Highlights
-
•
We tested the proposal that the Peptidyl Transferase Center (PTC) originated from proto-tRNAs.
-
•
Proto-tRNA ancestors were reconstructed.
-
•
Their sequences were compared to the sequences of the PTC from several organisms.
-
•
We conclude that fusions of proto-tRNAs may have given rise to the PTC.
Keywords: Ribosome, Peptidyl Transferase Center, Origin, tRNA
Abstract
We tested the hypothesis of Tamura (2011) [3] that molecules of tRNA gave origin to ribosomes, particularly to the Peptidyl Transferase Center (PTC) of the 23S ribosomal RNA. We reconstructed the ancestral sequences from all types of tRNA and compared them in their sequences with the current PTC of 23S ribosomal RNA from different organisms. We built an ancestral sequence of proto-tRNAs that showed a remarkable overall identity of 50.53% with the catalytic site of PTC. We conclude that the Peptidyl Transferase Center was indeed originated by the fusion of ancestral sequences of proto-tRNA.
1. Introduction
The transfer of information contained in the nucleic acids to proteins is considered to be a fundamental process in protein synthesis. The ancient origin of this system pertains to an era prior to the Last Universal Common Ancestor (LUCA) and it has evolved since then to modern organisms [1–3]. There is a panoply of molecules involved in this process, but the RNAs have a prominent role as the first informational molecules and later, part of this function came to be exercised, in most organisms by DNA [4–8]. Among the RNA molecules that participate in the translation process, the 23S ribosomal RNA stands up because the Peptidyl Transferase Center (PTC) is constituted around its domain V, with which the CCA ends of two tRNAs interact, and the peptide bond between amino acids occurs [9]. Despite the immense complexity of the large subunit rRNA, studies indicate that PTC would be the first portion of this molecule to arise, and later on other regions were gradually incorporated [2,10,11]. Currently, there are several crystallographic structures of the PTC [12–16]; however, its origin is still a matter of discussion in the scientific community. Studies reveal that the PTC has a symmetrical structure comprising approximately 180 nucleotides that are also found in other RNAs involved in protein synthesis such as the tRNAs [10–17]. Molecular structure models suggest that the catalytic portion of the 23S rRNA entities of the symmetrical region possesses the common stem-elbow-stem (SES) structural motif. This structure probably evolved by the dimerization of two SES motifs that stabilized and formed the ribozymic pocket, responsible for the peptidyl activity [3]. Davidovich et al. [10] showed that the dimerization process is a common phenomenon in RNA molecules that have the structural motifs proposed as proto-ribosome. Zenkin [18] proposed that the ribosomal activity was derived from RNAs with helicase activity. In this model it was proposed that peptidyl transferase activity had its origins in generating a unidirectional movement that emerged by interactions between tRNAs and mRNAs. Tamura [3] proposed a model based on PTC topology, where this region had been originated by the fusion of proto-tRNA molecules that could explain the topological similarity between them. The tRNA molecules, as well as the ribosomes, must have appeared very early in biological systems and, to present a functional relationship they must have had a common origin or one of them could have originated from the other. In this paper, we reconstructed ancestral sequences for tRNAs to test the model proposed by Tamura [3], where proto-tRNAs gave origin directly to ribosomes, particularly the 23S ribosomal RNA.
2. Materials and methods
2.1. Ancestral sequence reconstruction
The sequences of tRNA molecules were downloaded from the database http://trnadb.bioinf.uni-leipzig.de, accounting for a total of 9758 sequences that correspond to 361 organisms distributed in the three domains of life. The sequences of these molecules were separated according to the amino acid that transport and were aligned in ClustalW algorithm. Tests were performed for each group of tRNAs to choose the best evolutionary model, which pointed out to Kimura 2 parameters. Thirty five ancestral sequences were generated by Maximum Likelihood for 22 types of tRNA, 20 canonical tRNAs, an initiator tRNA, and a tRNA for selenocysteine. In the reconstruction of the phylogenetic trees, two ancestors were admitted if it was not possible to construct a single ancestor [19]. In the statistical analysis, the method of bootstrapping with 1000 replicates was carried out. These evaluations were done with MEGA5.05 program [20].
2.1.1. Analysis of sequences
The sequences obtained for each ancestral type of tRNA were aligned with the region V of the 23S rRNA from Thermus thermophilus. This organism was chosen because it is a model organism of hyperthermophiles and it could withstand similar conditions of Earth during the emergence of life. After the alignment of individual sequences of ancestral tRNAs, we built a single sequence with the tRNAs that aligned sequentially. The sequence derived from the tRNAs was aligned with the Peptidyl Transferase Center and the identity of the alignment was calculated. Analyzes were performed with the support of the program MEGA5 [20]. In addition, we generated 1000 random sequences with a length of 190 nucleotides each to further test the statistical significance of the alignment of our proposed ancestral sequence with the Peptidyl Transferase Center. The length of the generated random sequences was the same to the sequence that corresponds to the structure of PTC of the 23S ribosomal RNA from T. thermophilus. We used the version of Blastn with matrix 2–3 allowing the existence of gaps 2 penalties and 2 extension e word size of 7. The frequencies of the 16 dinucleotides were calculated with the support of the program Seqool (http://www.biossc.de/seqool/download.html). We also calculated the correlation coefficient between the frequencies of use of dinucleotides of the ancestor tRNA sequence with the PTC region. This value ranges from −1 to 1 and it was used to determine whether there is a relationship between these sequences.
3. Results
The structure of the 23S ribosomal RNA from T. thermophilus was defined in 2005 [21], consisting of 2902 nucleotides divided into 6 regions. The region V is located between the positions 2016 to 2624, with a length of about 609 nucleotides. In this region, we find the Peptidyl Transferase Center, located between the positions 2440 to 2620, equivalent to a length of 180 nucleotides. In our approach, we searched similarities between the sequences of ancestral tRNAs with positions in region V of the 23S ribosome RNA. The first analysis was performed by aligning each sequence to ancestral tRNA individually against region V. The results can be seen in Table 1.
Table 1.
Initial and final positions where each tRNA ancestor had alignment with the region V of the 23S ribosomal RNA. The 4th column indicates the percentage of the alignment. Two ancestors were admitted if it was not possible to construct a single ancestor. In all cases where two ancestors were accepted the identity was the same in both sequences.
| tRNA type | Initial position | Final position | % Match |
|---|---|---|---|
| Glu 1 | 27 | 78 | (20/50) 40% |
| Glu 2 | 27 | 78 | (20/50) 40% |
| Asp | 29 | 80 | (22/52) 42,3% |
| Leu | 54 | 92 | (17/39) 43,5% |
| Ser | 127 | 151 | (14/25) 56% |
| His 1 | 164 | 220 | (29/55) 52,7% |
| His 2 | 164 | 220 | (29/55) 52,7% |
| Pro 1 | 187 | 244 | (22/58) 37,9% |
| Pro 2 | 187 | 244 | (22/58) 37,9% |
| Tyr | 274 | 309 | (16/36) 44,4 |
| Tyr | 274 | 309 | (16/36) 44,4 |
| Ile 1 | 276 | 314 | (16/39) 41% |
| Ile 2 | 276 | 314 | (16/39) 41% |
| Trp 1 | 276 | 320 | (20/45) 44,4% |
| Trp 2 | 276 | 320 | (20/45) 44,4% |
| Asn 1 | 293 | 349 | (25/57) 43% |
| Asn 2 | 293 | 349 | (25/57) 43% |
| Cys 1 | 304 | 355 | (20/52) 38,4% |
| Cys 2 | 304 | 355 | (20/52) 38,4% |
| Phe 1 | 308 | 356 | (23/48) 47,9% |
| Phe 2 | 308 | 356 | (23/48) 47,9% |
| Ala | 319 | 357 | (18/39) 46% |
| InMet 1 | 325 | 388 | (33/61) 54% |
| InMet 2 | 325 | 388 | (33/61) 54% |
| Arg | 328 | 361 | (18/34) 52% |
| Met 1 | 336 | 388 | (24/50) 48% |
| Met 2 | 336 | 388 | (24/50) 48% |
| Gln 1 | 357 | 403 | (22/47) 46,8% |
| Gln 2 | 357 | 403 | (22/47) 46,8% |
| Sel | 358 | 402 | (21/45) 46,6% |
| Thr | 363 | 402 | (17/40) 42,5% |
| Val 1 | 368 | 414 | (19/47) 40.4% |
| Val 2 | 368 | 414 | (19/47) 40.4% |
| Gly | 474 | 521 | (17/47) 36,1% |
| Lys | 545 | 579 | (14/35) 40% |
We can see that different types of ancestral tRNAs have different alignment positions in region V. This result enabled us to build a sequence of the tRNA ancestors that we could verify its similarity to the PTC region. The sequence generated based on the ancestral tRNAs was aligned against PTC from T. themophilus and was composed by the following tRNAs ancestors in sequence: LeutRNA-SertRNA-HistRNA-ProtRNA-TyrtRNA-PhetRNA-GlntRNA-GlytRNA-LystRNA, ensuring the alignment of each sequence with the PTC moiety. In Fig. 1, we can see the result of the alignments that showed an overall identity of 50.53%. As described in Section 2.1.1., the alignment of the ancestral sequence proposed here was compared against 10,000 nucleotide sequences randomly generated. We observed that the alignment of the ancestral tRNAs against 5 out of 1000 sequences, attained identities varying between 7.48% and 10.16%, thus showing the relevance of identity of 50% of ancestral sequence proposed with PTC. Furthermore, Torarinsson et al. [22] argued that in the genomes of mouse and human there are more than 100,000 regions that are unalignable in relation to the primary sequence, but they show structural conservation. In that work, every consensus structure in the Rfam database indicated an average of ∼41% nucleotides involved in the base-pairing within the structures [22]. Finally, we evaluated the possibility that the observed identity value (50%) between the ancestral sequence and PTC could be achieved by any other random sequence. We analyzed the alignment again and the actual identity turned out to be 50.53%. To test the reliability of this result, we evaluated the possibility that the observed identity value (50%) between the ancestral sequence and PTC could be achieved by any other random sequence. Testing the hypothesis of finding at least 50% of 1000 random sequences with an identity value higher than 50%, when calculated with a Chi-square test and considering a degree of freedom = 2 and alpha = 0.05 was not significant. Indeed, the null hypothesis—that random tRNA sequences do align as our original proto-tRNA sequence—was rejected since the p-values were less than 0.00001. Therefore we were able to confirm that an ancestral sequence of proto-tRNAs has overall identity of 50.53% with the catalytic site of PTC. We also tested the same procedure in other thermophiles: Marinithermus hidrothermalis, Oceanithermus profundus and Thermus scotoductus. It has turned out that the matching between all these thermophiles organisms with the PTC ancestor is the same (50.53%) as the one obtained for T. thermophilus. This is not a surprising result given that these thermophiles organisms have 100% of identity among their PTC regions.
Fig. 1.
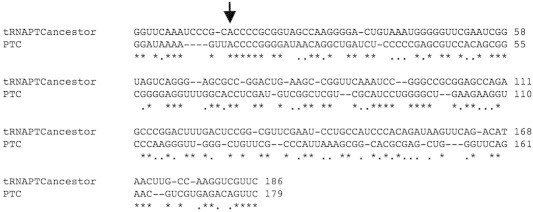
Alignment of the sequence of ancestral tRNAs (tRNAPTCancestor) against Peptidyl Transferase Center from T. thermophilus (PTC). The positions that have identity are marked in yellow. The arrow indicates the conserved catalytic position.
Fig. 2 shows the frequency of the 16 dinucleotides in the PTC from the proposed ancestor sequence and for the PTC from T. thermophilus, D. radiodurans, H. marismortui, E. coli, B. subtilis, C. jejuni and B. burgdoferi. Note that in all tested organisms the most frequent dinucleotide in PTC turned out to be the dinucleotide GG which is in agreement with the CCA universal motif of tRNA. When analyzing the PTC region (see Table 2) which performs the catalytic function of the 23S ribosomal RNA, and carries out the assembly of the amino acids, the correlation between the ancestor sequence of tRNA and all organisms analyzed here turned out to be positive, ranging from 0.51 as observed for E. coli to the high correlation of 0.87 in T. thermophilus.
Fig. 2.
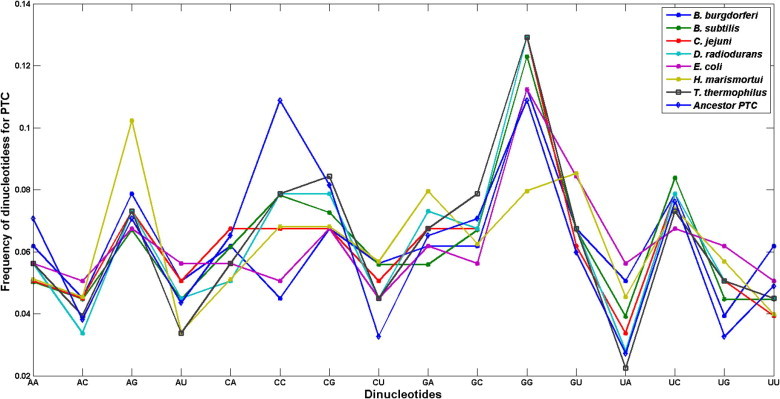
Frequency of the 16 dinucleotides for PTC in different organisms.
Table 2.
Correlation of the frequency values in the use of dinucleotides between the ancestral tRNA sequence herein proposed and the sequences of region PTC of the 23S subunit from various organisms.
| Organism | Correlation |
|---|---|
| Thermus thermophilus | 0.8734 |
| Halobacterium marismortui | 0.5404 |
| Escherichia coli | 0.5149 |
| Deinococcus radiodurans | 0.8662 |
| Campylobacter jejuni | 0.7840 |
| Bacillus subtilis | 0.8371 |
| Borrelia burgdoferi | 0.5996 |
4. Discussion
Tamura [3] proposed that the PTC originated as a consequence of dimerization of tRNA molecules based on the structural configuration of this portion of the rRNA that is the same as the one found in tRNA molecules. In our work, we reconstructed the ancestral sequence for all types of tRNA and compared them with the current PTC of 23S ribosomal RNA. Comparisons between individual ancestral tRNAs and PTC showed that different portions of the PTC had identity with different types of tRNA (Table 1). The latter may indicate a contribution of tRNA molecules in the appearance of the catalytic site of the 23S ribosomal RNA. From the points of the identity between tRNA ancestors and PTC, we built a sequence of the tRNAs that showed an outstanding overall identity of 50.53% with the PTC. Considering that the evolutionary process that resulted in 23S ribosomal RNA began about more than 3.5 billion years, and that in the early phases of this process, the error rates should be much greater than nowadays (the replication and repair systems were still in the process of optimization), we contend that the PTC maintains vestiges about its origins and that this process occurred by dimerization of molecules of proto-tRNAs. During peptide bond formation the CCA end of P-site tRNA and the PTC interact with conserved 23S bases A2451 and the P-loop, as it is shown in Fig. 1 [15]. Davidovich et al. [10] showed that molecules that have a structure like stem-elbow-stem tend to perform dimerization. This type of structure is typical of tRNA molecules, and is also found in the PTC, which may indicate that tRNA molecules gave rise to the ribosomal catalytic site. According to Tamura’s model [3] it seems likely that our proposed proto-tRNAs would conform to that of a minihelix region (acceptor stem plus T-stem). The PTC would be the first portion that appeared of the 23S ribosomal RNA [10,11]. Based on the evidence that the PTC was the initial portion of the 23S ribosomal RNA, it follows that the ribosome can be increased by duplications and fusions. In this model, the process that gave origin to ribosomal molecules as a whole, initially involved fusion of small molecules, the tRNAs, that gave origin to the PTC, which allowed the initiation of protein synthesis. The difference between our proposal and that of Fox [2] and Huang et al. [23] is that in our model the molecule that gave rise to the tRNA, gave origin to the PTC region as well. In the other proposals [2,23] the origin of the ribosome was due to a tRNA-like structure molecule. The data of the frequencies of dinucleotides showed that the information in modern sequences is the same information to that of our proposed ancestral sequence. Considering the evolutionary time of this molecule, the identity that they have with the molecules of tRNAs ancestors and the correlations between dinucleotides frequencies, we conclude that the PTC of the 23S ribosomal RNA originated indeed from the union of tRNA molecules and subsequent events of the insertion, deletion and mutation, which were responsible for functional adaptation. However, a group of original information still remains in the current molecules, thus, natural selection has preserved these positions for more than 3.5 billion years in order to maintain its structure and fundamental functionality.
Acknowledgment
MVJ was financially supported by PAPIIT-IN107112, UNAM, México.
Contributor Information
Sávio T. Farias, Email: stfarias@yahoo.com.br.
Marco V. José, Email: marcojose@biomedicas.unam.mx.
References
- 1.Woese C.R. Translation: in retrospect and prospect. RNA. 2001;7(8):1055–1067. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox G.E. Origin and evolution of the ribosome. Cold Spring Harbor Perspect, Biol. 2010;2(9):a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura K. Ribosome evolution: emergence of peptide synthesis machinery. J. Biosci. 2011;36(5):921–928. doi: 10.1007/s12038-011-9158-2. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz A.W. The RNA world and its origins. Planet Space Sci. 1995;43(1–2):161–165. doi: 10.1016/0032-0633(94)00166-o. [DOI] [PubMed] [Google Scholar]
- 5.Lazcano A., Miller S.L. The origin and early evolution of life: prebiotic chemistry, the pre-RNA world, and time. Cell. 1996;85(6):793–798. doi: 10.1016/s0092-8674(00)81263-5. [DOI] [PubMed] [Google Scholar]
- 6.Kua J., Bada J.L. Primordial ocean chemistry and its compatibility with the RNA world. Origins Life Evol. Biosphere. 2011;41(6):553–558. doi: 10.1007/s11084-011-9250-5. [DOI] [PubMed] [Google Scholar]
- 7.Kawamura K. Drawbacks of the ancient RNA-based life-like system under primitive earth conditions. Biochimie. 2012;94(7):1441–1450. doi: 10.1016/j.biochi.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Higgs P.G., Wu M. The importance of stochastic transitions for the origin of life. Origins Life Evol. Biosphere. 2012;42(5):453–457. doi: 10.1007/s11084-012-9307-0. [DOI] [PubMed] [Google Scholar]
- 9.Nissen P., Hansen J., Ban N. Moore PB and Steitz TA The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 10.Davidovich C., Belousoff M., Bashan A., Yonath A. The evolving ribosome: from non-coded peptide bond formation to sophisticated translation machinery. Res. Microbiol. 2009;160(7):487–492. doi: 10.1016/j.resmic.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Hsiao C., Lenz T.K., Peters J.K., Fang P.Y., Schneider D.M., Anderson E.J., Preeprem T., Bowman J.C., O’Neill E.B., Lie L., Athavale S.S., Gossett J.J., Trippe C., Murray J., Petrov A.S., Wartell R.M., Harvey S.C., Hud N.V., Williams L.D. Molecular paleontology: a biochemical model of the ancestral ribosome. Nucleic Acids Res. 2013;41(5):3373–3385. doi: 10.1093/nar/gkt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 13.Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 14.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 15.Selmer M., Dunham C.M., Murphy F.V., 4th, Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 16.Gabdulkhakov A., Nikonov S., Garber M. Revisiting the Haloarcula marismortui 50S ribosomal subunit model. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013;69(Pt6):997–1004. doi: 10.1107/S0907444913004745. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann J., Jossinet F., Gautheret D. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders. Nucleic Acids Res. 2013;41(10):5494–5502. doi: 10.1093/nar/gkt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zenkin N. Hypothesis: emergence of translation as a result of RNA helicase evolution. J. Mol. Evol. 2012;74(5–6):249–256. doi: 10.1007/s00239-012-9503-6. [DOI] [PubMed] [Google Scholar]
- 19.S.T. Farias, Suggested phylogeny of tRNAs based on the construction of ancestral sequence, J. Theor. Biol. 335 (2013) pp. 245-248 http://dx.doi.org/10.1016/j.jtbi.2013.06.033. [DOI] [PubMed]
- 20.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 22.Torarinsson E., Sawera M., Havgaard J.K., Fredholm M., Gorodkin J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L., Krupkin M., Bashan A., Yonath A., Massa L. Protoribosome by quantum kernel energy method. Proc. Natl. Acad. Sci. U.S.A. 2013 Sep 10;110(37):14900–14905. doi: 10.1073/pnas.1314112110. [DOI] [PMC free article] [PubMed] [Google Scholar]


