Abstract
Research on subterranean organisms has focused on the colonization process and some of the associated phenotypic changes, but little is known on the long-term evolutionary dynamics of subterranean lineages and the origin of some highly specialized complex characters. One of the most extreme modifications is the reduction of the number of larval instars in some Leptodirini beetles from the ancestral 3 to 2 and ultimately a single instar. This reduction is usually assumed to have occurred independently multiple times within the same lineage and geographical area, but its evolution has never been studied in a phylogenetic framework. Using a comprehensive molecular phylogeny, we found a low number of independent origins of the reduction in the number of instars, with a single transition, dated to the Oligocene–Miocene, from 3 to 2 and then 1 instar in the Pyrenees, the best-studied area. In the Pyrenees, the 1-instar lineage had a diversification rate (0.22 diversification events per lineage per million years) significantly higher than that of 3- or 2-instar lineages (0.10), and similar to that seen in other Coleopteran radiations. Far from being evolutionary dead-ends, ancient lineages fully adapted to subterranean life seem able to persist and diversify over long evolutionary periods.
Keywords: diversification, larval development, life cycle, speciation, subterranean environment
1. Introduction
Subterranean species are an ideal model for the study of phenotypic and ecological specialization, and the study of the changes associated with underground colonization have contributed many new insights into evolutionary biology and physiology in recent years [1–3]. Here, focus has usually been on the process of colonization, with less attention paid to macroevolutionary questions such as the origin of lineages with multiple subterranean species or their diversification dynamics. Despite recent work suggesting the possibility of evolutionary radiations of subterranean aquatic organisms [4], the prevailing view in the case of terrestrial organisms is still that once a lineage has adapted to the subterranean environment it becomes confined to a limited geographical area, being unable to expand or diversify other than at a very local scale [5–7]. The existence of ancient groups in which all species are subterranean and morphologically and ecologically highly specialized (i.e. strictly troglobitic) is traditionally interpreted as resulting from multiple independent colonizations, followed by the extinction of related surface species, supposedly driven by global climatic or ecological changes [5–9]. Although there is no doubt that in many disparate groups there were multiple independent subterranean colonization events, resulting in convergent phenotypes [10–13], recent work on some Pyrenean beetles proposed a single origin of subterranean adaptations in ancient, diverse and geographically widespread terrestrial lineages [14,15], even with the possibility of range expansions of subterranean species over non-carstified areas [16]. This single origin would imply that highly specialized, troglobitic lineages are able to disperse, diversify and persist in the subterranean environment for long evolutionary periods.
The reconstructed evolution of more complex characters that differ between fully subterranean species should allow one to distinguish between the two scenarios outlined above. When all species in a lineage share a complex character related to their subterranean habit, to assume their independent evolution from surface ancestors will not only require the extinction of all these surface relatives, but also of all subterranean species with intermediate degrees of development of the character. The alternative would be a concerted evolution of all surviving species to reach the same phenotype [17], which is equally unlikely.
One of these complex characters is the modification of the life cycle observed in some subterranean species of Coleoptera of the tribe Leptodirini, characterized by a reduction in the number of larval instars. The Leptodirini, with around 900 species, is one of the largest groups of insects in which most species are found in the subterranean environment [18]. Most of them show all the typical modifications associated to the subterranean life (lack of eyes and membranous wings, depigmentation and an elongation of body and appendixes), and have very restricted geographical distributions [18]. A number of species have also severely altered life cycles, with profound physiological changes [19,20]. While species living in forest litter or shallow soil have the typical life cycle of Coleoptera, with 3 larval instars [21], some cave species show a reduction to 2 or 1 larval instars. Species with 3 larval instars lay many small eggs and the larvae feed actively, growing and moulting until they pupate. In the so-called ‘intermediate’ life-cycle type [19], observed only in strictly subterranean species, females lay a reduced number of medium-sized eggs and the larval period is reduced to 2 larval instars with a mobile, free-living feeding phase. In the third and most modified type of life cycle (‘contracted’, also found exclusively in subterranean species) there is a single larval instar. The larva hatches from a large (macrolecital) egg, where the copious yolk provides sufficient nutrients to complete larval development without feeding. The mobility of such larva is also strongly limited, and a few days after hatching they build a solid chamber (‘logette’) of sand and gravel in which they pass the remainder of larval development, immobile.
These modifications are traditionally considered to result from multiple evolutionary processes originating from a typical 3-instar cycle and taking place in each species independently, even within the same genus. However, their evolution has never been studied in a phylogenetic context. Here, we use a comprehensive molecular phylogeny of the major lineages of Leptodirini, including most of the species for which life-cycle data are available (i) to trace their evolution and establish the number of independent origins of modified life cycles and (ii) to test the effect of changes in life cycle on diversifications rates. We focus on the Pyrenean clade of species, previously found to be monophyletic [15], and for which most data are available both for life cycle and the taxonomic coverage of the phylogeny.
2. Material and methods
(a). Life-cycle data, taxon sampling and phylogenetic analyses
Specimens were collected by direct search or with the use of baits and stored in pure ethanol. We obtained material from all but seven species for which the life cycle was known, with a special focus on the fauna of the Pyrenees (see the electronic supplementary material, tables S1 and S2). DNA extractions of single specimens were non-destructive, using either a phenol–chloroform method or commercial kits (mostly DNeasy Tissue Kit, Qiagen, Hilden, Germany) following the manufacturers’ instructions. Vouchers are kept at the Museo Nacional de Ciencias Naturales, Madrid (MNCN) and the Institute of Evolutionary Biology, Barcelona (IBE). We amplified and sequenced fragments of the mitochondrial (five genes: cox1—amplified in two fragments—cob, rrnL, trnL, nad1—the last three amplified in a continuous single fragment) and nuclear genomes (two genes: SSU and LSU) (see [16] for primers and PCR conditions). New sequences have been deposited in GenBank (EMBL) with accession numbers HG915308–HG915710 (electronic supplementary material, table S2).
Two datasets were compiled: (i) an extended dataset, including all species with known life cycles and a representative sample of 141 species of Leptodirini from their respective geographical areas, using the genus Platycholeus (Western North America) to root the tree [12]; (ii) the Pyrenean clade, with a comprehensive dataset of the Pyrenean lineage (according to [15] and the results of this study).
We aligned length variable (i.e. ribosomal) sequences with MAFFT v. 7 and the Q-INS-i algorithm, which considers the secondary structure [22] and analysed the extended dataset with Bayesian probabilities (as implemented in MrBayes v. 3.2 [23]) and a fast maximum-likelihood algorithm (as implemented in RAxML v. 7.0 [24]). In both cases, we used a partition by gene fragments, pooling the sequence of the genes rrnL and trnL, and using a GTR + I + G model with unlinked parameters for each partition. We used the best of 100 RAxML replicates as our preferred ML topology. Node support was measured with 1000 fast bootstrap replicates using the CAT approximation [24]. To obtain an ultrametric tree we used BEAST v. 1.7 [25] with an uncorrelated lognormal clock and a Yule speciation process. We used the same partition and evolutionary model as in the phylogenetic analyses, but linked the evolutionary rates of the mitochondrial protein coding genes and the two nuclear ribosomal genes respectively to reduce the number of free parameters. We used the tectonic separation between the Corso-Sardinian plate and the European continent as a calibration point, with a normal distribution for the prior of the age of the node separating the Sardinian clade from its continental sister (as obtained in the MrBayes analyses) with an average of 33 Ma and a standard deviation of 1 Myr, following [15]. The analyses were run for 100 million generations, with a burn-in of 10%. In all Bayesian analyses convergence was assessed with the effective sample size in TRACER v. 1.5 [25].
(b). Ancestral state reconstruction and rates of diversification
We restricted the detailed analyses of the evolution of the life cycle to the Pyrenean clade of Leptodirini, as it included the highest number of species for which the life cycle was known and the group for which we had the most comprehensive sampling (electronic supplementary material, table S1). We reconstructed the ancestral character states for the number of larval instars using BEAST to account for topological uncertainty in the phylogenetic tree. We rooted and dated the tree according to the results of the extended dataset, using the estimated average age of the two basal nodes as calibration points. We used an asymmetrical matrix of character change rates and applied other settings as in the analyses of the extended dataset.
To estimate diversification rates within the Pyrenean clade, we associated the species not included in the phylogeny to the less inclusive clade of our phylogeny according to their current taxonomy. We then considered these clades as unresolved terminals in a backbone tree (electronic supplementary material, figure S3). We did not consider subspecies owing to their uncertain taxonomic status, although the number of subspecies is higher among the 1-instar taxa [26], so any potential bias would be against our conclusions. We analysed the backbone tree (with missing species and species with missing information on the life cycle; electronic supplementary material, tables S1 and S2, and figure S3) with MEDUSA from the GEIGER 1.3-1 package in R [27]. MEDUSA fits a birth–death model to different clades of an ultrametric tree, optimizing the complexity and fit of the model with the Akaike information criterion (AIC) and identifying the branches with significant shifts in diversification rates. To account for topological uncertainty, we resampled 1000 trees from the stationary set of trees found in the analysis of the Pyrenean dataset in BEAST using LOGCOMBINER [25], pruned the terminals to fit the unresolved backbone tree using R and ran MEDUSA on them.
For the estimation of the speciation and extinction rates associated with each type of larval development and the rates of character state change we used the binary state speciation and extinction method (BiSSE), which unites trait evolution and species diversification in the same birth–death models [28]. We used the modification of FitzJohn et al. [29] implemented in the R package DIVERSITREE [30] to allow the inclusion of incompletely resolved terminals and missing character state data. We obtained Bayesian estimates of the parameters using MCMC as implemented in DIVERSITREE, with runs of 10 000 steps and a burn-in of 5000. To account for topological uncertainty, we ran the selected optimal model of BiSSE on the same set of 1000 trees used for MEDUSA above. As BiSSE uses only binary data, we pooled the character states in all three possible combinations for comparison (1 instar versus 2 and 3 instars; 2 versus 1 and 3; and 3 versus 1 and 2).
FitzJohn [30] extended the BiSSE method to allow the use of multistate characters (MuSSE, multistate speciation and extinction), also with the possibility of using incompletely resolved terminals and missing data. In preliminary tests using MuSSE the high number of unresolved terminals and missing data prevented convergence of the models, so we built 1000 trees with randomly resolved terminal clades using MESQUITE and analysed them in MuSSE. In order to try to reduce the number of parameters, we compared the full model with a model with no extinction and assuming the impossibility of reversals in the evolution of the life cycle, and used this reduced model when differences were not significant. To test for the effect of missing life-cycle data we used, in addition to the matrix with known character states only, two matrices with different levels of missing data (electronic supplementary material, table S4).
3. Results
(a). Phylogenetic origin of the modifications in the life cycle
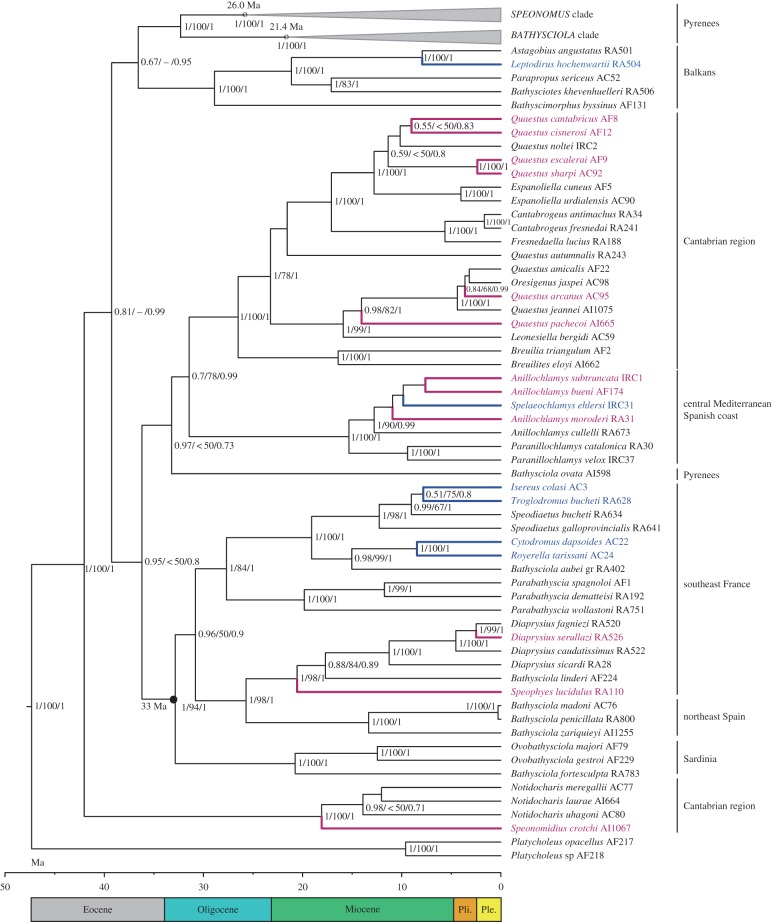
The monophyly of the Mediterranean Leptodirini (i.e. with the exclusion of the American Platycholeus) and of the lineages of each of the main geographical regions were very well supported, although the relationships among these lineages were not (figure 1). Topological differences between the trees obtained by different methods (Bayesian probabilities and ML) were limited to poorly supported nodes, mostly at the backbone of the tree (figure 1; electronic supplementary material, figure S1).
Figure 1.
Calibrated ultrametric tree obtained with BEAST with the extended dataset. Black circle, node used for calibration (vicariant split between the Sardinian and the continental clade). Numbers at nodes, posterior probability of BEAST/maximum-likelihood bootstrap support in RAxML/posterior probability in MrBayes. Species of the Pyrenean lineage were collapsed in the Speonomus and Bathysciola clades (see figure 2). Species with known life cycle are coloured: in blue, species with larvae with a 1-instar cycle; in purple, a 2-instar cycle.
For the calibration of the tree in BEAST, we used the node linking the Sardinian clade with Leptodirini from the Mediterranean coast of France (including one species in northwest Catalonia; figure 1), which resulted in mean rates for the protein coding, ribosomal mitochondrial and ribosomal nuclear genes of 0.015, 0.006 and 0.004 substitutions per site per Myr, respectively. The Mediterranean Leptodirini were estimated to have an Early Eocene origin (42 ± 5 Ma), and the main lineages in each of the geographical areas a late Eocene–Oligocene origin (figure 1).
In all the main lineages, corresponding to well-defined geographical areas, there was at least one transition to a 2-instar or a 1-instar cycle, but there was no evidence of more than 1 (figure 1).
(b). Ancestral state reconstruction in the Pyrenean lineage
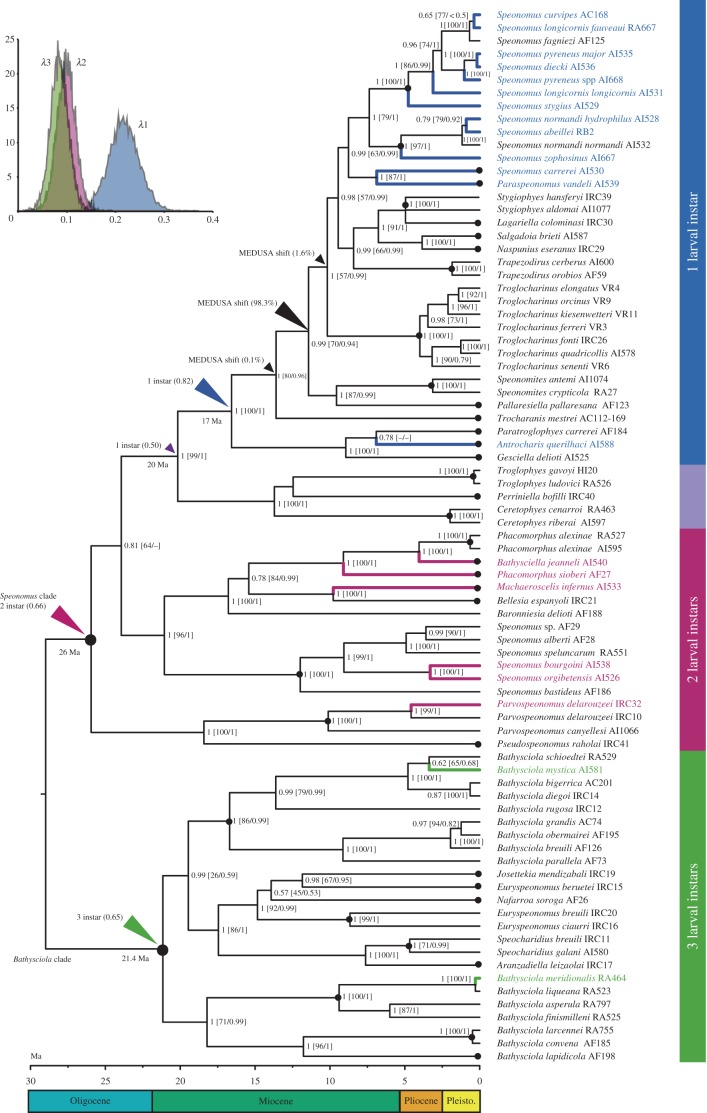
We included 81 taxa of the Pyrenean lineage, with examples of all known genera and main species groups (electronic supplementary material, table S2). To root and calibrate the tree we constrained the two main clades, referred to as the Speonomus and Bathysciola clades, according to the results of the previous analysis (figures 1 and 2). The Bathysciola clade included the only two species confirmed to have 3 larval instars, and was reconstructed to have a 3-instar ancestor. The Speonomus clade was reconstructed to have a 2-instar larval ancestor, with a single transition to a 1-instar larvae around 20 Ma. The Speonomus clade was reconstructed to have a 2-instar larval ancestor, with a single transition to a 1-instar larvae at either 17 or 20 Ma. The clade between the 2- and 1-instar species, with three genera without life-cycle data, was reconstructed to have a 1-instar ancestor at 20 Ma, but this was with a probability (0.50) very similar to that of a 2-instar ancestor (0.38). These results were essentially unaltered by uncertain life-cycle data, with only one additional transition if Parvospeonomus canyellesi was considered to have 1 instar, affecting only this species (see below; see also electronic supplementary material, table S1 and figure S2).
Figure 2.
Calibrated ultrametric tree obtained with BEAST with the Pyrenean dataset. Large black circles, nodes used for the calibration (see figure 1). Small black circles, nodes collapsed in the backbone tree used for the diversification analyses. Numbers at nodes, posterior probability of BEAST/(maximum-likelihood bootstrap support in RAxML/posterior probability in MrBayes, both with the extended dataset). Black arrows, reconstructed shifts in the diversification rate with MEDUSA, with percentage of the 1000 post-burn-in trees in which the transition was found. Coloured arrows, shifts in the ancestral reconstruction of the number of larval instars with BEAST, with the associated probabilities. Insert: Frequency distribution of the estimated speciation rates with MuSSE associated to the three types of development. In blue, species with larvae with 1 instar; purple, 2 instars; green, 3 instars.
(c). Shifts in diversification in the Pyrenean lineage
To test for differences in diversification rates within the Pyrenean lineage we constructed an unresolved backbone tree with 38 terminals to which the 141 known species of the lineage could be associated. These terminals corresponded to monophyletic genera plus groups of species for para- or polyphyletic genera (figure 2; electronic supplementary material, figure S3). Using maximum-likelihood contrasts, we found a single significant change in diversification rate in a selection of 1000 trees from the stationary phase of a BEAST run. This shift was always within the clade having 1-instar larvae, and in the same node in 983 of the trees (figure 2). Of the remaining trees, 16 had a significant shift in the same node but excluding two genera, and one also in the same node but including the genus Trocharanis (electronic supplementary material, figure S4). The diversification rate estimated for the fast part of the tree was always more than twice that of the slowest part (table 1).
Table 1.
Estimated values of the parameters of the preferred diversification models tested. λ1, λ2, λ3 and μ1, μ2, speciation (λ) and extinction (μ) rates associated to the lineages with different numbers of larval instars, respectively (BiSSE and MuSSE) or the section of the tree with higher and lower diversification rates (MEDUSA); q, transition rates between different states of the trait ‘number of instars’. In the BiSSE models, λ2, μ2 and q2 combine species with 2 and 3 larval instars. In square brackets, 95% CIs of the 1000 resampled trees in BEAST (MEDUSA and BiSSE), 1000 randomly resolved trees of the unresolved backbone (MuSSE) or the 10 000 MCMC generations used for parameter estimation using the single, optimal tree (BiSSE and MuSSE). See electronic supplementary material, tables S3 and S4 for model selection statistics.
| λ3 | λ2 | λ1 | μ2 | μ1 | q3–2 | q2–1 | |
|---|---|---|---|---|---|---|---|
| MEDUSA1000 | n.a. | 0.11 [0.10–0.12] | 0.28 [0.24–0.31] | <0.0001 | <0.0001 | n.a. | n.a. |
| BiSSE-MCMC | n.a. | 0.13 [0.09–0.18] | 0.27 [0.18–0.38] | 0.027 [0–0.08] | 0.058 [0–0.16] | n.a. | 0.008 [0.000–0.020] |
| BiSSE-1000 | n.a. | 0.12 [0.11–0.13] | 0.24 [0.21–0.27] | <0.0001 | <0.0001 | n.a. | 0.004 [0.004–0.005] |
| MuSSE-MCMC | 0.09 [0.06–0.13] | 0.11 [0.08–0.14] | 0.24 [0.18–0.29] | n.a. | n.a. | 0.006 [0.0001–0.0136] | 0.006 [0.0001–0.0133] |
| MuSSE-1000 | 0.09 [0.087–0.093] | 0.11 [0.10–0.11] | 0.23 [0.22–0.23] | n.a. | n.a. | 0.003 [0.0028–0.0030] | 0.003 [0.0028–0.0029] |
(d). Association between number of instars and diversification rates
All BiSSE models including differences in diversification rates among species with different life cycles were significantly better than the assumption of no differences, with the exception of the model pooling species with 1 and 2 versus 3 instars (electronic supplementary material, table S3). Of the different combinations of species with different cycles, the model with the best likelihood grouped species with 3 and 2 instars versus species with 1 instar. The significance of this model was not affected by the topological uncertainty of the tree (mean of the 1000 BEAST trees p = 0.012 ± 0.0002, with only three trees with p > 0.05) or the inclusion of uncertain life-cycle data. For this comparison, there were also no significant differences between the models with reversible or irreversible changes, as measured with a likelihood ratio test (electronic supplementary material, table S3), again irrespective of the topologies (p > 0.95 in all 1000 trees). In all cases, both speciation and extinction rates of the fast diversifying lineages (always including species with 1 instar) was twice that of the slow-diversifying lineages (always including species with 3 instars). The 95% CIs of the Bayesian estimates of the two speciation rates with the best model (1 versus 2 and 3 instars, with irreversible character state transitions) did not overlap, and were very robust to topological variations in the tree or the inclusion of uncertain life-cycle data (table 1; electronic supplementary material, table S3).
For the implementation of multistate character models (MuSSE), we first used the backbone phylogeny with randomly resolved terminals. MuSSE models were not significantly different when only known life-cycle data were included or when species were assumed to have the character state as reconstructed with Bayesian probabilities in BEAST. For all comparisons there were also no significant differences between a full model (i.e. with 12 parameters: three speciation rates, three extinction rates and six transitions between the three character states) and a model with no extinction and irreversible change (five parameters; electronic supplementary material, table S4). To reduce the level of missing data we then used 1000 trees with randomly resolved terminals and with the reconstructed number of instars, except for the intermediate clade between two and three instars, which were left as unknown. Again, in all trees the full and reduced models (no extinction, irreversible change) were not significantly different (p > 0.9 for all 1000 trees). The reduced model with three speciation rates was, however, always significantly better than a model with equal probability of speciation (p < 0.0001 for all 1000 trees). The estimated speciation rate of the 1-instar clade (0.23 species per lineage per Myr) was higher than that of the lineages with 2 and 3 instars (0.11 and 0.09, respectively), with no significant difference between the latter (figure 2, table 1; electronic supplementary material, table S4).
4. Discussion
Our results support a single origin of life-cycle modification in the Pyrenean lineage of subterranean Leptodirini beetles, with a single transition from the ancestral 3-instar to a 2-instar cycle in the Late Oligocene, and a subsequent single transition to 1 instar in the Early Miocene. Our results are also compatible with a single origin of such modifications in all the other major lineages of the tribe, although the limited knowledge of their life cycles does not allow this to be tested more robustly. Contrary to expectations, the transition to a 1-instar life cycle, the most modified and specialized phenotype, was associated in the Pyrenean lineage with a highly significant increase in diversification rate.
(a). Single origin of the life cycle modifications in the Pyrenean lineage
We found clear geographical structuring within the Leptodirini, in agreement with previous results for the same group and for other subterranean organisms [11,14,15,31]. Even if within each of these geographically restricted lineages there were species known to have a modified life cycle, the required minimum number of independent transitions according to our phylogeny was surprisingly low. In the best-known area, the Pyrenees, we reconstructed a single transition from 3 to 2 and another from 2 to 1 instars. In southern France, and despite the high level of missing data, all species known to have a contracted cycle form a monophyletic lineage, also suggesting a single transition. The progressive reduction of the number of instars through the phylogeny of the Pyrenean Leptodirini is strong evidence of a single, ancient origin of their subterranean adaptations, as suggested by Ribera et al. [15] and contrary to the common view hypothesizing multiple origins, followed by extinction of epigean lineages [5–9].
There are several caveats to be considered in evaluating the robustness of our conclusions, in particular the amount of missing data and the uncertainty in some of the life-cycle information. By adding more data it would always be possible to find additional transitions, but these would most probably affect terminal species, and have no consequences for our estimated diversification rates. There is also indirect evidence supporting our reconstructed evolution of larval development. While it is common to find larvae of the species reconstructed as having a 2-instar cycle (in fact, many were known before their cycles could be studied in the laboratory, e.g. [32]; electronic supplementary material, table S1), we are not aware of any instance in which a single larva of a species of the reconstructed 1-instar clade has ever been found in a cave, despite a continuous search for decades in caves in which some of these species are very common [38] (J.F. 2013, unpublished observations; A. Faille & C. Bourdeau 2013, personal communication). This strongly suggests that the larvae of these species do not have an active phase, as is characteristic of the 1-instar cycle.
For the detailed reconstruction of the life cycle in the Pyrenean lineage we used only data from papers reporting the complete life cycle, from egg to adult, which are basically those produced at the Laboratoire Souterrain in Moulis (electronic supplementary material, table S1). There are other incomplete reports of life-cycle data, and with a single exception they agree with our reconstruction. This exception is the species Parvospeonomus canyellesi, reconstructed as having a 2-instar cycle in our phylogeny but reported as having 1 instar [33]. This was mainly based on the fact that the first instar larva was relatively large, inactive and non-feeding. The type of life cycle could not be established experimentally, and the size of the first instar larvae is actually of little value to predict the number of instars (I.R. & A.C. 2013, unpublished data). In any case, the consideration of P. canyellesi as having a 1-instar cycle would only add an additional terminal transition within the Pyrenean lineage, leaving the reconstruction of ancestral states and the estimation of diversification rates unaltered (electronic supplementary material, table S5).
There could also be the possibility of taxonomic bias if there was an over-splitting of the 1-instar clade in comparison with 3- and 2-instar species. In Leptodirini species delimitation is traditionally based on external morphology, and in particular on the male aedeagus [18] (as common in many groups of Coleoptera). The general morphology and structure of the genitalia is very similar in all species of Leptodirini and there is no reason to assume differences in taxonomic treatment. In the cases in which molecular data are available the general trend is the discovery of cryptic diversity within recognized species, frequently in agreement with named subspecies [16]. As noted in the Material and methods section, the number of subspecies was not considered in the species counts, but it is higher in 1-instar than in 2- or 3-instar species [18,26]. In other words, accounting for cryptic diversity is only likely to increase the observed difference in diversification rates.
(b). Diversification rates
The estimated diversification rate for the 1-instar Pyrenean clade (approx. 0.2–0.3 species per lineage per Myr) is well above the overall Coleopteran rate (0.05–0.07 [34]), and fully comparable with the estimated rates of a number of Coleopteran radiations, also ranging from 0.2 to 0.3 [35–38]. The estimated rate was very consistent across the different methods, analytical conditions and uncertainties in the life-cycle data. It is also likely to be conservative, as there seems to be a high degree of cryptic diversity among the Pyrenean Leptodirini [15,16], as is generally the case in subterranean species [39].
Our results do not allow us to unambiguously link the increase in speciation and extinction rates and the development of a 1-instar cycle, both because the transition to a contracted life cycle happened previous to the shift in diversification and since it is a single evolutionary event. There are a number of uncertainties in the identification of drivers of diversification shifts using phylogenies [40], and a temporal delay may be observed for different reasons. In any case, the evolution of a contracted life cycle, usually considered an extreme specialization to the subterranean environment, was not an evolutionary dead-end for the lineage, but instead allowed its long-term persistence and diversification.
The reduction of the number of larval instars has been interpreted as a strategy to escape predation at the most vulnerable developmental stage [41], which may result in a reduced extinction probability and an increased diversification rate. The building of a refuge of sand and gravel would offer a high degree of protection to an immobile larva freed from the need to search for food, although there are no data on the rates of predation in caves compared with the surface, or between caves occupied by 2- or 1-instar species.
An alternative possibility is that a non-feeding larva may allow the species to survive in more extreme environments, as the more mobile adult can forage over a larger area and provide enough resources to complete the whole development [5]. In the Pyrenees, scarcer resources are usually associated with caves at higher altitudes or with more isolated, deeper areas within the same subterranean system. The increased diversification may then be linked to the colonization of these deeper regions or of new areas, requiring the geographical expansion of the species. Highly modified species in the deep subterranean environment have usually low dispersal abilities and very restricted, allopatric distributions (most often they are found in a single cave or in nearby caves of the same geological system [18,26]), but the global distribution of the Pyrenean 1-instar clade, including the central Pyrenees and some coastal areas near Barcelona [15], demonstrates that there were at least occasional range expansions to nearby areas [16].
Subterranean species have frequently been used as examples of evolutionary dead-ends (Darwin's ‘wrecks of ancient life’ [42], or Jeannel's ‘fossiles vivants’ [8]), locked in their restricted habitat by their strong morphological and physiological specializations. This work shows that, far from being an evolutionary dead-end, the transition to a 1-instar cycle in the Pyrenean Leptodirini did not lead to isolated forms but to a highly diverse and relatively widespread radiation with a very dynamic evolutionary history.
Acknowledgements
We thank V. Assing, M. Baena, C. Bourdeau, A. Casale, A. Castro, P. Déliot, F. Fadrique, A. Faille, C. Hernando, A. Kilian and I. Zabalegui for sending material for study or information; A. Izquerdo, A. Faille and R. Alonso for laboratory work; and C. Bourdeau, A. Casale, A. Faille and V. Rizzo for multiple discussions on the evolution of the subterranean fauna. We also thank A. Minelli, X. Bellés, D. T. Bilton and two anonymous referees for comments to the manuscript. Laboratory work was conducted in the MNCN (Madrid) and IBE (Barcelona).
Data accessibility
All new sequences have been submitted to the EMBL database. All other primary data are included either in the main text or in the electronic supplementary material.
Funding statement
This work was funded by projects CGL2007-61943 and CGL2006-11403 from the Ministerio de Ciencia e Innovación (MICINN, Spain) to A.C.
References
- 1.Yamamoto Y, Stock DW, Jeffery WR. 2004. Hedgehog signalling controls eye degeneration in blind cavefish. Nature 431, 844–847 (doi:10.1038/nature02864) [DOI] [PubMed] [Google Scholar]
- 2.Duboué ER, Keene AC, Borowsky RL. 2011. Evolutionary convergence on sleep loss in cavefish populations. Curr. Biol. 21, 671–676 (doi:10.1016/j.cub.2011.03.020) [DOI] [PubMed] [Google Scholar]
- 3.Protas ME, Trontelj P, Patel NH. 2011. Genetic basis of eye and pigment loss in the cave Crustacean, Asellus aquaticus. Proc. Natl Acad. Sci. USA 108, 5702–5707 (doi/10.1073/pnas.1013850108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holsinger JR. 2000. Ecological derivation, colonization, and speciation. In Ecosystems of the world. 30. Subterranean ecosystems (eds Wilkens H, Culver DC, Humphreys WF.), pp. 399–415 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- 5.Poulson TL, White WB. 1969. The cave environment. Science 165, 971–981 (doi:10.1126/science.165.3897.971) [DOI] [PubMed] [Google Scholar]
- 6.Barr TC, Holsinger JR. 1985. Speciation in cave faunas. Annu. Rev. Ecol. Syst. 16, 313–337 (doi:10.1146/annurev.es.16.110185.001525) [Google Scholar]
- 7.Culver DC, Pipan T. 2009. The biology of caves and other subterranean habitats. Oxford, UK: Oxford University Press [Google Scholar]
- 8.Jeannel R. 1943. Les fossiles vivants des cavernes. Paris, France: Gallimard [Google Scholar]
- 9.Zhang Y, Li S. 2013. Ancient lineage, young troglobites: recent colonization of caves by Nesticella spiders. BMC Evol. Biol. 13, 183 (doi:10.1186/1471-2148-13-183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christiansen K. 1961. Convergence and parallelism in cave Entomobryinae. Evolution 15, 288–301 (doi:10.2307/2406229) [Google Scholar]
- 11.Faille A, Casale A, Ribera I. 2011. Phylogenetic relationships of Western Mediterranean subterranean Trechini groundbeetles (Coleoptera: Carabidae). Zool. Scr. 40, 282–295 (doi:10.1163/187631201X00191) [Google Scholar]
- 12.Fresneda J, Grebennikov VV, Ribera I. 2011. The phylogenetic and geographic limits of Leptodirini (Insecta: Coleoptera: Leiodidae: Cholevinae), with a description of Sciaphyes shestakovi sp.n. from the Russian Far East. Arthropod Syst. Phylo. 69, 99–123 [Google Scholar]
- 13.Trontelj P, Blejec A, Fišer C. 2012. Ecomorphological convergence of cave communities. Evolution 66, 3852–3865 (doi:10.1111/j.1558-5646.2012.01734.x) [DOI] [PubMed] [Google Scholar]
- 14.Faille A, Ribera I, Deharveng L, Bourdeau C, Garnerye L, Quéinnec E, Deuve TA. 2010. A molecular phylogeny shows the single origin of the Pyrenean subterranean Trechini ground beetles (Coleoptera: Carabidae). Mol. Phylogenet. Evol. 54, 97–106 (doi:10.1016/j.ympev.2009.10.008) [DOI] [PubMed] [Google Scholar]
- 15.Ribera I, Fresneda J, Bucur R, Izquerdo A, Vogler AP, Salgado JM, Cieslak A. 2010. Ancient origin of a western Mediterranean radiation of subterranean beetles. BMC Evol. Biol. 10, 1–14 (doi:10.1186/1471-2148-10-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizzo V, Comas J, Fadrique F, Fresneda J, Ribera I. 2013. Early Pliocene range expansion of a clade of subterranean Pyrenean beetles. J. Biogeogr. 40, 1861–1873 (doi:10.1111/jbi.12139) [Google Scholar]
- 17.Jeannel R. 1950. La marche de l’évolution. Paris, France: Publications du Muséum national d'histoire naturelle [Google Scholar]
- 18.Salgado JM, Blas M, Fresneda J. 2008. Fauna Iberica, vol. 31 Coleoptera: Cholevidae Madrid, Spain: Editorial CSIC [Google Scholar]
- 19.Deleurance S. 1963. Recherches sur les Coléoptères troglobies de la sous-famille Bathysciinae. Ann. Sci. Nat. Zool. Paris Ser. 12 1, 1–172 [Google Scholar]
- 20.Delay B. 1978. Milieu souterrain et écophysiologie de la reproduction et du développement des Coléoptères Bathysciinae hypogés. Mém. Biosp. 5, 1–349 [Google Scholar]
- 21.Minelli A, Fusco G. 2013. Arthropod post-embryonic development. In Arthropod biology and evolution (eds Minelli A, Boxshall G, Fusco G.), pp. 91–122 Berlin, Germany: Springer [Google Scholar]
- 22.Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298 (doi:10.1093/bib/bbn013) [DOI] [PubMed] [Google Scholar]
- 23.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 24.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 25.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (doi:10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perreau M. 2000. Catalogue des Coléoptères Leiodidae et Platypsyllinae. Mém. Soc. Ent. France 4, 1–460 [Google Scholar]
- 27.Alfaro ME, Santini F, Brock C, Alamillo H, Dornburg A, Rabosky DL, Carnevale G, Harmon LJ. 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13 410–13 414 (doi:10.1073/pnas.0811087106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710 (doi:10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 29.FitzJohn RG, Maddison WP, Otto SP. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611 (doi:10.1093/sysbio/syp067) [DOI] [PubMed] [Google Scholar]
- 30.FitzJohn RG. 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092 (doi:10.1111/j.2041-210X.2012.00234.x) [Google Scholar]
- 31.Fišer C, Sket B, Trontelj P. 2008. A phylogenetic perspective on 160 years of troubled taxonomy of Niphargus (Crustacea: Amphipoda). Zool. Scr. 37, 665–680 (doi:10.1111/j.1463-6409.2008.00347.x) [Google Scholar]
- 32.Glaçon S. 1955. Remarques sur la morphologie et la biologie de quelques larves de Bathysciinae cavernicoles. C.R. Acad. Sci. Paris 240, 679–681 [Google Scholar]
- 33.Bellés X, Delay B, Juberthie-Jupau L. 1986. Speonomus canyenesi, un Bathysciinae (Coleoptera, Catopidae) molt evolucionat endèmic del Montseny. J. Nat. Montseny 1, 69–73 [Google Scholar]
- 34.Hunt T, et al. 2007. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318, 1913–1916 (doi:10.1126/science.1146954) [DOI] [PubMed] [Google Scholar]
- 35.Barraclough TG, Vogler AP. 2002. Recent diversification rates in North American tiger beetles estimated from a dated mtDNA phylogenetic tree. Mol. Biol. Evol. 19, 1706–1716 (doi:10.1093/oxfordjournals.molbev.a003993) [DOI] [PubMed] [Google Scholar]
- 36.Trizzino M, Audisio PA, Antonini G, Mancini E, Ribera I. 2011. Molecular phylogeny and diversification of the ‘Haenydra’ lineage (Hydraenidae, genus Hydraena), a north-Mediterranean endemic-rich group of rheophilic Coleoptera. Mol. Phylogenet. Evol. 61, 772–783 (doi:10.1016/j.ympev.2011.07.020) [DOI] [PubMed] [Google Scholar]
- 37.Gómez-Zurita J, Sassi D, Cardoso A, Balke M. 2012. Evolution of Cryptocephalus leaf beetles related to C. sericeus (Coleoptera: Chrysomelidae) and the role of hybridization in generating species mtDNA paraphyly. Zool. Scr. 41, 47–67 (doi:10.1111/j.1463-6409.2011.00500.x) [Google Scholar]
- 38.Papadopoulou A, Cardoso A, Gómez-Zurita J. 2013. Diversity and diversification of Eumolpinae (Coleoptera: Chrysomelidae) in New Caledonia. Zool. J. Linn. Soc. 168, 473–495 (doi:10.1111/zoj.12039) [Google Scholar]
- 39.Trontelj P, Douady CJ, Fiser C, Gibert J, Goricki S, Lefebure T, Sket B, Zaksek V. 2009. A molecular test for cryptic diversity in ground water: how large are the ranges of macro-stygobionts? Freshw. Biol. 54, 727–744 (doi:10.1111/j.1365-2427.2007.01877.x) [Google Scholar]
- 40.Moore BR, Donoghue MJ. 2007. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. Am. Nat. 170(S2), S28–S55 (doi:10.1086/519460) [DOI] [PubMed] [Google Scholar]
- 41.Culver DC. 1982. Cave life: evolution and ecology. Cambridge, MA: Harvard University Press [Google Scholar]
- 42.Darwin C. 1859. On the origin of species. London, UK: John Murray [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All new sequences have been submitted to the EMBL database. All other primary data are included either in the main text or in the electronic supplementary material.