Abstract
Data on long-term circulation of pathogens in wildlife populations are seldom collected, and hence understanding of spatial–temporal variation in prevalence and genotypes is limited. Here, we analysed a long-term surveillance series on influenza A virus (IAV) in mallards collected at an important migratory stopover site from 2002 to 2010, and characterized seasonal dynamics in virus prevalence and subtype diversity. Prevalence dynamics were influenced by year, but retained a common pattern for all years whereby prevalence was low in spring and summer, but increased in early autumn with a first peak in August, and a second more pronounced peak during October–November. A total of 74 haemagglutinin (HA)/neuraminidase (NA) combinations were isolated, including all NA and most HA (H1–H12) subtypes. The most common subtype combinations were H4N6, H1N1, H2N3, H5N2, H6N2 and H11N9, and showed a clear linkage between specific HA and NA subtypes. Furthermore, there was a temporal structuring of subtypes within seasons based on HA phylogenetic relatedness. Dissimilar HA subtypes tended to have different temporal occurrence within seasons, where the subtypes that dominated in early autumn were rare in late autumn, and vice versa. This suggests that build-up of herd immunity affected IAV dynamics in this system.
Keywords: influenza A virus, mallards, prevalence, diversity, disease dynamics, host–pathogen interactions
1. Introduction
Influenza A viruses (IAV) infect a range of animal species, including humans, bats, swine, horses and seals [1]. However, most virus subtypes and the largest genetic variation are found in wild birds [1]. Birds associated with wetlands, such as waterfowl (Anseriformes), and shorebirds and gulls (Charadriiformes), are more commonly infected, while terrestrial birds, such as songbirds are seldom infected [1,2]. The vast majority of IAVs that circulate among wild birds cause mild infections in their natural hosts, and are termed low-pathogenic avian influenza (LPAI). However, these viruses may spill over to other species, including our domestic animals, where they can evolve increased pathogenicity, including highly pathogenic avian influenza (HPAI) or fowl plague in poultry [3], and pandemic influenza in humans [1]. The classification of IAVs is based on two antigenically important surface proteins, haemagglutinin (HA) and neuraminidase (NA). Currently, 16 HA and 9 NA protein variants have been detected in birds [4]. The IAV genome is segmented, and the eight RNA segments can be exchanged between co-infecting viruses in a process called reassortment, creating a potential 144 different HA/NA subtype combinations to occur [4]. A current challenge is to understand the spatial and temporal dynamics of the different subtypes and their maintenance in natural hosts.
The importance of waterfowl and shorebirds as hosts for LPAI viruses was recognized in the late 1960s [5], when the first IAV isolations and serological studies were carried out in wild birds. In waterfowl, especially in dabbling ducks of the genus Anas, nearly all HA and all NA variants have now been detected [2,6]. A striking property of IAV ecology is the immense variation at the subtype level in Anas hosts, where large-scale screening studies report large subtype diversity both within and between seasons [7–16].
Long-term studies of IAV in wild bird populations are rare, and most studies have either been of short duration, sampled several locations but with relatively few samples from each site, or had high effort at only a single time point. However, there seem to be repeatable seasonal patterns of occurrence of IAV across the Northern Hemisphere, with similar peaks in prevalence in North America, Europe and Asia [7,9,10,13].
Temporal prevalence patterns correspond with immunological status and breeding phenology of ducks; specifically, when a large proportion of immunologically naive hatch-year individuals are recruited to the population there is a peak of IAV. Consequently, IAV prevalence in ducks increases at premigratory gatherings during the autumn migration, and subsequently drops during winter once most individuals have experienced infections and developed an immune response [9,10,17]. Given the great antigenic subtype diversity and the large spatial–temporal variations commonly detected in IAV studies, longer time series conducted in a standardized way are needed to address epidemiological questions with higher accuracy.
In the present study we used data from a 9 year study of IAV occurrence in a migratory population of mallard ducks, Anas platyrhynchos, at a stopover site in southern Sweden to investigate long-term patterns in IAV prevalence and subtype variation. This study system has generated a number of targeted studies [4,9,10,18–20], which have advanced the understanding of LPAI and host–pathogen interactions. Here, we investigate central questions of LPAI epidemiology and use our long-term dataset consisting of more than 22 000 samples to characterize some fundamental epidemiological factors. Specifically, the aims of the study were to analyse the dynamics of IAV prevalence and diversity, and how these are related to seasonal variation in host biology and phenology of migration.
2. Material and methods
(a). Sample collection
The study site is situated at the southern-most point of the island Öland in the Baltic Sea, in southeast Sweden (56o12′ N 16o24′ E). At this site, a funnel live-duck trap operated by staff at the Ottenby Bird Observatory was used to trap and sample waterfowl. The time series started on 29 September 2002, and samples collected until 30 November 2010 were included in this study. Additionally, daily visual counts of birds staying in the Ottenby Nature Reserve were performed in 2009. The sampling protocol was approved by Linköping Animal Research Ethics Board (permit numbers 8-06, 34-06, 80-07, 111-11, 112-11).
The field season started in spring each year following ice melt (March–April) and continued until mid-December when the ice returned. Two different sampling methodologies were used: fresh faeces or cloacal swabs. The ducks were placed in single-use cardboard boxes, and if the duck defecated in the box, faecal material was collected with a sterile cotton-tipped applicator [9,21]. The time in the box varied for each individual depending on the number of trapped and handled birds on a given day, with a range from a few minutes up to 3 h in extreme cases. Cloacal swabs were taken from birds that did not defecate in boxes, by swirling a sterile swab in the cloaca. Swabs were stored in virus transport medium (Hanks’ balanced salt solution containing 0.5% lactalbumin, 10% glycerol, 200 U ml−1 penicillin, 200 µg ml−1 streptomycin, 100 U ml−1 polymyxin B sulfate, 250 µg ml−1 gentamycin and 50 U ml−1 nystatin; Sigma) at −70°C within 1–4 h of collection.
(b). Virus detection, isolation and characterization
Methods for screening, isolation and subtyping of samples can be found in previous publications [9]. Briefly, RNA from cloacal and faecal samples was extracted with two different automated systems, either the M48 robot (Qiagen, Germantown, MD, USA) (for the years 2006–2009), or the MagNA Pure 96 Extraction robot (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) for samples collected in 2010. Samples were screened by real-time reverse transcriptase polymerase chain reaction (RRT-PCR) assays targeting the IAV matrix gene using either LightCycler 1.5 (Roche) or StepOnePlus instrument (Applied BioSystems, NJ, USA) (see electronic supplementary material, table S1 and [9] for a summary). H5- and H7-specific RRT-PCRs were run for IAV matrix-gene positive samples, and the cleavage site of the HA was sequenced for H5 or H7 positive samples to screen for potential HPAI viruses, according to EC recommendations. Specific pathogen-free embryonated chicken eggs were inoculated according to standard methods for IAV propagation. A haemagglutination inhibition (HI) assay was used to characterize the HA subtype from isolates, while the NA subtype was obtained by partial sequencing of the NA gene, or by a SYBR-green-based RRT-PCR screening method for the Eurasian N3–N8 NA lineages [22]. New primers were designed for the N1, N2 and N9 subtypes owing to poor performance (electronic supplementary material, table S2, [20]).
(c). Seasonal trends in influenza A virus prevalence and migration
The R software [23] was used to compute all statistical tests and models. The sampling period was divided into weeks to depict migration intensity and IAV prevalence. All collected samples were included in the prevalence analyses, including data from recaptured individuals. Smoothed curves depicting seasonal variation in trapping and prevalence were obtained fitting trapping data and IAV matrix RRT-PCR results, respectively, using general additive models (GAMs) with the mgcv package, and including spline functions of week. Three models were considered: one where distinct spline functions were included for each year and where consequently a seasonal pattern was allowed to differ among years; a second model including a single spline under the assumption that the seasonal pattern was similar in all years; finally, in a third model, the pattern was considered constant over the season. In addition, all three models included a fixed effect of year as a categorical variable to account for inter-annual variation for each year separately, and for the seasonal trend smooth curve fitted to the data pooled over all years. An analysis of deviance (ANODEV) statistic was computed from the three models mentioned above to assess the proportion of seasonal variation in trapping and IAV prevalence that was accounted for the common pattern across years. The ANODEV statistic is thus an indicator of the consistency of seasonal patterns across years.
Immigration and site usage patterns of mallards were also characterized. First, the average number of newly ringed ducks per week was determined. The proportion of newly ringed birds (i.e. at first capture) among all captures was calculated per week, and smooth functions were fitted for each year, and for all years combined using GAMs as described earlier. Second, the time between the first and the last capture of individual mallards within season (i.e. in the same autumn, August–December) was used to approximate the time that mallards used the stopover site. This term, ‘length of stay’, was log-transformed to achieve normality and to test differences in mean values among years using ANOVA.
(d). Temporal haemagglutinin diversity
Putative seasonal effects on HA subtype diversity were investigated using vector generalized linear models (VGLMs) [24], within the vgam package in R [24]. VGLMs are generalized linear models dealing with polytomous responses. The response variable was the proportion of a specific subtype k (Prk) in infected individuals which could have k categorical outcomes. Owing to the high diversity of HA subtypes, we grouped all the HA subtypes detected into three classes based on their phylogeny [4]. The H1 class included the H1 clade (comprising the H1, H2, H5 and H6 subtypes) and the H9 clade (H8, H9 and H12 subtypes), the H3 class included the H3 clade and the H7 clade (basically the subtypes that belong to HA group 2: H3, H4, H10 and H7), and finally the H11 class included the H11 subtype [20]. This grouping allowed investigation of dynamics between major HA antigenic classes. At the same time it reduced the number of outcomes, preventing convergence problems. To further reduce convergence problems, seasonal variation was depicted using two week long time intervals and was included in the model as a continuous explanatory variable starting at 1 August of each year, as data were sparse in spring and summer months. The seasonal trend was modelled as a first-order polynomial. The modelling approach aimed to test whether there was: (i) a common seasonal variation of each of the HA classes between years, (ii) a difference in amplitude between years despite common seasonal variation of each HA class or (iii) no common pattern across years. The model with best fit to the data was selected using the Akaike information criterion corrected (AICc) for small sample sizes [25]; we considered models as having equivalent support for the data when the difference in their AICc values was less than 2. In this case, we followed the principle of parsimony [25] and retained the model with the least number of parameters. Details on model building can be found in the electronic supplementary material, methods S1 and table S4.
(e). Haemagglutinin and neuraminidase linkage
To evaluate the linkage between HA and NA variants, a contingency table with fully typed virus isolates, including only one isolate per infection for each individual bird, was tested for independence with the Fisher's exact test (implemented in the ‘fisher.test’ function of R) [26,27]. As the resulting table was large (12 × 9), the test algorithm was computationally intractable. The p-value for the null hypothesis was instead determined using a Monte Carlo approach (option ‘simulate.p.value = TRUE’ in the R fisher.test function). To examine the specific patterns of departure from independence, the standardized Pearson's residuals were used (electronic supplementary material, methods S2). In this analysis, positive residuals reflect an overrepresentation of observed cases compared with the number of expected cases under the assumption of independence between HA and NA combinations. Reciprocally, negative residuals reflect an underrepresentation of some combinations compared with the expected frequency.
3. Results
(a). Mallard migration and seasonal variation in influenza A virus prevalence
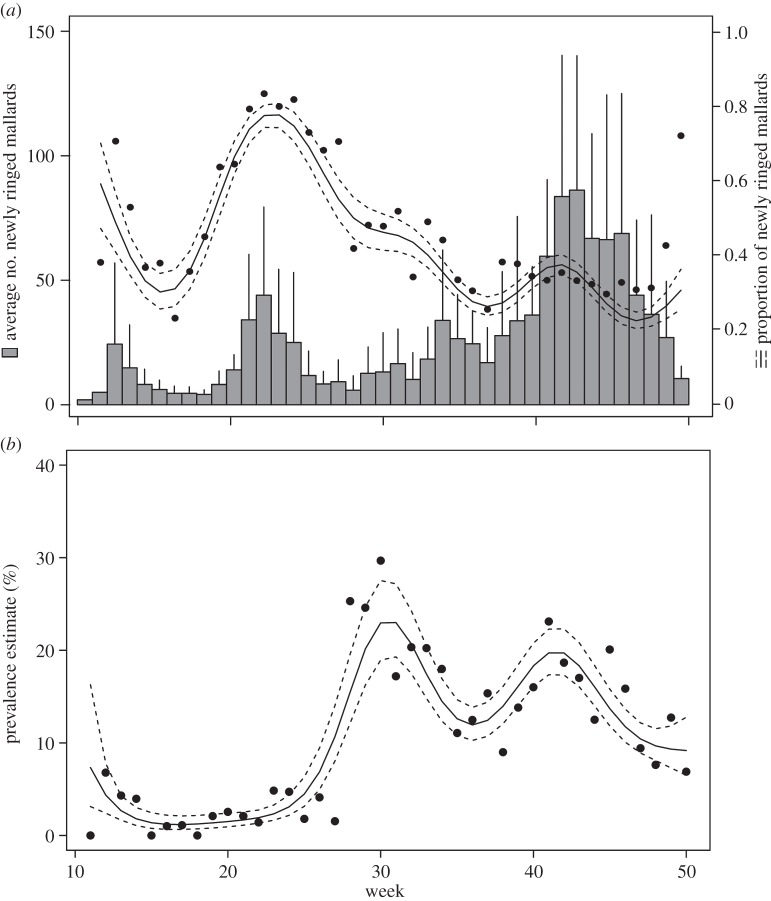
Trapping of mallards showed a distinct seasonal pattern that accounted for 44% of the overall seasonal deviation in trapping numbers among years (figure 1a). The daily number of trapped birds was correlated with the number of staging mallards in the reserve (Spearman rank correlation, R = 0.526, p < 0.001, n = 102), meaning that trapping numbers could be used as a proxy for migration phenology. At the onset of spring (weeks 10–12), most birds were trapped for the first time, but the proportion of newly ringed individuals decreased with the progress of spring. During the breeding season (weeks 22–30), when relatively few birds were captured, the proportion of new birds was large, probably reflecting recruitment in the local population. The proportion of newly ringed individuals during autumn migration had a bimodal shape, with an initial influx of birds in August (weeks 31–34), and a secondary, larger influx in October–November (weeks 40–44, figure 1a). In autumn, the average estimated length of stay ranged from 8.1 to 17.8 days in different years (ANOVA, F8, 2804 = 20.53, p < 0.001). In individual years, the timing of autumn migration differed, and subsequently the timing of the IAV peak prevalence (see below, and electronic supplementary material, figure S1). For instance, in the autumns of 2002–2005 and 2007, arrival of ducks was fairly constant, measured as number of birds captured for the first time per week. Other years with very mild climatic conditions during late autumn, such as 2006 and 2008–2010, migration was delayed. In such years, trapping data were dominated by a high amplitude peak in late autumn with many birds arriving within a short period. In December (weeks 48–50), trapping numbers went down, but the site continued to harbour ducks (figure 1a).
Figure 1.
Seasonal variation in (a) trapping of mallards and (b) IAV prevalence 2002–2010. In (a) the y-axis depicts the average number of newly trapped birds, and data are presented as bar plots with error bars. The secondary y-axis shows the variation of newly trapped birds compared with total for all years with 95% CI. The circles show the raw estimates for influx computed on data pooled across years, stratified by week. In (b), the y-axis gives the seasonal variation in prevalence, where the raw estimates of prevalence are given by filled circles, and the continuous line represents the estimated prevalence by the spline model, and the discontinuous lines the 95% CI. In both panels, the x-axis depicts the annual time scale in weeks.
The smoothed curve depicting the variation in IAV prevalence across years showed a strong seasonal pattern (figure 1b). The smooth curve was fitted to the data pooled over years based on RRT-PCR results (22 229 samples with a total IAV prevalence of 16.49% across 8529 individuals (electronic supplementary material, table S3)) and accounted for 48% of the total temporal variation in IAV prevalence. IAV prevalence was low during spring migration and the breeding season, but showed a marked increase in August (weeks 31–34) and later in October–November (weeks 41–46, figure 1b), coinciding with an increase of immigrating mallards (figure 1a). Similar to the temporal pattern of bird immigration, IAV prevalence showed a distinct bimodal pattern in autumn, with one prevalence peak in August and a second peak spanning October and November (weeks 40–44, figure 1b). In late autumn and early winter, the prevalence dropped to 10%, but still remained elevated compared with the spring, when the levels were lower than 5%. Although the common seasonal pattern accounted for a considerable fraction of the total seasonal variation across years, there was also variation in the prevalence levels and timing of IAV peaks for individual years (electronic supplementary material, figure S1).
(b). Influenza A virus diversity and temporal variation of subtypes
A total of 1081 viruses were isolated from 2451 RRT-PCR positive samples (44% isolation success) and subtyped, representing the majority of HA (H1–H12), and all NA (N1–N9) variants in a total of 74 HA/NA combinations (table 1). All viruses, including the H5 and H7 viruses were low-pathogenic, LPAI. The majority of isolates were from the autumn migration period, with only 19 isolates retrieved from samples taken during spring or early summer. However, these few isolates represented at least 10 different HA/NA subtypes: H2N3, H2N5, H3N8, H4N6, H7N4, H8N4, H10N4, H10N5, H10N7 and H10N8.
Table 1.
Distribution of IAV subtypes combinations (total number of isolates) in mallards sampled at Ottenby Bird Observatory, in 2002–2009. The most common subtype combinations (more than 20 isolates) are shown in bold.
| haemagglutinin | neuraminidase |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | N3 | N4 | N5 | N6 | N7 | N8 | N9 | unknown | total | |
| H1 | 94 | 6 | 5 | 1 | 4 | 1 | 30 | 141 | |||
| H2 | 6 | 5 | 58 | 2 | 1 | 1 | 6 | 7 | 10 | 96 | |
| H3 | 2 | 14 | 2 | 1 | 1 | 13 | 32 | 9 | 74 | ||
| H4 | 6 | 35 | 5 | 1 | 5 | 198 | 1 | 2 | 1 | 37 | 291 |
| H5 | 1 | 53 | 27 | 1 | 8 | 9 | 99 | ||||
| H6 | 9 | 53 | 6 | 2 | 14 | 3 | 8 | 1 | 9 | 105 | |
| H7 | 2 | 1 | 1 | 3 | 32 | 1 | 1 | 41 | |||
| H8 | 1 | 1 | 12 | 5 | 19 | ||||||
| H9 | 1 | 5 | 1 | 2 | 9 | ||||||
| H10 | 4 | 2 | 8 | 4 | 4 | 5 | 3 | 7 | 26 | 63 | |
| H11 | 3 | 36 | 7 | 1 | 3 | 2 | 1 | 51 | 14 | 118 | |
| H12 | 1 | 6 | 3 | 4 | 14 | ||||||
| unknown | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 11 | |||
| total | 125 | 217 | 116 | 26 | 35 | 231 | 41 | 54 | 80 | 156 | 1081 |
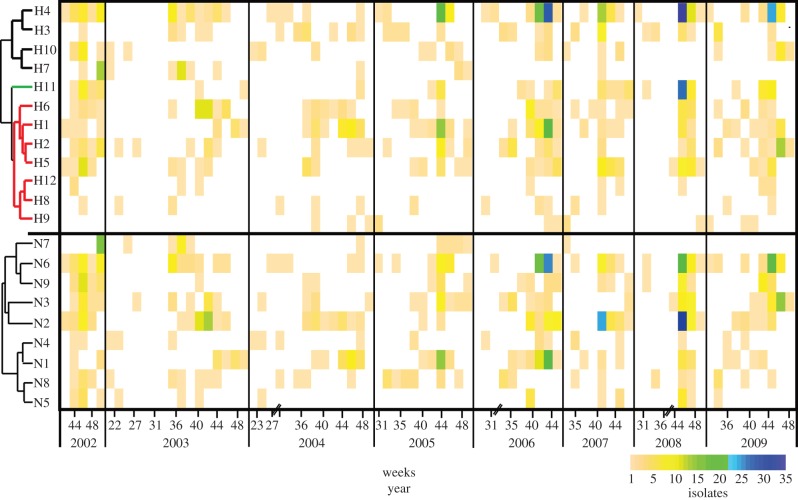
The six most common subtypes were H4N6, H1N1, H2N3, H5N2, H6N2 and H11N9, respectively, which together accounted for 46.9% of all isolated viruses. The number of subtype combinations per year varied from 18 to 30. The HA subtypes H1–H6 and H11 were isolated each year, while H7–H10 and H12 were isolated in only some years (figure 2). The two rarest HA subtypes were H9 and H12, and were only found in 6 out of the 9 years of the study. The H7 viruses were common in autumn 2002 and 2003 and isolated also in 2004–2005 and 2007, although H7s remained virtually absent in the last years of the time series (figure 2). In contrast, H5 viruses were common in the mallard population (figure 2).
Figure 2.
Temporal diversity of HA and NA (years 2002–2009). Filled boxes show the number of virus isolates within a two week period, where a topographic colour scale was used to indicate subtype abundance, with increasing numbers of isolates depicted as green and blue in the scale. The y-axis indicates the subtypes, the dendrogram shows subtype phylogenetic relatedness and thicker coloured branches indicate the HA classes. The H1 class is represented in red, the H3 class in black and the H11 class in green. Long periods of time without retrieved isolates were not included and are indicated with diagonal lines on the temporal x-axis to indicate discontinuous time.
Some subtype combinations detected in specific years had patterns suggesting outbreaks and clonal expansion of particular subtypes. These occurrences were typically manifested as high isolation frequencies in short periods of time. For instance, repeated isolation of H1N1 late in November 2004 and 2006, H6N2 in November 2003 and September 2006 and H2N3 in November 2008 (figure 2). By contrast, the H4N6 subtype was found in high numbers during the entire study period. Some subtypes consistently occurred early in the season, such as H3N8, compared with others that appeared at the end of the season such as H11; in general subtype presence was seasonal, whereby the same combination was not isolated continuously. Common HA/NA subtype combinations could be isolated for periods of three to five weeks during the autumn (figure 2). Deviation from this pattern occurred in years with delayed migration and arrival of birds, such as 2008, resulting in a peak of prevalence and IAV diversity concentrated in a few weeks in November (weeks 44–48).
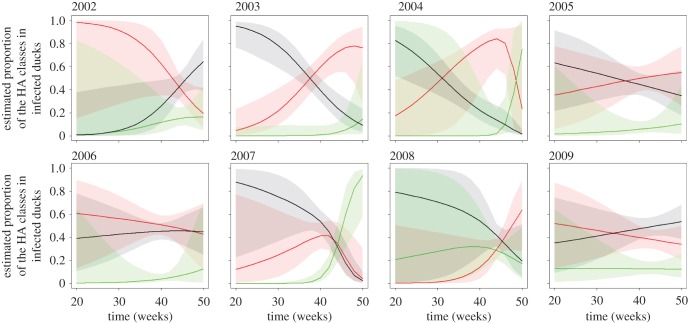
A modelling approach was used to determine specific patterns of diversity between years and within season by grouping HA subtypes (n = 1070) into three classes based on their phylogenetic relationships. The model with strongest statistical support (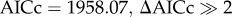 ) showed a robust year effect, with seasonal trends that included the interaction between year and season (electronic supplementary material, table S4). The visualization of the model (figure 3) suggests emergence of H3 class viruses earlier in autumn (weeks 20–35, subtypes in group 2: H3, H4, H7 and H10) compared with later emergence of H1 class viruses (H1 clade: H1, H2, H5 and H6; and H9 clade: H8, H9 and H12) and the H11 subtype.
) showed a robust year effect, with seasonal trends that included the interaction between year and season (electronic supplementary material, table S4). The visualization of the model (figure 3) suggests emergence of H3 class viruses earlier in autumn (weeks 20–35, subtypes in group 2: H3, H4, H7 and H10) compared with later emergence of H1 class viruses (H1 clade: H1, H2, H5 and H6; and H9 clade: H8, H9 and H12) and the H11 subtype.
Figure 3.
Seasonal variation in the estimated proportions of the three HA classes in infected individuals as a function of time (in weeks) and across years (2002–2009). Continuous lines represent the estimated proportions of each class in infected individuals, the 95% CI are represented by the shaded areas. The H1 class is represented in red, the H3 class in black and the H11 class in green.
(c). Haemagglutinin and neuraminidase linkage
The observed frequencies of HA/NA subtype combinations were significantly different from expected frequencies (Fisher exact test with Monte Carlo simulated p-value, 12 × 9 contingency table, n = 805, p < 0.001), indicating dependence between specific HA and NA variants. The standardized Pearson's residuals showed that H1N1, H11N9, H10N4, H12N5, H2N3, H3N8, H4N6, H5N2, H6N2 and H7N7 were isolated more often than their respective HA and NA abundances would indicate. Other subtypes, including H4N1, H1N2, H4N3, H1N6 and H6N6, among others, were isolated less frequently than expected (electronic supplementary material, table S5).
4. Discussion
(a). Seasonal variation in influenza A virus prevalence and host migratory behaviour
Seasonal environmental changes are a driving force for periodic life cycle events in many organisms, influencing timing of reproduction, migration and many other behaviours [28]. The regularity in these events also impacts host–pathogen interactions [28] and has consequences for spatial–temporal variations in incidence and prevalence of pathogens. Consequently, it is essential to determine factors and processes in the ecology of the host species that may influence pathogen perpetuation [29–31]. The main reservoir hosts for IAV in the Northern Hemisphere are waterfowl, gulls and shorebirds, of which most species are migratory and hence should induce spatial and temporal variation in IAV transmission and persistence factors. Long-term datasets on IAV in wild birds are restricted to a few sites in Europe and North America [8,10,13–15,17,32,33], and only recently emerged in other areas, such as Africa and Australia [6,34,35]. Using trapping and virology data collected from a single site across 9 years, we show that, although there is a considerable interannual variation in IAV prevalence in mallards during migration, there were still trends correlated to host ecology, and possibly to immune processes at individual and population scales.
IAV prevalence and migration intensity were associated in late autumn, when the second IAV prevalence peak coincided with a period during which many mallards arrived at the study site. In summer, the influx probably consisted of local recruitment and not migrants. During spring migration IAV prevalence was low, but possibly sufficient to maintain IAV circulation in the reservoir host. Spring migration in this species is faster than autumn migration [36], as the ducks are heading towards breeding areas. This was reflected in the low numbers of ducks trapped during the spring period and the highest trapping intensity in the end of May (week 22). The dataset from the autumn period was much larger, and depicted a strong seasonal pattern with pronounced IAV peaks in August and October–November, the second one corresponding to a main peak in mallard migration.
(b). What is governing the seasonal trends in influenza A virus prevalence and subtype predominance?
The factors driving seasonal trends in IAV prevalence and subtype diversity are not fully characterized. One hypothesis is that the temporal variation is driven primarily through host population immunity. Population immunity drives the dynamics of different human pathogens [37,38], including replacement of influenza strains in the human population [39]. Similarly in mallards, the development of population immunity could explain changes in the IAV viral population. At present, little knowledge is available on how IAV variation at the individual or population levels is related to host immune functions. Experimental infections have shown that a primary infection can give protection against infections with the same strain (homosubtypic immunity) [40,41]. In addition, the development of partial protection against other strains, defined as heterosubtypic or cross-protective immunity, may also occur [40,42–45]. This seems to be true for sentinel mallards in nature [46]. Recently, we showed that this study population develops homo- and heterosubtypic immunity from natural infections [20], and that the heterosubtypic immunity at the HA clade level persists for at least a month. This cross-protective immunity could affect the dynamics of virus transmission in the population (i.e. changes in infection probabilities and in subtype fitness as the proportion of resistant hosts against a particular subtype/s increases over time), the circulation of certain virus subtypes [20] and probable patterns of reassortment [47]. The degree of immunity to IAV is probably related to exposure, and under the assumption that all birds have an equal exposure to viruses, population immunity should increase with seasonal progression, and be higher in adult birds than in juvenile birds.
Interestingly, the two peaks in IAV prevalence in autumn tended to be dominated by different virus subtypes. At the HA diversity level, there was a year effect and a seasonal trend with the interaction of year and season. The modelling suggested an interplay or a succession pattern between the different HA classes. The two major classes, the H1 class (H1 and H9 clades) and the H3 class (HA group 2) appeared to have different trends, where the proportion of H3 class viruses in infected individuals was higher in the early autumn compared with the H1 and H11 classes which in turn appeared in late autumn.
Another hypothesis is that seasonal trends are caused by different populations of mallards, each having different viral populations, migrating through an area with different phenology. As a consequence, temporal variation could arise due to continuous seeding and import of viruses to staging birds [18,29,31]. The success of these introductions at the site could, in turn, depend on population immunity. At our study site, there is temporal succession in the arrival of mallards from different breeding areas, where early autumn migration is dominated by birds from the Baltic states and Finland, which are gradually replaced with birds originating in northwestern Russia [9,18]. Through repeated recaptures of individuals we know that there is ongoing virus transmission at our study site [20], and it is probable that the baited trap used for catching the birds may amplify transmission [48].
Further, timing of mallard migration coincides with the migrations of many other bird species, some of which may also be hosts for the virus. Thus, coexistence of different hosts at the stopover site may affect transmission frequency and subtype circulation. For instance, the first peak of IAV prevalence in mallards in autumn coincides with the main migration period of terns, gulls and waders from the Siberian taiga and tundra. During the later IAV prevalence peak in mallards, the area also harbours several thousand geese: mainly barnacle geese Branta leucopsis and brent geese Branta bernicla. Other Anas species, including Eurasian teal A. crecca, northern shoveller A. clypeata, Eurasian wigeon A. penelope and northern pintail A. acuta, have slightly different migration phenologies compared with mallards, which could potentially affect IAV prevalence and transmission in the system [49].
(c). Subtype diversity maintenance and distribution
Over the total study period, HA subtypes from H1 to H12 and all NA subtypes were isolated from mallards, representing 74 different HA/NA combinations. Most HA and NA subtypes were found each year, but the number of different combinations varied between 18 and 30. The H1 and H6 subtypes were the dominant HA subtypes throughout the study period. Subtype diversity levels were comparable between years, even if within-season diversity varied depending on the timing of migration. Overall, our study site at Ottenby harbours the highest IAV subtype diversity reported to date for a single site. The Genbank database records report a total of 102 subtype combinations isolated from Anseriformes. Most large-scale IAV studies conducted show seasonal patterns in prevalence levels and large subtype diversity in IAV collections from ducks, regardless of locations in North America, in Europe, or elsewhere [7–11,13,16,17,32–33].
Some subtypes, such as H4 and H6, are abundant globally and distributed in both the Eurasian and the North American continents, while others tend to be rarely detected (e.g. H8 or H7 subtype), or are variable in their abundance between years, or studies (e.g. H10 and H12 subtypes) [13,14]. An enigma in the IAV system is how subtype diversity is maintained, especially for rare subtypes that would be sensitive to stochastic events. Balancing selection could favour rare variants, but at the same time rare variants are more prone to be affected by bottleneck effects, for instance in winter when prevalence is low and population immunity may be highest. Sharp et al. [14] described prevalence of some subtypes in Canada as either consistent or sporadic, while some other subtypes followed a 3 year cycle: peak the first year, decrease the second year and decrease more dramatically the third year. Clear cyclic patterns were not observed at Ottenby; however H7, H8, H9 and H12 were found at low prevalence and were absent some years of the study. A low prevalence could increase randomness of detection probabilities, but could also indicate that these viruses are maintained in other host species and are transmitted to the mallards when different bird species come in contact during migration or wintering. Interestingly, the H7 subtype was commonly isolated in 2002 and 2003 [50], but has remained rare in our series since then. H7 viruses have been detected in other countries in Europe during the study period, including outbreaks of HPAI. Apart from H7 viruses, we also isolated the notifiable LPAI subtypes H5 and H9, which pose risks of transmission into poultry [3]. However, no HPAI viruses were detected at the site, including HPAI H5N1, which circulated in wild birds and poultry in Europe in 2005–2006. This suggests that HPAI viruses are not maintained in wild waterfowl populations in Europe.
Additionally, specific HA subtypes were mainly associated with certain NA combinations, such as H4N6 and H6N2. This suggests advantageous linkage between particular HA and NA, and possibly the existence of particularly fit waterfowl viruses as HA and NA could have coevolved resulting in more stable genome constellations, while rarer combinations may be more adapted to other hosts. Co-infections are probably common in the mallard population [11,12,51] owing to the high prevalence and diversity, and therefore new gene constellations are expected to arise from reassortment. If all the different HA and NA were functionally exchangeable, and could randomly be associated in different combinations without a fitness consequence, one would expect a relatively high number of reassortants between common HA/NA subtypes. However, the H1N1 and H4N6 subtypes are both common and co-circulate, while H1N4 or H4N1 reassortants were rare. One explanation could be that some reassortants have lower fitness, although a recent study showed that natural reassortants may have similar fitness levels when contrasting viral load, duration of shedding and survival in the environment [52].
The present long-term study gives new insights into the diversity dynamics at a stopover site and represents a step forward in understanding IAV epidemiology in one of the major reservoir hosts for avian influenza. Further phylogenetic analysis on isolates across years could shed light on the evolutionary dynamics of subtypes, lineages and the impact of reassortment events in IAV viral population dynamics.
Acknowledgements
We thank the staff at Ottenby Bird Observatory who trapped, sampled and measured all mallards in this study. We also thank former and present staff in the laboratories for technical assistance and advice: P. Griekspoor, L. Svensson, J. Olofsson, D. Axelsson-Olsson, A. Jawad, A. Wallensten, G. Gunnarsson, G. Orozovic, C. Baas, P. Lexmond, E. Jourdain, P. Ellström, J. Wahlgren, M. Blomqvist and M. Karlsson.
Funding statement
This study was supported by grants from the Swedish Environmental Protection Agency (V-124-01 and V-98-04), the Swedish Research Council (2008-58, 2010-3067, 2011-48), the Swedish Research Council Formas (2007-297, 2009-1220) and the Sparbanksstiftelsen Kronan. The surveillance at Ottenby was part of the European Union wild bird surveillance and has received support from the Swedish Board of Agriculture and from the EU NP6-funded New Flubird project. This is contribution no. 278 from Ottenby Bird Observatory.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus ADME, Fouchier RAM. 2006. Global patterns of influenza A virus in wild birds. Science 312, 384–388 (doi:10.1126/science.1122438) [DOI] [PubMed] [Google Scholar]
- 3.Alexander DJ, Capua I. 2008. Avian influenza in poultry. World Poultry Sci. J. 64, 513–531 (doi:10.1017/S0043933908000184) [Google Scholar]
- 4.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79, 2814–2822 (doi:10.1128/JVI.79.5.2814-2822.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Easterday BC, Trainer DO, Tumova B, Pereira HG. 1968. Evidence of infection with influenza viruses in migratory waterfowl. Nature 219, 523–524 (doi:10.1038/219523a0) [DOI] [PubMed] [Google Scholar]
- 6.Gaidet N, et al. 2011. Understanding the ecological drivers of avian influenza virus infection in wildfowl: a continental-scale study across Africa. Proc. R. Soc. B 279, 1131–1141 (doi:10.1098/rspb.2011.1417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinshaw VS, Webster RG, Turner B. 1980. The perpetuation of Orthomyxoviruses and Paramyxoviruses in Canadian waterfowl. Can. J. Microbiol. 26, 622–629 (doi:10.1139/m80-108) [DOI] [PubMed] [Google Scholar]
- 8.Suss J, Schafer J, Sinnecker H, Webster RG. 1994. Influenza virus subtypes in aquatic birds of eastern Germany. Arch. Virol. 135, 101–114 (doi:10.1007/BF01309768) [DOI] [PubMed] [Google Scholar]
- 9.Wallensten A, et al. 2007. Surveillance of influenza A virus in migratory waterfowl in northern Europe. Emerg. Infect. Dis. 13, 404–411 (doi:10.3201/eid1303.061130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munster VJ, et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3, 630–638 (doi:10.1371/journal.ppat.0030061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dugan VG, et al. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 4, e1000076 (doi:10.1371/journal.ppat.1000076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang RX, Soll L, Dugan V, Runstadler J, Happ G, Slemons RD, Taubenberger JK. 2008. Examining the hemagglutinin subtype diversity among wild duck-origin influenza A viruses using ethanol-fixed cloacal swabs and a novel RT-PCR method. Virology 375, 182–189 (doi:10.1016/j.virol.2008.01.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krauss S, Walker D, Pryor SP, Niles L, Li CH, Hinshaw VS, Webster RG. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector-Borne Zoonotic Dis. 4, 177–189 (doi:10.1089/vbz.2004.4.177) [DOI] [PubMed] [Google Scholar]
- 14.Sharp GB, Kawaoka Y, Wright SM, Turner B, Hinshaw V, Webster RG. 1993. Wild ducks are the reservoir for only a limited number of influenza A-subtypes. Epidemiol. Infect. 110, 161–176 (doi:10.1017/S0950268800050780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng MC, Lee MS, Ho YH, Chyi WL, Wang CH. 2010. Avian influenza monitoring in migrating birds in Taiwan during 1998–2007. Avian Dis. 54, 109–114 (doi:10.1637/8960-061709-Reg.1) [DOI] [PubMed] [Google Scholar]
- 16.Wilcox BR, et al. 2011. Influenza-A viruses in ducks in northwestern Minnesota: fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS ONE 6, e24010 (doi:10.1371/journal.pone.0024010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinshaw VS, Wood JM, Webster RG, Deibel R, Turner B. 1985. Circulation of influenza-viruses and paramyxoviruses in waterfowl originating from 2 different areas of North America. Bull. World Health Organ. 63, 711–719 [PMC free article] [PubMed] [Google Scholar]
- 18.Gunnarsson G, Latorre-Margalef N, Hobson KA, Van Wilgenburg SL, Elmberg J, Olsen B, Fouchier RAM, Waldenström J. 2012. Disease dynamics and bird migration—linking mallards Anas platyrhynchos and subtype diversity of the influenza A virus in time and space. PLoS ONE 7, e35679 (doi:10.1371/journal.pone.0035679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latorre-Margalef N, et al. 2009. Effects of influenza A virus infection on migrating mallard ducks. Proc. R. Soc. B 276, 1029–1036 (doi:10.1098/rspb.2008.1501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latorre-Margalef N, Grosbois V, Wahlgren J, Munster VJ, Tolf C, Fouchier RAM, Osterhaus ADME, Olsen B, Waldenström J. 2013. Heterosubtypic immunity to influenza A virus infections in mallards may explain existence of multiple virus subtypes. PLoS Pathog. 9, e1003443 (doi:10.1371/journal.ppat.1003443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus RHS, van Hooft P, Waldenstrom J, Latorre-Margalef N, Ydenberg RC, Prins HHT. 2011. Avian influenza surveillance with FTA cards: field methods, biosafety, and transportation issues solved. J. Vis. Exp. 54, e2832 (doi:10.3791/2832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukamoto K, et al. 2012. SYBR green-based real-time reverse transcription-PCR for typing and subtyping of all hemagglutinin and neuraminidase genes of avian influenza viruses and comparison to standard serological subtyping tests. J. Clin. Microbiol. 50, 37–45 (doi:10.1128/jcm.01195-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Development Core Team. R version 2.14.0 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 24.Yee TW. 2010. The VGAM package for categorical data analysis. J. Stat. Softw. 32, 1–34 [Google Scholar]
- 25.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach, p. 488, 2nd edn New York: Springer Science+Business Media [Google Scholar]
- 26.Agresti A. 2001. Exact inference for categorical data: recent advances and continuing controversies. Stat. Med. 20, 2709–2722 (doi:10.1002/sim.738) [DOI] [PubMed] [Google Scholar]
- 27.Mehta CR, Patel NR. 1998. Exact inference for categorical data. In Encyclopedia of biostatistics (eds Armitage P, Colton T.), pp. 1411–1422 Chichester, UK: Wiley [Google Scholar]
- 28.Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. 2006. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 9, 467–484 (doi:10.1111/j.1461-0248.2005.00879.x) [DOI] [PubMed] [Google Scholar]
- 29.Altizer S, Bartel R, Han BA. 2011. Animal migration and infectious disease risk. Science 331, 296–302 (doi:10.1126/science.1194694) [DOI] [PubMed] [Google Scholar]
- 30.Fuller T, Bensch S, Müller I, Novembre J, Pérez-Tris J, Ricklefs R, Smith T, Waldenström J. 2012. The ecology of emerging infectious diseases in migratory birds: an assessment of the role of climate change and priorities for future research. EcoHealth 9, 80–88 (doi:10.1007/s10393-012-0750-1) [DOI] [PubMed] [Google Scholar]
- 31.Hill NJ, Takekawa JY, Ackerman JT, Hobson KA, Herring G, Cardona CJ, Runstadler JA, Boyce WM. 2012. Migration strategy affects avian influenza dynamics in mallards (Anas platyrhynchos). Mol. Ecol. 21, 5986–5999 (doi:10.1111/j.1365-294X.2012.05735.x) [DOI] [PubMed] [Google Scholar]
- 32.Krauss S, et al. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 3, 1684–1693 (doi:10.1371/journal.ppat.0030167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. 1995. Perpetuation of influenza-A viruses in Alaskan waterfowl reservoirs. Arch. Virol. 140, 1163–1172 (doi:10.1007/BF01322743) [DOI] [PubMed] [Google Scholar]
- 34.Hansbro PM, et al. 2010. Surveillance and analysis of avian influenza viruses, Australia. Emerg. Infect. Dis. 16, 1896–1904 (doi:10.3201/eid1612.100776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caron A, et al. 2012. Persistence of low pathogenic avian influenza virus in waterfowl in a southern African ecosystem. EcoHealth 8, 109–115 (doi:10.1007/s10393-010-0356-4) [DOI] [PubMed] [Google Scholar]
- 36.Cramp SS, Simmons, KEL 1977. Handbook of the birds of Europe, the Middle East and North Africa, p. 714 Oxford, UK: Oxford University Press [Google Scholar]
- 37.Recker M, Blyuss KB, Simmons CP, Hien TT, Wills B, Farrar J, Gupta S. 2009. Immunological serotype interactions and their effect on the epidemiological pattern of dengue. Proc. R. Soc. B 276, 2541–2548 (doi:10.1098/rspb.2009.0331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitzer VE, Patel MM, Lopman BA, Viboud C, Parashar UD, Grenfell BT. 2011. Modeling rotavirus strain dynamics in developed countries to understand the potential impact of vaccination on genotype distributions. Proc. Natl Acad. Sci. USA 108, 19 353–19 358 (doi:10.1073/pnas.1110507108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palese P, Wang TT. 2011. Why do influenza virus subtypes die out? A hypothesis. mBio 2, e00150-11 (doi:10.1128/mBio.00150-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jourdain E, et al. 2010. Influenza virus in a natural host, the mallard: experimental infection data. PLoS ONE 5, e8935 (doi:10.1371/journal.pone.0008935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kida H, Yanagawa R, Matsuoka Y. 1980. Duck influenza lacking evidence of disease signs and immune response. Infect. Immun. 30, 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa TP, Brown JD, Howerth EW, Stallknecht DE. 2010. Effect of a prior exposure to a low pathogenic avian influenza virus in the outcome of a heterosubtypic low pathogenic avian influenza infection in mallards (Anas platyrhynchos). Avian Dis. 54, 1286–1291 (doi:10.1637/9480-072210-Reg.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa TP, Brown JD, Howerth EW, Stallknecht DE, Swayne DE. 2011. Homo- and heterosubtypic low pathogenic avian influenza exposure on H5N1 highly pathogenic avian influenza virus infection in wood ducks (Aix sponsa). PLoS ONE 6, e15987 (doi:10.15910.11371/journal.pone.0015987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fereidouni SR, et al. 2009. Highly pathogenic avian influenza virus infection of mallards with homo- and heterosubtypic immunity induced by low pathogenic avian influenza viruses. PLoS ONE 4, e6706 (doi:10.1371/journal.pone.0006706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pepin KM, VanDalen KK, Mooers NL, Ellis JW, Sullivan HJ, Root JJ, Webb CT, Franklin AB, Shriner SA. 2012. Quantification of heterosubtypic immunity between avian influenza subtypes H3N8 and H4N6 in multiple avian host species. J. Gen. Virol. 93, 2575–2583 (doi:10.1099/vir.0.045427-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolf C, et al. 2013. Individual variation in influenza A virus infection histories and long-term immune responses in mallards. PLoS ONE 8, e61201 (doi:10.1371/journal.pone.0061201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wille M, Tolf C, Avril A, Latorre-Margalef N, Wallerstrom S, Olsen B, Waldenstrom J. 2013. Frequency and patterns of reassortment in natural influenza A virus infection in a reservoir host. Virology 443, 150–160 (doi:10.1016/j.virol.2013.05.004) [DOI] [PubMed] [Google Scholar]
- 48.Soos C, Parmley EJ, McAloney K, Pollard B, Jenkins E, Kibenge F, Leighton FA. 2012. Bait trapping linked to higher avian influenza virus detection in wild ducks. J. Wildl. Dis. 48, 444–448 (doi:10.7589/0090-3558-48.2.444) [DOI] [PubMed] [Google Scholar]
- 49.Stallknecht DE, Brown JD. 2007. Wild birds and the epidemiology of avian influenza. J. Wildl. Dis. 43(Suppl. 3), S15–S20 [Google Scholar]
- 50.Munster VJ, Wallensten A, Baas C, Rimmelzwaan GF, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. 2005. Mallards and highly pathogenic avian influenza ancestral viruses, northern Europe. Emerg. Infect. Dis. 11, 1545–1551 (doi:10.3201/eid1110.050546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharp GB, Kawaoka Y, Jones DJ, Bean WJ, Pryor SP, Hinshaw V, Webster RG. 1997. Coinfection of wild ducks by influenza A viruses: distribution patterns and biological significance. J. Virol. 71, 6128–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lebarbenchon C, Sreevatsan S, Lefèvre T, Yang M, Ramakrishnan MA, Brown JD, Stallknecht DE. 2012. Reassortant influenza A viruses in wild duck populations: effects on viral shedding and persistence in water. Proc. R. Soc. B 279, 3967–3975 (doi:10.1098/rspb.2012.1271) [DOI] [PMC free article] [PubMed] [Google Scholar]