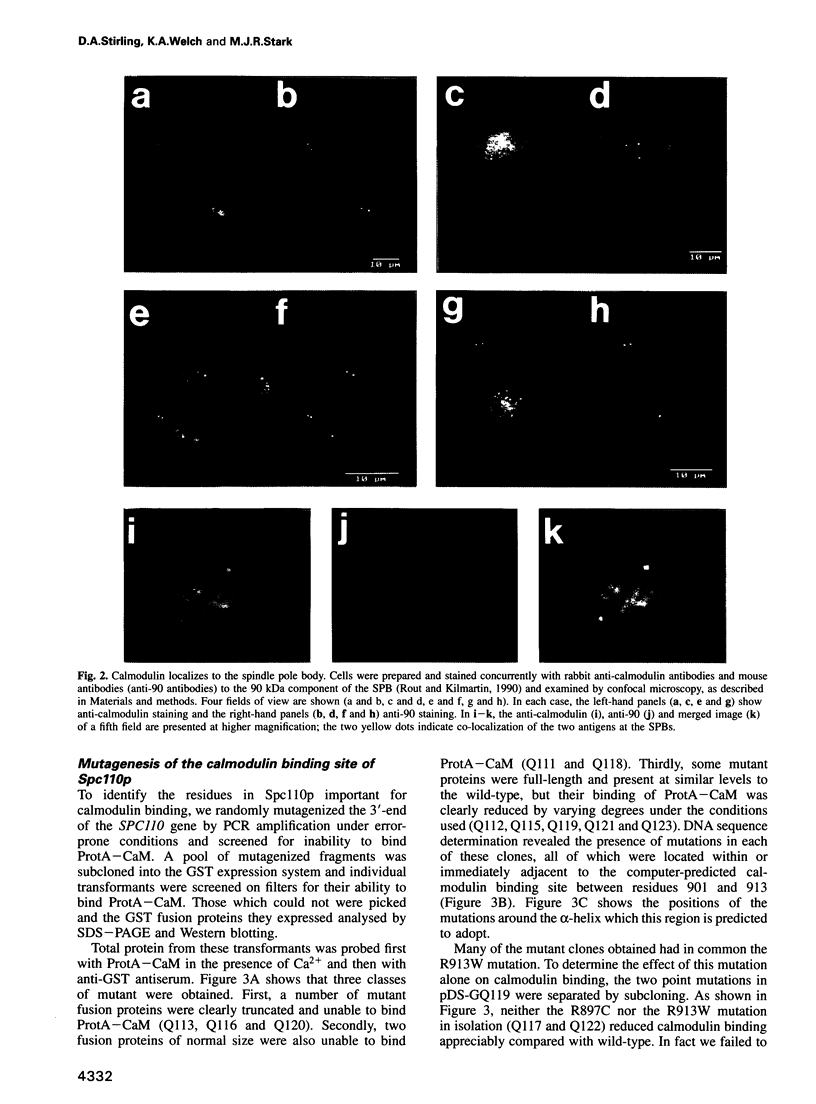
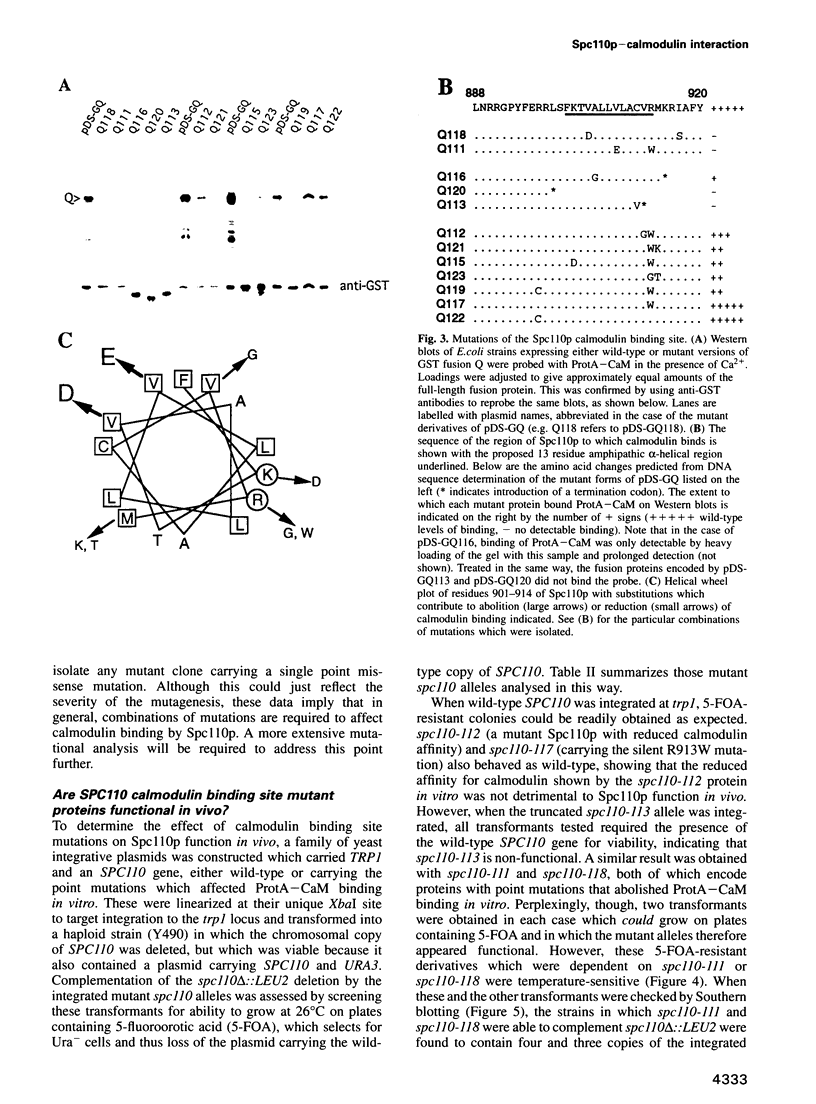
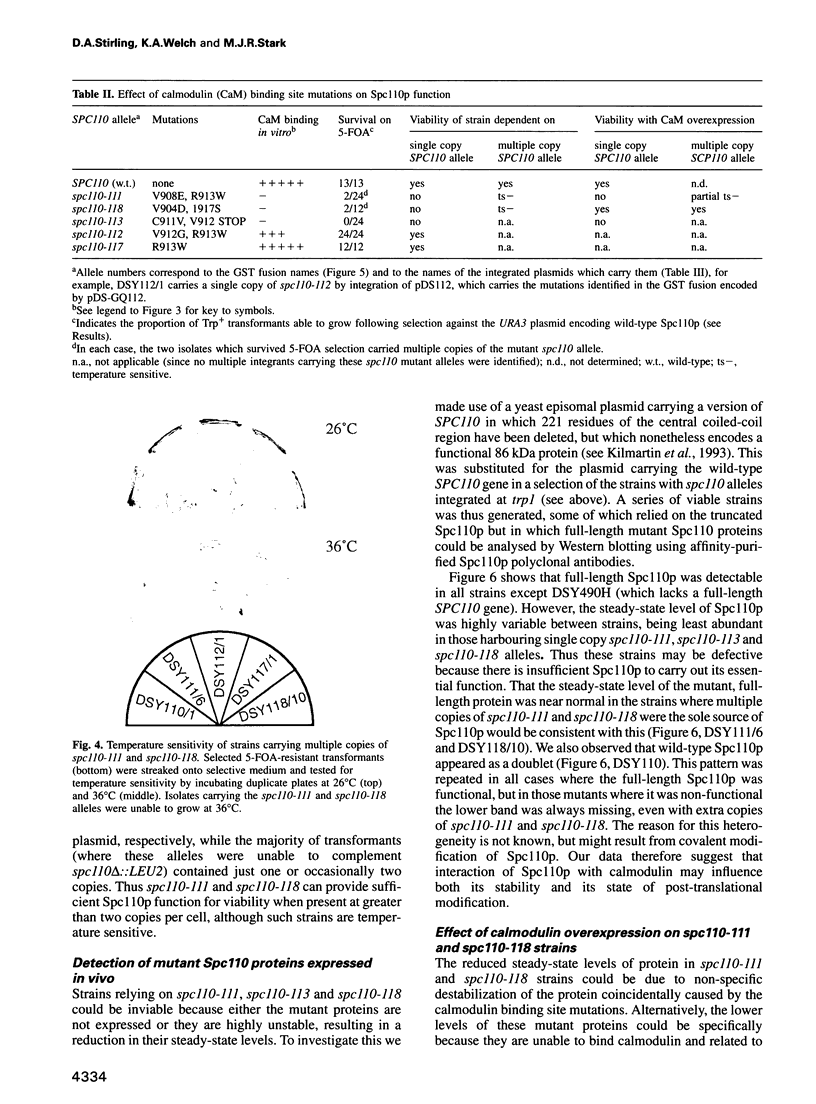
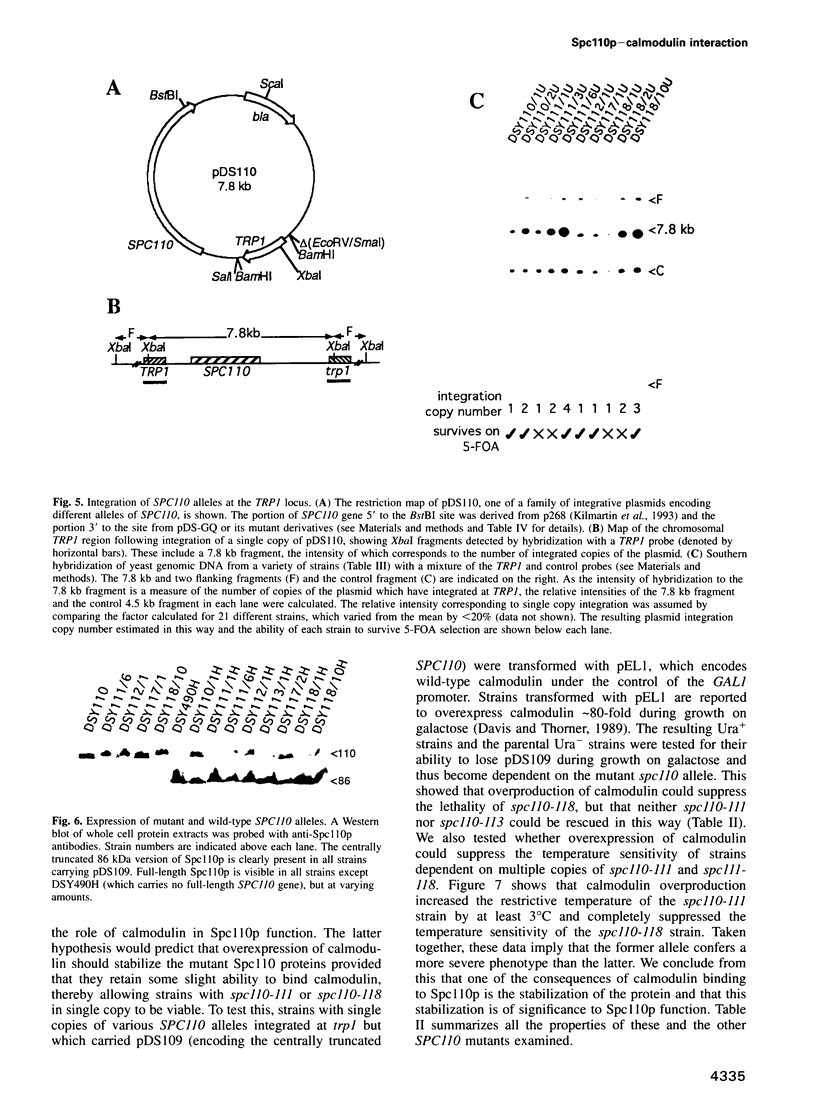







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander K. A., Wakim B. T., Doyle G. S., Walsh K. A., Storm D. R. Identification and characterization of the calmodulin-binding domain of neuromodulin, a neurospecific calmodulin-binding protein. J Biol Chem. 1988 Jun 5;263(16):7544–7549. [PubMed] [Google Scholar]
- Berben G., Dumont J., Gilliquet V., Bolle P. A., Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991 Jul;7(5):475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Brockerhoff S. E., Davis T. N. Calmodulin concentrates at regions of cell growth in Saccharomyces cerevisiae. J Cell Biol. 1992 Aug;118(3):619–629. doi: 10.1083/jcb.118.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E. R., Au D., Alexander K. A., Nicolson T. A., Storm D. R. Characterization of the calmodulin binding domain of neuromodulin. Functional significance of serine 41 and phenylalanine 42. J Biol Chem. 1991 Jan 5;266(1):207–213. [PubMed] [Google Scholar]
- Cheney R. E., Mooseker M. S. Unconventional myosins. Curr Opin Cell Biol. 1992 Feb;4(1):27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M. S., Kunisawa R., Kaim D., Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. N. A temperature-sensitive calmodulin mutant loses viability during mitosis. J Cell Biol. 1992 Aug;118(3):607–617. doi: 10.1083/jcb.118.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. N., Thorner J. Vertebrate and yeast calmodulin, despite significant sequence divergence, are functionally interchangeable. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7909–7913. doi: 10.1073/pnas.86.20.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. N., Urdea M. S., Masiarz F. R., Thorner J. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell. 1986 Nov 7;47(3):423–431. doi: 10.1016/0092-8674(86)90599-4. [DOI] [PubMed] [Google Scholar]
- DeGrado W. F., Erickson-Viitanen S., Wolfe H. R., Jr, O'Neil K. T. Predicted calmodulin-binding sequence in the gamma subunit of phosphorylase b kinase. Proteins. 1987;2(1):20–33. doi: 10.1002/prot.340020104. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P. L., Yeh S. H., Yang Y., Kilburn A. E., Lee W. H., Elledge S. J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993 Apr;7(4):555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Erickson-Viitanen S., DeGrado W. F. Recognition and characterization of calmodulin-binding sequences in peptides and proteins. Methods Enzymol. 1987;139:455–478. doi: 10.1016/0076-6879(87)39106-2. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fitzsimons D. P., Herring B. P., Stull J. T., Gallagher P. J. Identification of basic residues involved in activation and calmodulin binding of rabbit smooth muscle myosin light chain kinase. J Biol Chem. 1992 Nov 25;267(33):23903–23909. [PMC free article] [PubMed] [Google Scholar]
- Geiser J. R., Sundberg H. A., Chang B. H., Muller E. G., Davis T. N. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser J. R., van Tuinen D., Brockerhoff S. E., Neff M. M., Davis T. N. Can calmodulin function without binding calcium? Cell. 1991 Jun 14;65(6):949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992 Mar 25;20(6):1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna R., Anderson W. B. Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem Biophys Res Commun. 1982 Jan 29;104(2):830–836. doi: 10.1016/0006-291x(82)90712-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Haiech J., Kilhoffer M. C., Lukas T. J., Craig T. A., Roberts D. M., Watterson D. M. Restoration of the calcium binding activity of mutant calmodulins toward normal by the presence of a calmodulin binding structure. J Biol Chem. 1991 Feb 25;266(6):3427–3431. [PubMed] [Google Scholar]
- Hakes D. J., Dixon J. E. New vectors for high level expression of recombinant proteins in bacteria. Anal Biochem. 1992 May 1;202(2):293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992 May 1;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Dyos S. L., Kershaw D., Finch J. T. A spacer protein in the Saccharomyces cerevisiae spindle poly body whose transcript is cell cycle-regulated. J Cell Biol. 1993 Dec;123(5):1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohse K. P., Heilmeyer L. M., Jr The effects of Mg2+ on the Ca2+-binding properties and Ca2+-induced tyrosine-fluorescence changes of calmodulin isolated from rabbit skeletal muscle. Eur J Biochem. 1981 Jul;117(3):507–513. doi: 10.1111/j.1432-1033.1981.tb06366.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ishii S., Tokai M., Tsutsumi H., Ohki O., Akada R., Tanaka K., Tsuchiya E., Fukui S., Miyakawa T. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol Gen Genet. 1991 May;227(1):52–59. doi: 10.1007/BF00260706. [DOI] [PubMed] [Google Scholar]
- Luan Y., Matsuura I., Yazawa M., Nakamura T., Yagi K. Yeast calmodulin: structural and functional differences compared with vertebrate calmodulin. J Biochem. 1987 Dec;102(6):1531–1537. doi: 10.1093/oxfordjournals.jbchem.a122201. [DOI] [PubMed] [Google Scholar]
- Mirzayan C., Copeland C. S., Snyder M. The NUF1 gene encodes an essential coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J Cell Biol. 1992 Mar;116(6):1319–1332. doi: 10.1083/jcb.116.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T., Nakano A., Nomura K., Sekimizu K., Natori S. Purification, gene cloning, and gene disruption of the transcription elongation factor S-II in Saccharomyces cerevisiae. J Biol Chem. 1992 Jul 5;267(19):13200–13204. [PubMed] [Google Scholar]
- Ohya Y., Anraku Y. Functional expression of chicken calmodulin in yeast. Biochem Biophys Res Commun. 1989 Jan 31;158(2):541–547. doi: 10.1016/s0006-291x(89)80083-x. [DOI] [PubMed] [Google Scholar]
- Ohya Y., Botstein D. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science. 1994 Feb 18;263(5149):963–966. doi: 10.1126/science.8310294. [DOI] [PubMed] [Google Scholar]
- Ohya Y., Kawasaki H., Suzuki K., Londesborough J., Anraku Y. Two yeast genes encoding calmodulin-dependent protein kinases. Isolation, sequencing and bacterial expressions of CMK1 and CMK2. J Biol Chem. 1991 Jul 5;266(19):12784–12794. [PubMed] [Google Scholar]
- Olwin B. B., Storm D. R. Calcium binding to complexes of calmodulin and calmodulin binding proteins. Biochemistry. 1985 Dec 31;24(27):8081–8086. doi: 10.1021/bi00348a037. [DOI] [PubMed] [Google Scholar]
- Pausch M. H., Kaim D., Kunisawa R., Admon A., Thorner J. Multiple Ca2+/calmodulin-dependent protein kinase genes in a unicellular eukaryote. EMBO J. 1991 Jun;10(6):1511–1522. doi: 10.1002/j.1460-2075.1991.tb07671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R., Adams A. E., Drubin D. G., Haarer B. K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rasmussen C. D., Means R. L., Lu K. P., May G. S., Means A. R. Characterization and expression of the unique calmodulin gene of Aspergillus nidulans. J Biol Chem. 1990 Aug 15;265(23):13767–13775. [PubMed] [Google Scholar]
- Rout M. P., Kilmartin J. V. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990 Nov;111(5 Pt 1):1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman H., Hanson P. I., Meyer T. Decoding calcium signals by multifunctional CaM kinase. Cell Calcium. 1992 Jun-Jul;13(6-7):401–411. doi: 10.1016/0143-4160(92)90053-u. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sneddon A. A., Cohen P. T., Stark M. J. Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J. 1990 Dec;9(13):4339–4346. doi: 10.1002/j.1460-2075.1990.tb07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starovasnik M. A., Davis T. N., Klevit R. E. Similarities and differences between yeast and vertebrate calmodulin: an examination of the calcium-binding and structural properties of calmodulin from the yeast Saccharomyces cerevisiae. Biochemistry. 1993 Apr 6;32(13):3261–3270. doi: 10.1021/bi00064a008. [DOI] [PubMed] [Google Scholar]
- Stirling D. A., Petrie A., Pulford D. J., Paterson D. T., Stark M. J. Protein A-calmodulin fusions: a novel approach for investigating calmodulin function in yeast. Mol Microbiol. 1992 Mar;6(6):703–713. doi: 10.1111/j.1365-2958.1992.tb01519.x. [DOI] [PubMed] [Google Scholar]
- Sun G. H., Hirata A., Ohya Y., Anraku Y. Mutations in yeast calmodulin cause defects in spindle pole body functions and nuclear integrity. J Cell Biol. 1992 Dec;119(6):1625–1639. doi: 10.1083/jcb.119.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet S. C., Rogers C. M., Welsh M. J. Calmodulin is associated with microtubules forming in PTK1 cells upon release from nocodazole treatment. Cell Motil Cytoskeleton. 1989;12(2):113–122. doi: 10.1002/cm.970120206. [DOI] [PubMed] [Google Scholar]
- Takeda T., Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török K., Whitaker M. Taking a long, hard look at calmodulin's warm embrace. Bioessays. 1994 Apr;16(4):221–224. doi: 10.1002/bies.950160402. [DOI] [PubMed] [Google Scholar]
- Welsh M. J., Dedman J. R., Brinkley B. R., Means A. R. Tubulin and calmodulin. Effects of microtubule and microfilament inhibitors on localization in the mitotic apparatus. J Cell Biol. 1979 Jun;81(3):624–634. doi: 10.1083/jcb.81.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., Araki Y., Hales S. J., Tanaka M., Ikebe M. Boundary of the autoinhibitory region of smooth muscle myosin light-chain kinase. Biochemistry. 1993 Nov 16;32(45):12054–12061. doi: 10.1021/bi00096a016. [DOI] [PubMed] [Google Scholar]
- Zueco J., Boyd A. Protein A fusion proteins as molecular weight markers for use in immunoblotting. Anal Biochem. 1992 Dec;207(2):348–349. doi: 10.1016/0003-2697(92)90023-z. [DOI] [PubMed] [Google Scholar]