Abstract
A major goal of modern evolutionary biology is to understand the causes and consequences of phenotypic plasticity, the ability of a single genotype to produce multiple phenotypes in response to variable environments. While ecological and quantitative genetic studies have evaluated models of the evolution of adaptive plasticity, some long-standing questions about plasticity require more mechanistic approaches. Here, we address two of those questions: does plasticity facilitate adaptive evolution? And do physiological costs place limits on plasticity? We examine these questions by comparing genetically and plastically regulated behavioural variation in sailfin mollies (Poecilia latipinna), which exhibit striking variation in plasticity for male mating behaviour. In this species, some genotypes respond plastically to a change in the social environment by switching between primarily courting and primarily sneaking behaviour. In contrast, other genotypes have fixed mating strategies (either courting or sneaking) and do not display plasticity. We found that genetic and plastic variation in behaviour were accompanied by partially, but not completely overlapping changes in brain gene expression, in partial support of models that predict that plasticity can facilitate adaptive evolution. We also found that behavioural plasticity was accompanied by broader and more robust changes in brain gene expression, suggesting a substantial physiological cost to plasticity. We also observed that sneaking behaviour, but not courting, was associated with upregulation of genes involved in learning and memory, suggesting that sneaking is more cognitively demanding than courtship.
Keywords: plasticity, Poecilia latipinna, transcriptome, mating behaviour, courtship
1. Introduction
Phenotypic plasticity, the ability of a genotype to produce different phenotypes in different environments, is widespread and has been considered a potential adaptation for thriving in variable environments (reviewed in [1–3]). A variety of approaches have demonstrated that plasticity can be adaptive [4], but several long-standing controversies are likely to be resolved only through a mechanistic understanding of phenotypic expression [5–7]. One unanswered question is whether plasticity facilitates adaptation to new environments. The traditional view is that plasticity retards adaptation by shielding organisms from natural selection [8–12]. A competing hypothesis is that plasticity precedes and facilitates adaptive evolution by allowing organisms to persist in novel environments to which they are initially poorly adapted [3,13–17]. Plasticity might also facilitate adaptation through genetic assimilation or by producing evolvable genetic architectures [6,18,19]. This idea that plasticity might be the first step in adaptive evolution has proved to be controversial [6,20–22]. While some models of this hypothesis have supported it [18,19], others have shown that plasticity might have minor effects on the rate of adaptation [6,12]. A potential resolution to these mixed results is the idea that plasticity is most likely to facilitate adaptive evolution when genetic and plastic changes to the phenotype depend on the same underlying proximate mechanisms [3,23–25].
A second unresolved issue is the nature and magnitude of the cost of plasticity [26,27]. There must be a net fitness cost of plasticity or plasticity would be unlimited. Costs are posited to arise through a fitness mismatch between a plastically produced phenotype and its environment [28], energetic demands of sensory and physiological systems that support plasticity [29], or fitness costs of pleiotropic effects of systems necessary to support plasticity [30]. While ecological experiments can detect fitness costs and mismatches, detecting the energetic costs requires mechanistic understanding [31,32].
Both these fundamental questions are particularly relevant to understanding behavioural plasticity and evolution. Strong arguments have been made that plasticity facilitates behavioural evolution [3,33,34], and considerable effort has been devoted to understanding the mechanistic control of plasticity to illuminate this issue [7,35]. Costs of behavioural plasticity are often overlooked, but they are central to understanding animal personalities; the existence of personalities [36] and behavioural syndromes [37,38] reflects the fact that plasticity is limited. Whether ecological or energetic costs are most important in limiting behavioural plasticity is a question that can only be answered if both kinds of costs can be quantified in the same system.
To address these questions, we exploited a system in which the same phenotypic transition is accomplished either by plasticity or by genetic polymorphism, and both these strategies coexist in the same population. Male sailfin mollies (Poecilia latipinna) vary widely in body size and mating behaviour based on a Y-linked polymorphism [39–44] (details in electronic supplementary material). Males can exhibit courtship and ‘sneaking’ mating behaviour, but the degree of plasticity in behaviour depends on genetically determined male body size [44,45]. Large males typically perform courtship displays (raising and spreading of the dorsal fin towards a female, often accompanied by a C-shaped curving of the body), and small males adopt a ‘sneaker’ strategy: they attempt copulation without apparent female cooperation by thrusting the gonopodium at the female's gonopore [40]. In contrast, intermediate-size males are plastic in behaviour, adopting a courting strategy in the absence of other males and a sneaking strategy in the presence of other males. The courtship/sneaking dichotomy is thus genetically determined between males at the extremes of the size distribution but environmentally regulated in intermediate-size males.
This system is especially useful for investigating mechanisms underlying plasticity because we can separate responses to an environmental change from responses directly associated with a change in the phenotype. That is, we can expose both ‘fixed’ and ‘plastic’ genotypes to the same range of environments, but only the plastic genotype will respond to the environment by altering its phenotype. This separation has not been possible in other investigations of phenotypic plasticity [24,46,47], including studies of alternative reproductive tactics [48–50], because these studies did not have access to morphs that do and do not display plasticity under the same environmental conditions.
To investigate the molecular basis of variation in plasticity, we used data on brain transcriptional state. Although transcriptional data do not provide a complete description of the molecular and genomic mechanisms that regulate behaviour, they have several advantages for our study. First, environmental effects on the phenotype are likely to be mediated by changes in gene expression [51,52]. Second, gene expression measures provide an assay of ‘first-order phenotypes’ that underlie variation in ‘higher-order’ observable phenotypes [7]. Third, recent gene-network models of plasticity are explicitly based on interactions between gene products in transcriptional networks [6,18,19]. Finally, high-throughput technologies deliver reliable information about gene expression that does not depend on prior knowledge of specific gene networks [53,54]. We therefore used RNA-sequencing (RNA-Seq) to assay gene expression in the brains of male mollies that varied in their degree of plasticity for mating behaviour.
We tested two hypotheses derived from our broader questions. First, plastic and genetic regulation of behaviour should be associated with similar changes in brain gene expression patterns. This hypothesis emerges from the theoretical demonstration that plasticity is more likely to facilitate adaptive evolution if genetic and plastic changes are underlain by the same mechanisms [3,23–25]. Second, the plastic expression of mating behaviour in intermediate males should be associated with greater change in brain gene expression patterns than seen in small males exposed to the same environmental shift. This hypothesis derives from the idea that behavioural plasticity is limited because it incurs energetic or metabolic costs that are absent when individuals exhibit non-plastic behaviour over the same range of environments [36,38]. In addition to testing these hypotheses, we sought to identify unique transcriptional profiles associated with courtship and sneaking behaviour because these alternative reproductive tactics are common, and sneaking strategy has previously been associated with upregulation of genes involved in learning and memory [50].
2. Material and methods
(a). Experimental animals
We used wild-caught males from the salt marsh population at Bald Point, Florida, USA (W 84°20′27″, N 29°54′21″). We classified males by size following Travis & Woodward [45] with slight modifications; small males were less than 3.4 cm standard length (SL), intermediate males were 3.4–4.8 cm SL and large males were more than 4.8 cm SL (see electronic supplementary material). Large males were rare, so we used them only as social context fish and restricted focal fish to either small or intermediate-sized males. We tested 31 males (17 small, 14 intermediate) in two different social environments (single-male and multimale groups). To establish appropriate size distributions and sex ratios for social treatments, context fish were collected from nearby sites but were not used for any molecular analyses. Whenever possible all female context fish in a given trial were from the same population and context males were from another population (electronic supplementary material, table S1). All focal males were collected on either 6 May or 20 August 2010; capture date did not significantly affect any behaviour (see Results). Fish were kept on a 14 L : 10 D schedule and fed crushed flake fish food (Tetramin) ad libitum twice daily.
(b). Mating trials
We used a protocol similar to that of Travis & Woodward [45]. In one ‘social context’ the focal male was tested alone with three females. In the alternative context, a focal male was tested with two other males (such that all three size classes of males were present) and six females. In most cases, focal intermediate males were tested with a small and a large context male, and small focal males were tested with an intermediate and a large context male. Owing to limited availability of large males, in three trials the focal intermediate male was the largest male in the trial, but this had no effect on behaviour (all p > 0.50 in one-way ANOVA models, for all behaviours described below). In total, four experimental groups were used: intermediate males tested alone (IA; n = 7, SL (mean ± s.e.) = 4.12 ± 0.16), intermediate males in multimale groups (IM; n = 7, SL = 4.04 ± 0.16), small males tested alone (SA; n = 9, SL = 2.76 ± 0.14) and small males in multimale groups (SM; n = 8, SL = 2.83 ± 0.14). IA and IM males differed only in the social context used in the mating trials—a short-term difference that nonetheless produced dramatic variation in behaviour (see the Results section). By contrast, IA and SA males represent animals that behave differently even when they are reared and tested under identical conditions [55]; behavioural differences can therefore be attributed to genotypic effects.
Prior to mating trials, all animals were housed in group tanks (10–114 days, mean 80 days). Focal and context males were isolated and placed in separate tanks the night before the trial. Females were taken from their home tanks immediately before the trial. Focal males were used only once, while context males and females were used in multiple trials. Focal males, context males and context females used in the same trial were all from different home tanks (electronic supplementary material, table S1). Context males were chosen so that size difference between males was more than or equal to 0.5 cm SL; prior work [56] indicates that males can distinguish between females more than or equal to 0.6 cm different so we are confident that males can perceive body length differences among males of at least 0.5 cm. Females were chosen to be approximately the same size (within 1 cm SL) and to have not given birth within the previous 3 days to minimize differences in attractiveness to males [57]. All males were placed into the observation tank (30 × 75 × 30 cm with opaque sides) simultaneously and left to acclimate for 5 min. Females were then added to the tank, and behavioural observations began immediately and continued for 30 min. All fish were fed at least 3 h before the trial commenced. All trials took place between 11.00 and 13.00, 27 July–14 October 2010, within the mating season of the species [39]. Temperature and salinity were kept constant at approximately 26°C and 6 ppt, respectively. Each tank had its own full spectrum light source.
We quantified three sexual behaviours as instantaneous events [58] in accordance with Travis & Woodward [45]: (i) courtship displays involve raising and spreading of the dorsal fin towards a female, often accompanied by a C-shaped curving of the body; (ii) nibbles are oral contact by a male of the female's gonopore; (iii) gonoporal thrusting is an attempt to insert the gonopodium into the gonopore of the female. We also quantified male–male aggression: the number of chases and the number of times being chased. Total activity was calculated as the sum of activity in each of these categories.
(c). RNA sequencing
Immediately after the trial, focal males were euthanized in cold water. A coarse dissection removed the braincase, which was placed in RNAlater (Ambion, Austin, TX), stored in 4°C for 24 h, then stored at −80°C. Whole brains were later dissected completely from the brain case. To minimize variation associated with seasonality, we evaluated gene expression only on males collected on 6 May 2010 and used in mating trials that took place between 27 July and 3 August 2010. Eleven males met these criteria: three from each size class and treatment, except for IM, which had two males. We prepared RNA-Seq libraries from whole brains of each individual (see electronic supplementary material). To control for lane effects, each of three sequencing lanes contained an aliquot of all 11 samples.
Sequencing reads (100 bp, single-end) were mapped to a high-quality brain-specific reference transcriptome of the congener, Poecilia reticulata (Trinidad guppy). The assembly contains 41 347 contigs, N50 = 2548, and recovers 63% of Tilapia (Oreochromis niloticus) Ensembl proteins (Release 70). See electronic supplementary material, table S2 for assembly and mapping details. We aligned sequencing reads to the reference using Bowtie2 v. 2.0.0 retaining mappings with quality scores more than 30 (less than 0.001 probability that the read maps elsewhere in the reference), keeping only contigs represented by at least 1 count per million reads in at least three samples, resulting in 31 869 contigs in the dataset.
(d). Data analysis
We used general linear models to analyse phenotypic variation in behaviour, with size and social context as crossed fixed effects, and date collected, trial date and observation tank as covariates. Number of displays, thrusts, nibbles and aggressive interactions were standardized by overall activity, and these proportions were arc-sine square-root transformed to improve normality and homoscedasticity of residuals.
We assessed differential gene expression using generalized linear models (GLMs) with number of sequencing reads mapping to a contig as the dependent variable and log-transformed TMM-normalized library size as a model offset [59]. Size, social context and their interaction were fixed effects. To test the hypothesis that plastic and genetic variation in behaviour are underlain by the same molecular processes, we determined whether transcripts that were differentially expressed (DE) in intermediate males across social contexts (IA versus IM) were also DE between different genotypes tested in the single-male context (IA versus SA). We identified these two groups of transcripts by first selecting transcripts with a significant size-by-context interaction in the GLM, then testing for a significant pairwise difference in the appropriate model contrast (IA–IM or IA–SA) and selecting those with FDR < 0.05. To determine whether more transcripts than expected appeared on both lists and had DE in the same direction, we used a χ2-test of association. To account for potential bias in this association test (because variation in IA contributes to both comparisons), we also evaluated the overlap between lists using permutation tests (electronic supplementary material).
To identify transcripts with expression patterns that changed across social contexts in one size class but not the other, we applied three criteria: (i) a significant size × context interaction; (ii) a significant linear contrast between contexts in one size class; (iii) similar expression in the other size class. Significance cutoffs for criteria (i) and (ii) were the same as above. Criterion (iii) is equivalent to accepting the null hypothesis that expression does not differ across contexts, so we used the least-squares method of identifying true null hypotheses implemented in SAS Proc Multtest [60] to evaluate this criterion. We identified transcripts with expression patterns that were associated directly with behaviour by first filtering on a significant size × context interaction, and then on a significant linear contrast in expression between IA males (the only ones to display courtship) and mean expression in the other groups of males (IM, SA and SM).
We used gene ontology (GO) classification to evaluate enrichment of functional classes, using GO IDs derived from GI-to-GO mappings (UniProt, downloaded 3 October 2013). We evaluated over-representation of biological process, molecular function and cellular component ontologies using the topGo package and the elim algorithm [61] to account for correlation in the graph topology (eliminating genes from ancestor terms if they are significantly enriched in a child term). Beyond setting a stringent p-value cutoff, we did not apply formal correction for multiple testing. When p-values of GO terms are conditioned on neighbouring terms, tests are not independent and multiple testing theory does not strictly apply [61].
3. Results
(a). Intermediate and small males differ in behavioural plasticity
Intermediate males were highly plastic in mating behaviour, but small males were not (figure 1). Intermediate males devoted about 75% of their actions to courtship displays when in single-male trials, but devoted about 75% of their actions to gonoporal thrusts or nibbles when the social environment contained other males. In contrast, about 80% of the actions of small males were gonoporal thrusts or nibbles, irrespective of their social environment. For both displays and thrusts, the size × social environment interaction was significant (displays: F1,21 = 7.4, p = 0.01; thrusts: F1,21 = 10.1, p = 0.005). For nibbles, the interaction was non-significant (F1,21 = 3.9, p = 0.06). Total activity did not differ among groups (all p > 0.1), so variation in behavioural repertoire was not confounded with differences in activity (full model results for all behaviours in electronic supplementary material, table S3).
Figure 1.
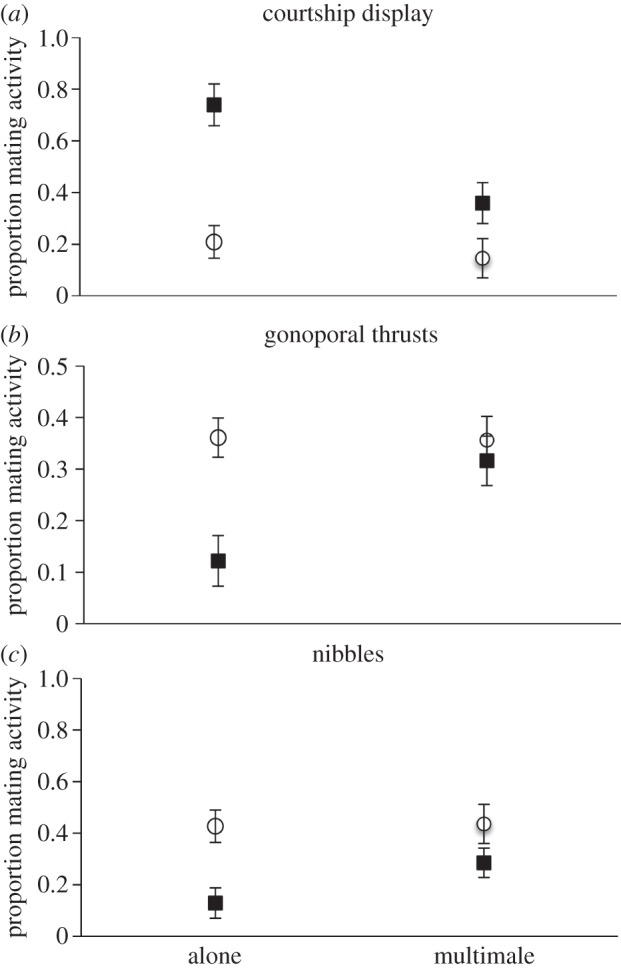
(a–c) Courtship behaviour of focal males. Behaviour in mating trial including only the focal male (alone), and when trials included small, intermediate and large males (multimale). Open circles represent least-square means of untransformed data for small males. Filled squares represent intermediate males. Bars are standard errors calculated from the GLM. Note the differences in the scale of the y-axis.
Male–male aggression (which could only be measured in multimale trials) did not differ between small and intermediate males. Neither the number of chases nor the number of times chased differed significantly between groups, although there was a trend for small males to be chased more often than intermediate males (electronic supplementary material, figure S1 and table S3).
(b). Transcriptional differences associated with plasticity are also associated with genotypic variation
To test the hypothesis that plastic and genetic variation in behaviour are underlain by the same molecular processes, we determined whether transcriptional differences associated with a plastic behavioural response to social environment in intermediate males (IA compared with IM males) were also DE between intermediate and small males when tested in the same single-male social context (IA and SA). Recall that behavioural differences between IA and SA males persist when reared in a common environment and are therefore regulated by genotype. Of the transcripts with a significant size×social context interaction in the GLM, 2466 were concordantly DE in both comparisons (i.e. the difference between IA and IM males was in the same direction as that between IA and SA males), significantly more than expected if the two lists were independent (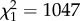 , p < 0.0001). Only four transcripts appeared on both lists and had discordant expression differences. To account for potential bias due to correlated errors in the above test, we performed permutation tests to call individual transcripts DE and to determine the number of transcripts expected to meet our criteria if the two lists were independent. Based on these tests, 2190 transcripts were significantly and concordantly DE in both the IA versus IM and IA versus SA contrasts, while only three transcripts were discordantly DE (figure 2). The number concordantly DE is much larger than the median of 118 (interquartile range = 70–232) in the 100 permuted datasets (electronic supplementary material, figure S2). Thus, both the conventional tests and the permutation tests indicated that many more transcripts overlapped between these two lists than expected. Concordantly DE transcripts tended to exhibit larger genetically regulated change in expression than socially regulated change (Wilcoxon's paired Z = 7.5 × 105, p < 0.0001; figure 2).
, p < 0.0001). Only four transcripts appeared on both lists and had discordant expression differences. To account for potential bias due to correlated errors in the above test, we performed permutation tests to call individual transcripts DE and to determine the number of transcripts expected to meet our criteria if the two lists were independent. Based on these tests, 2190 transcripts were significantly and concordantly DE in both the IA versus IM and IA versus SA contrasts, while only three transcripts were discordantly DE (figure 2). The number concordantly DE is much larger than the median of 118 (interquartile range = 70–232) in the 100 permuted datasets (electronic supplementary material, figure S2). Thus, both the conventional tests and the permutation tests indicated that many more transcripts overlapped between these two lists than expected. Concordantly DE transcripts tended to exhibit larger genetically regulated change in expression than socially regulated change (Wilcoxon's paired Z = 7.5 × 105, p < 0.0001; figure 2).
Figure 2.
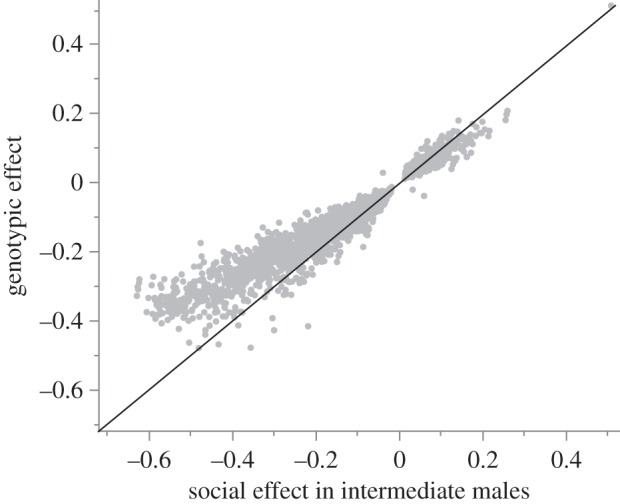
Social and genetic regulation of gene expression. The x-axis shows the difference in transcript abundance across social contexts in intermediate males, illustrating social regulation; the y-axis shows difference in transcript abundance between intermediate and small males when tested alone, illustrating genotypic regulation. Expression differences are standardized by mean abundance of each transcript such that values represent a proportion of mean transcript abundance. Grey points are transcripts significantly DE in both contrasts, based on permutation tests. Black line indicates equivalent differential expression in both contrasts. Most points fall above this line, illustrating the tendency for individual transcripts to show larger effects of genotypic than of social regulation.
(c). Intermediate and small males exhibit very different brain transcriptional responses to the social environment
Twenty-eight percent of all transcripts in the dataset (8838) displayed a significant size × social context interaction, suggesting that intermediate and small males had different transcriptional responses to change in the social environment. Indeed, 6% of tested transcripts (1830) responded to the social environment only in intermediate males, 4% (1287) responded only in small males and another 4% (1357) responded robustly but in opposite directions in the two kinds of males (electronic supplementary material, figure S3). Only 0.3% (102) responded significantly and in the same direction in both genotypes. Overall, more transcripts changed across contexts in intermediate than in small males, and the magnitude of change tended to be larger in intermediate than in small males (15.78 ± 0.86 and 13.48 ± 0.65 counts per million reads, respectively; Wilcoxon's Z = 3.1 × 105, p < 0.0001).
(d). Sneaking behaviour is associated with upregulation of genes involved in neurotransmission, learning and mechanosensory processes
In the comparison between IA males, which performed mainly courtship, and the other groups of males, which performed mainly sneaking, 1520 transcripts were significantly upregulated in sneakers and 500 transcripts were upregulated in the IA courting males. Surprisingly, transcripts upregulated in ‘sneaker’ males were highly significantly enriched for GO terms relating to neurotransmission, learning and locomotory behaviour. In contrast, transcripts upregulated in courting males were enriched for functions related to mRNA processing and translation (electronic supplementary material, table S4).
4. Discussion
We investigated mating behaviour and brain gene expression patterns of sailfin mollies that exhibited both genotypic and plastic regulation of behaviour. Intermediate-size males changed from a courting to a sneaking mating strategy depending on their social environment. Small males exhibited a sneaking strategy in both environments. This behavioural variation was associated with a broad neurogenomic response; one-third of brain transcripts we examined were associated with genotypically determined male size, the social environment or an interaction between the two. This transcriptional response is nearly as large as differences seen between behavioural castes in honeybees [62]. Aubin-Horth et al. [50] also reported a substantial neurogenomic difference between alternative Atlantic salmon male morphs, where approximately 15% of the interrogated brain transcripts differed between age-matched morphs collected from the same natural population.
Although behavioural trials lasted only 30 min, it is probable that small and intermediate males differed in behaviour and brain transcriptional patterns during most of their adult lives; transcriptional variation related to the main effect of male size probably reflects these long-standing differences. However, the transcriptional response to the social environment must have been triggered by the experiment, because all males inhabited similar physical and social environments before their mating trials. The rapid and divergent transcriptional responses in small and intermediate males suggests that their brains are ‘primed’ to respond differently to changing social conditions because of differences in neural architecture or physiology. Variation in transcriptional response is a product of these underlying differences.
Several authors have argued that plasticity is likely to facilitate adaptive evolution when genetic and plastic changes to the phenotype depend on the same underlying mechanisms [3,23–25], and gene network models that predict this facilitation implicitly make this assumption [18,19]. Using a system in which both plastic and genetically regulated phenotypes occur in the same population, we found that these two types of variation did significantly overlap in their transcriptional patterns, supporting the idea of shared molecular mechanisms. The overlap we observed was not complete, however, with about half of the transcripts in each contrast not being found in the other, and with generally larger magnitude of genetic regulation compared with social regulation. The recent models of phenotypic plasticity do not address how quantitative variation in the level of ‘transcriptional similarity’ affects the likelihood that plasticity facilitates subsequent evolution. Development of such models would allow more refined tests of the hypothesis.
In small males, a large number of transcripts were DE across social environments, despite displaying very similar behaviour. That is, transcriptional plasticity was not directly proportional to the behavioural plasticity we observed, although small males might have changed behaviour or physiology in ways that we did not measure. Nevertheless, both the number of transcripts responding and the magnitude of change were larger in intermediate than in small males. If transcriptional regulation incurs a fitness cost [63–65] (but see [66]), these results imply that the plasticity we observed in this experiment incurs a cost. This cost of plasticity per se is one of the avenues through which costs can limit the evolution of plasticity [29,32]. Reviews of the cost of plasticity [26,27] have found limited evidence for this type of cost because few studies have been designed to find it; there is little relevant information outside of model systems [67] and few studies of the cost of behavioural plasticity in contexts other than learning [68–70]. Our results suggest that there are transcriptional costs of behavioural plasticity and that it is possible to estimate the magnitude of these costs in a variety of empirical systems. If the behavioural plasticity can be linked to fitness variation, then the cost incurred by phenotype–fitness mismatches [28] could also be estimated.
In addition to these general patterns, we observed a dramatic enrichment of gene products related to cognitive and locomotory functions in males that performed mainly sneaking behaviour (across both size morphs) compared with males that performed courtship. Strikingly, this enrichment occurred among transcripts that were upregulated in sneakers, but not in those upregulated in courting males. This pattern suggests that sneaking involves the active regulation of many neural functions, whereas courtship behaviour does not. A similar pattern has been reported in a comparison between mature sneaker males and age-matched immature Atlantic salmon, Salmo salar [50]. Those authors proposed that mature sneaker males experienced greater cognitive demands than did immature males that were destined for an anadromous life history. Our results suggest that these differences might be a general feature of alternative male reproductive tactics.
Studies of other taxa with alternative male morphs are needed to determine whether the neurogenomic patterns observed by Aubin-Horth et al. [50] and by us are general. Nonetheless, sailfins should be a useful model for investigating mechanisms underlying this pattern. Males of all sizes mature in less than a year, sneaking and courting are almost mutually exclusive between males at the extremes of the body size distribution [40], and the conditions that induce a shift from courting to sneaking in intermediate males are straightforward to reproduce. Moreover, populations of sailfins differ in the relationship between size and behaviour [43], and these differences could be exploited to investigate the genetic and environmental regulation of both traits.
Acknowledgements
We thank Mia Adreani, Joseph Apodaca, Alex Landy and Pamela MacRae for field collections, and Cory Smith for help with animal husbandry.
All procedures were conducted in accordance with Florida State University policies and were approved in Protocol no. 1001.
Data accessibility
Morphological and behavioural data used in this paper are available in Dryad (doi:10.5061/dryad.4qn6n). RNA-Seq data are available in NCBI Short Read Archive (accession SRP036185).
Funding statement
This work was funded by NSF grants IOS-0934451 and DEB-0848337 to K.A.H. and by funds provided by Florida State University.
References
- 1.Travis J. 1994. Evaluating the adaptive role of morphological plasticity. In Ecological morphology: integrative organismal biology (eds Wainwright PC, Reilly SM.), pp. 99–122 Chicago, IL: University of Chicago Press [Google Scholar]
- 2.Schlichting CD, Pigliucci M. 1998. Phenotypic evolution: a reaction norm perspective. Sunderland, MA: Sinauer Associates Incorporated [Google Scholar]
- 3.West-Eberhard MJ. 2003. Developmental plasticity and evolution. Oxford, UK: Oxford University Press [Google Scholar]
- 4.Schmitt J, Stinchcombe JR, Heschel MS, Huber H. 2003. The adaptive evolution of plasticity: phytochrome-mediated shade avoidance responses. Integr. Comp. Biol. 43, 459–469 (doi:10.1093/icb/43.3.459) [DOI] [PubMed] [Google Scholar]
- 5.Pigliucci M, Murren CJ, Schlichting CD. 2006. Phenotypic plasticity and evolution by genetic assimilation. J. Exp. Biol. 209, 2362–2367 (doi:10.1242/jeb.02070) [DOI] [PubMed] [Google Scholar]
- 6.Draghi JA, Whitlock MC. 2012. Phenotypic plasticity facilitates mutational variance, genetic variance, and evolvability along the major axis of environmental variation. Evolution 66, 2891–2902 (doi:10.1111/j.1558-5646.2012.01649.x) [DOI] [PubMed] [Google Scholar]
- 7.Renn SCP, Schumer ME. 2013. Genetic accommodation and behavioural evolution: insights from genomic studies. Anim. Behav. 85, 1012–1022 (doi:10.1016/j.anbehav.2013.02.012) [Google Scholar]
- 8.Grant V. 1977. Organismic evolution. San Francisco, CA: Freeman [Google Scholar]
- 9.Falconer DS. 1981. Introduction to quantitative genetics. New York, NY: Ronald Press Co [Google Scholar]
- 10.Levin DA. 1988. Plasticity, canalization and evolutionary stasis in plants. In Plant population ecology (eds Davy AJ, Hutchings MJ, Watkinson AR.), pp. 35–45 Oxford, UK: Blackwell Scientific Publications [Google Scholar]
- 11.Anderson RW. 1995. Learning and evolution: a quantitative genetics approach. J. Theor. Biol. 175, 89–101 (doi:10.1006/jtbi.1995.0123) [DOI] [PubMed] [Google Scholar]
- 12.Ancel LW. 2000. Undermining the Baldwin expediting effect: does phenotypic plasticity accelerate evolution? Theor. Popul. Biol. 58, 307–319 (doi:10.1006_tpbi.2000.1484) [DOI] [PubMed] [Google Scholar]
- 13.Baldwin JM. 1896. A new factor in evolution. Am. Nat. 30, 441–451 (doi:10.1086/276408) [Google Scholar]
- 14.Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150, 563–565 (doi:10.1038/150563a0) [DOI] [PubMed] [Google Scholar]
- 15.Schmalhausen II. 1949. Factors of evolution. Philadelphia, PA: Blakiston [Google Scholar]
- 16.Hinton GE, Nowlan SJ. 1987. How learning can guide evolution. Complex Syst. 1, 495–502 (doi:10.1177/089443938900700128) [Google Scholar]
- 17.Price TD, Qvarnström A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. Lond. B 270, 1433–1440 (doi:10.1098/rspb.2003.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fierst JL. 2011. A history of phenotypic plasticity accelerates adaptation to a new environment. J. Evol. Biol. 24, 1992–2001 (doi:10.1111/j.1420-9101.2011.02333.x) [DOI] [PubMed] [Google Scholar]
- 19.Espinosa-Soto C, Martin OC, Wagner A. 2011. Phenotypic plasticity can facilitate adaptive evolution in gene regulatory circuits. BMC Evol. Biol. 11, 5 (doi:10.1186/1471-2148-11-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West-Eberhard MJ. 2005. Phenotypic accommodation: adaptive innovation due to developmental plasticity. J. Exp. Zool. 304B, 610–618 (doi:10.1002/jez.b.21071.PHENOTYPIC) [DOI] [PubMed] [Google Scholar]
- 21.De Jong G. 2005. Evolution of phenotypic plasticity: patterns of plasticity and the emergence of ecotypes. New Phytol. 166, 101–117 (doi:10.1111/j.1469-8137.2005.01322.x) [DOI] [PubMed] [Google Scholar]
- 22.Ghalambor CK, McKay JK, Carroll SP, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407 (doi:10.1111/j.1365-2435.2007.01283.x) [Google Scholar]
- 23.Mayley G. 1996. Landscapes, learning costs and genetic assimilation. Evol. Comput. 4, 213–234 (doi:10.1162/evco.1996.4.3.213) [Google Scholar]
- 24.Alaux C, et al. 2009. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl Acad. Sci. USA 106, 15 400–15 405 (doi:10.1073/pnas.0907043106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scoville AG, Pfrender ME. 2010. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proc. Natl Acad. Sci USA 107, 4260–4263 (doi:10.1073/pnas.0912748107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Buskirk J, Steiner U. 2009. The fitness costs of developmental canalization and plasticity. J. Evol. Biol. 22, 852–860 (doi:10.1111/j.1420-9101.2009.01685.x) [DOI] [PubMed] [Google Scholar]
- 27.Auld JR, Agrawal AA, Relyea RA. 2010. Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proc. R. Soc. B 277, 503–511 (doi:10.1098/rspb.2009.1355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman RA. 1992. Adaptive plasticity in amphibian metamorphosis. BioScience 42, 671–678 (doi:10.2307/1312173) [Google Scholar]
- 29.DeWitt TJ, Sih A, Wilson DS. 1998. Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 13, 77–81 (doi:10.1111/j.1558-5646.2009.00647.x) [DOI] [PubMed] [Google Scholar]
- 30.Van Kleunen M, Fischer M. 2005. Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytol. 166, 49–60 (doi:10.1111/j.1469-8137.2004.01296.x) [DOI] [PubMed] [Google Scholar]
- 31.Pigliucci M. 2001. Phenotypic plasticity: beyond nature and nurture. Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 32.Callahan HS, Maughan H, Steiner UK. 2008. Phenotypic plasticity, costs of phenotypes, and costs of plasticity: toward an integrative view. Ann. NY Acad. Sci. 1133, 44–66 (doi:10.1196/annals.1438.008) [DOI] [PubMed] [Google Scholar]
- 33.Ghalambor CK, Angeloni L, Carrol SP. 2010. Behavior as phenotypic plasticity. In Evolutionary behavioral ecology (eds Fox C, Westneat D.), pp. 90–107 New York, NY: Oxford University Press [Google Scholar]
- 34.Foster SA. 2013. Evolution of behavioural phenotypes: influences of ancestry and expression. Anim. Behav. 85, 1061–1075 (doi:10.1016/j.anbehav.2013.02.008) [Google Scholar]
- 35.Bell AM, Robinson GE. 2011. Behavior and the dynamic genome. Science 332, 1161–1162 (doi:10.1126/science.1203295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamps J, Groothuis TGG. 2010. The development of animal personality: relevance, concepts and perspectives. Biol. Rev. Camb. Phil. Soc. 85, 301–325 (doi:10.1111/j.1469-185X.2009.00103.x) [DOI] [PubMed] [Google Scholar]
- 37.Sih A, Bell A, Johnson JC. 2004. Behavioral syndromes: an ecological and evolutionary overview. Trends Ecol. Evol. 19, 372–378 (doi:10.1016/j.tree.2004.04.009) [DOI] [PubMed] [Google Scholar]
- 38.Sih A, Bell AM. 2008. Insights for behavioral ecology from behavioral syndromes. Adv. Study Behav. 38, 227–281 (doi:10.1016/S0065-3454(08)00005-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travis J. 1994. Size dependent behavioral variation and its genetic control within and among populations. In Quantitative genetic studies of behavioral evolution (ed. Boake BRC.), pp. 165–187 Chicago, IL: University of Chicago Press [Google Scholar]
- 40.Farr JA, Travis J, Trexler JC. 1986. Behavioural allometry and interdemic variation in sexual behaviour of the sailfin molly, Poecilia latipinna (Pisces: Poeciliidae). Anim. Behav. 34, 497–509. (10.1016/S0003-3472(86)80118-X) [Google Scholar]
- 41.Trexler JC, Travis J, Trexler M. 1990. Phenotypic plasticity in the sailfin molly, Poecilia latipinna (Pisces: Poeciliidae). II. Laboratory experiment. Evolution 44, 157–167 (doi:10.2307/2409531) [DOI] [PubMed] [Google Scholar]
- 42.Trexler JC, Travis J. 1990. Phenotypic plasticity in the sailfin molly, Poecilia latipinna (Pisces: Poeciliidae). I. Field experiments. Evolution 44, 143–156 (doi:10.2307/2409530) [DOI] [PubMed] [Google Scholar]
- 43.Ptacek MB, Travis J. 1996. Inter-population variation in male mating behaviours in the sailfin mollie, Poecilia latipinna. Anim. Behav. 52, 59–71 (doi:10.1006/anbe.1996.0152) [Google Scholar]
- 44.Seda JB, Childress MJ, Ptacek MB. 2012. Individual variation in male size and behavioral repertoire in the Sailfin Molly Poecilia latipinna. Ethology 118, 411–421 (doi:10.1111/j.1439-0310.2012.02025.x) [Google Scholar]
- 45.Travis J, Woodward BD. 1989. Social context and courtship flexibility in male sailfin mollies, Poecilia latipinna (Pisces: Poeciliidae). Anim. Behav. 38, 1001–1011 (doi:10.1016/S0003-3472(89)80139-3) [Google Scholar]
- 46.Whitehead A, Roach JL, Zhang S, Galvez F. 2011. Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proc. Natl Acad. Sci. USA 108, 6193–6198 (doi:10.1073/pnas.1017542108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt BG, Ometto L, Wurm Y, Shoemaker D, Yi SV, Keller L. 2011. Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc. Natl Acad. Sci. USA 108, 15 936–15 941 (doi:10.1073/pnas.1104825108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burmeister SS, Jarvis ED, Fernald RD. 2005. Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 3, 1996–2004 (doi:10.1371/journal.pbio.0030363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snell-Rood EC, Cash A, Han MV, Kijimoto T, Andrews J, Moczek AP. 2011. Developmental decoupling of alternative phenotypes: insights from the transcriptomes of horn-polyphenic beetles. Evolution 65, 231–245 (doi:10.1111/j.1558-5646.2010.01106.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aubin-Horth N, Landry CR, Letcher BH, Hofmann HA. 2005. Alternative life histories shape brain gene expression profiles in males of the same population. Proc. R. Soc. B 272, 1655–1662 (doi:10.1098/rspb.2005.3125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibson G. 2008. The environmental contribution to gene expression profiles. Nat. Rev. Genet. 9, 575–581 (doi:10.1038/nrg2383) [DOI] [PubMed] [Google Scholar]
- 52.Aubin-Horth N, Renn SCP. 2009. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol. Ecol. 18, 3763–3780. (:10.1111/j.1365-294X.2009.04313.x) [DOI] [PubMed] [Google Scholar]
- 53.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (doi:10.1038/nmeth.1226) [DOI] [PubMed] [Google Scholar]
- 54.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517 (doi:10.1101/gr.079558.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farr JA, Travis J. 1989. The effect of ontogenetic experience on variation in growth, maturation, and sexual behavior in the sailfin molly, Poecilia latipinna (Pisces: Poeciliidae). Environ. Biol. Fish. 26, 39–48 (doi:10.1007/BF00002474) [Google Scholar]
- 56.Ptacek MB, Travis J. 1997. Mate choice in the Sailfin molly, Poecilia latipinna. Evolution 51, 1217–1231 (doi:10.1006/anbe.2001.1982) [DOI] [PubMed] [Google Scholar]
- 57.Sumner IT, Travis J, Johnson CD. 1994. Methods of female fertility advertisement and variation among males in responsiveness in the sailfin molly (Poecilia latipinna). Copeia 1994, 27–34 (doi:10.2307/1446667) [Google Scholar]
- 58.Altmann J. 1974. Observational study of behavior: sampling methods. Behaviour 49, 227–267 (doi:10.2307/4533591) [DOI] [PubMed] [Google Scholar]
- 59.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (doi:10.1186/gb-2010-11-3-r25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inc SI. 2011. SAS/STAT 9.3 User‘s guide. Cary, NC: SAS Institute [Google Scholar]
- 61.Alexa A, Rahnenführer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (doi:10.1093/bioinformatics/btl140) [DOI] [PubMed] [Google Scholar]
- 62.Whitfield CW, Cziko A-M, Robinson GE. 2003. Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296–299 (doi:10.1126/science.1086807) [DOI] [PubMed] [Google Scholar]
- 63.Wagner A. 2005. Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 22, 1365–1374 (doi:10.1093/molbev/msi126) [DOI] [PubMed] [Google Scholar]
- 64.Stoebel DM, Dean AM, Dykhuizen DE. 2008. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178, 1653–1660 (doi:10.1534/genetics.107.085399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lang GI, Murray AW, Botstein D. 2009. The cost of gene expression underlies a fitness trade-off in yeast. Proc. Natl Acad. Sci. USA 106, 5755–5760 (doi:10.1073/pnas.0901620106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eames M, Kortemme T. 2012. Cost–benefit tradeoffs in engineered lac operons. Science 336, 911–915 (doi:10.1126/science.1219083) [DOI] [PubMed] [Google Scholar]
- 67.Krebs RA, Feder ME. 1997. Deleterious consequences of Hsp70 overexpression in Drosophila melanogaster larvae. Cell Stress Chaperones 2, 60–71 (doi:10.1379/1466-1268(1997)002<0060:DCOHOI>2.3.CO;2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kotrschal A, Rogell B, Bundsen A, Svensson B, Zajitschek S, Brännström I, Immler S, Maklakov AA, Kolm N. 2013. Artificial selection on relative brain size in the guppy reveals costs and benefits of evolving a larger brain. Curr. Biol. 23, 168–171 (doi:10.1016/j.cub.2012.11.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mery F, Kawecki TJ. 2004. An operating cost of learning in Drosophila melanogaster. Anim. Behav. 68, 589–598 (doi:10.1016/j.anbehav.2003.12.005) [Google Scholar]
- 70.Mery F, Kawecki TJ. 2003. A fitness cost of learning ability in Drosophila melanogaster. Proc. R. Soc. Lond. B 270, 2465–2469 (doi:10.1098/rspb.2003.2548) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Morphological and behavioural data used in this paper are available in Dryad (doi:10.5061/dryad.4qn6n). RNA-Seq data are available in NCBI Short Read Archive (accession SRP036185).


