Abstract
Episodic ataxias (EAs) are rare neurological channelopathies that are characterized by spells of imbalance and a lack of co-ordination. There are seven clinically recognized EAs and multiple isolated cases. Five disease-causing genes have been identified to date. We describe a novel form of autosomal dominant EA in a large three-generation Irish family. This form of EA presents in early childhood with periods of unsteadiness generalized weakness and slurred speech during an attack, which may be triggered by physical tiredness or stress. Linkage analysis undertaken in 13 related individuals identified a single disease locus (1p36.13-p34.3) with a LOD score of 3.29. Exome sequencing was performed. Following data analysis, which included presence/absence within the linkage peak, two candidate variants were identified. These are located in the HSPG2 and UBR4 genes. UBR4 is an ubiquitin ligase protein that is known to interact with calmodulin, a Ca2+ protein, in the cytoplasm. It also co-localizes with ITPR1 a calcium release channel that is a major determinant of mammal co-ordination. Although UBR4 is not an ion channel gene, the potential for disrupted Ca2+ control within neuronal cells highlights its potential for a role in this form of EA.
Keywords: episodic ataxia, exome sequencing, brain, linkage analysis, UBR4, HSPG2
Introduction
Episodic ataxias (EAs) are rare neurological channelopathies that are characterized by spells of imbalance and a lack of co-ordination. Affected individuals may also experience nystagums, myokymia, epilepsy and/or tinnitus. Clinical and genetic heterogeneity is not uncommon, and a number of EA subtypes have been described (see Table 1 for further information). The most common EA subtypes (1 and 2) have been described in multiple families. They are caused by mutations in the potassium and calcium channel genes, KCNA1 and CACNA1A.1, 2 Mutations in CACNB4 and SLC1A3 cause EA5 and EA6.3, 4 Further, mutations in PRRT2 have been identified in a single patient who also presents with hemiplegic migraines5 and in two brothers with familial paroxysmal kinesigenic dystonia in addition to learning disabilities, absences and EA.6 The PRRT2 protein has been shown to interact with the SNAP25 protein that has a major role in synaptic exocytosis.7 This observation demonstrates that ion channel dysfunction is not the sole mechanism for developing EA. Novel loci have also been identified for EA3 (1q42) and EA7 (19q13), although the defective genes remain to be identified. Additional forms of EA with phenotypes that are not consistent with those reported with the previously identified genes/loci have also been reported (EA4 and isolated EA cases).8 Many patients do not have mutations in the known EA genes. It is clear that new genes remain to be discovered.
Table 1. Comparison of Irish EA to previously reported EA.
| EA1 | EA2 | EA3 | EA4 | EA5 | EA6 | EA7 | Other EAs | Irish Familial EA | |
|---|---|---|---|---|---|---|---|---|---|
| OMIM | 160120 | 108500 | 606554 | 606552 | 613855 | 612656 | 611907 | unassigned | unassigned |
| Attack duration | seconds to min | hours | 1 min to 6 h | brief | hours | hours-days | hours-days | hours to days | min to 24 h |
| Age of onset | 2–15 | 2–20 | 1–42 | 23–60 | 3 to teen | 5 | <20 years | After 30 | Early infancy |
| Myokymia | Usual | No | Usual | No | No | No | No | No | Occasionally |
| Nystagmus | No | Usual | Occasional | Usual | Usual | No | No | Usual | Occasionally |
| Slurred speech (needs checking) | Occasional | No | No | No | Occasional | No | NA | Yes | |
| Epilepsy | Occasional | Infrequent | Occasional | Occasional | Usual | Yes | No | No | No |
| Tinnitus | Infrequent | No | Usual | Occasional | No | No | No | No | No |
| Acetazolamide | Occasional | Usual | Usual | No | Transient | No | Unknown | Occasional | No |
| Inheritance | AD | AD | AD | AD | AD | Sporadic | AD | Multiple | AD |
| Chr locus | 12q13 | 19p13 | 1q42 | Unknown | 2q22-q23 | 5p13.2 | 19q13 | Unknown | 1p36.13-p34.3 |
| Mutated gene | KCNA1 | CACNA1A | Unknown | Unknown | CACNB4 | SLC1A3 | Unknown | Unknown | Unknown |
| Mutated protein | Kv1.1 | Cav2.1 | Unknown | Unknown | Cav2.1 | EAATI | Unknown | Unknown | Unknown |
Abbreviations: AD, autosomal dominant; EA, episodic ataxia.
Although a number of similarities exist between this form of EA and more common forms of EA, some differences do occur. These include the early age of onset and long duration of attacks. Slurred speech also occurs in all affected individuals within this pedigree and treatment with acetazolamide is of no effect. Interictal findings vary between individuals.
In this report, we describe an autosomal dominant EA that differs in clinical and genetic features from previously described EAs. Parametric linkage analysis ruled out all previously loci reported for EA. A single novel locus was identified on chromosome 1p36.13-p34.3. This peak spanned 18.5 Mb, 281 RefSeq genes and 10 miRNAs. This linkage finding confirms that differences in clinical presentation are due to a genetic defect in a previously unreported gene, highlighting the existence of yet another form of episodic ataxia.
Patients and methods
Patients
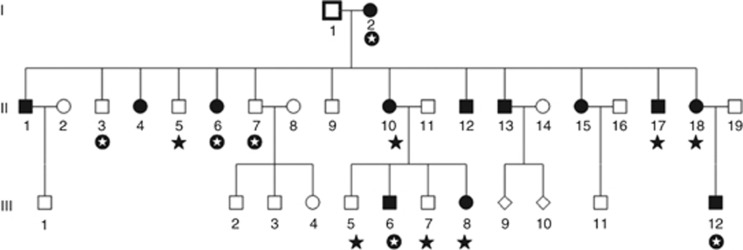
This family was referred to the clinical genetic service for genetic assessment at the National Centre of Medical Genetics, Ireland. Thirteen family members in three generations are affected by EA (see Figure 1). In total, 13 of these individuals (I-2, II-3, II-5, II-6, II-7, II-10, II-18, III-5, III-6, III-7, III-8 and III-12) attended either the Adelaide and Meath Hospital, incorporating the National Children's Hospital or the National Children's Hospital for Sick Children for full clinical assessment. A DNA sample was also obtained for individual II-17. Clinical mutation screening of KCNA1, CACNA1A, and (CAG)n repeats in SCA1, SCA2, SCA3 and SCA6 revealed no causative mutation. Ethical approval for this study was granted by the Adelaide and Meath Hospital, incorporating the National Children's Hospital and informed consent was received for each individual involved.
Figure 1.
Irish family presenting with novel form of autosomal dominant episodic ataxia. DNA from the 13 family members identified above was available for molecular analysis (I-2, II-3, II-5, II-6, II-7, II-10, II-17, II-18, III-5, III-6, III-7, III-8 and III-12, marked by a black star or a white star on a black background). Exome sequencing was performed on six of these samples (I-2, II-3, II-6, II-7, III-6 and III-12, represented by a white star on a black background).
Clinical presentation
The symptoms of EA become apparent in this family in the second year of life as the children learn to walk. Attacks are characterized by unsteadiness, generalized weakness and slurred speech, which may be triggered by physical tiredness or stress. They do not suffer tinnitus or epilepsy although two of the affected individuals have migraine without aura. The number of attacks varies between family members and indeed throughout life. Some family members suffer up to two attacks per day, whereas others can go months without an attack. In some cases, the frequency and severity of attacks appear to lessen with increasing age and, in one case, lessened during pregnancy. Attacks are eased by clonazepam. None of the affected individuals responded to acetazolamide. The disorder is not progressive in nature, although most have mild intention tremor and impaired tandem gait interictally, which remains asymptomatic; I-2 is now in her 70 s and remains asymptomatic between attacks despite mild abnormalities on examination. One individual (II-12) had a MRI brain scan twice, at age 19 and again at age 29 years. Both were normal. Specifically, there were no cerebellar abnormalities observed. Information describing the family is presented in Table 2, and one affected individual is reported here in detail.
Table 2. Clinical presentation of patients with EA in Irish pedigree.
| I-2 | II-6 | II-10 | II-17 | II-18 | II-1 | II-4 | |
|---|---|---|---|---|---|---|---|
| Attended clinical review | Y | Y | Y | Y | Y | N/A | N/A |
| Attack duration | minutes to hours | 10 min to 12 h | 30 min to 6 h | 2 min to 10 h | <2 h | ||
| Attack frequency | Varies from 0/month to 3 or 4 per week in clusters | 2/day | 1/day | 1-2/day | daily | Daily | U |
| Characteristics of attack | |||||||
| Muscle weakness | Y | Y | Y | Y | Y | Y | Y |
| Slurring of speech | Y | Y | Y | Y | Y | Y | Y |
| Poor balance | Y | Y | Y | Y | Y | Y | Y |
| Twitching around eyes | Y | Y | Y | ||||
| Response to clonazepam | Y | Y | Y | Y | U | Y | U |
| Additional information | Frequency of episodes is decreasing with age | Frequency of episodes has reduced with age | |||||
| EA during pregnancy | U | No pregnancies | Marked improvement | Marked improvement | no pregnancies | ||
| Interictal examination findings | N/A | N/A | |||||
| Nystagmus | Y | N | Y | N | N | ||
| Gait ataxia | Y | Y | Y | N | Y | ||
| Myokymia | N | Y | N | N | N | ||
| Dysarthria | N | Y | Y | N | Y | ||
| Tremor | Y | Y | Y | Y | Y | ||
| Additional interictal symptoms | N | Myokimia, migraine, depression | Migraine | N | Depression | N | N |
| II-12 | II-13 | II-15 | III-6 | III-8 | III-12 | |
|---|---|---|---|---|---|---|
| Attended clinical review | N/A | N/A | N/A | Y | Y | Y |
| Attack duration | 10 min–1 day | 10–15 min | ||||
| Attack frequency | 2/day | U | Several/day | 2/day | Weekly | 2/month |
| Characteristics of attack | ||||||
| Muscle weakness | Y | Y | Y | Y | Y | Y |
| Slurring of speech | Y | Y | Y | Y | Y | Y |
| Poor balance | Y | Y | Y | Y | Y | Y |
| Twitching around eyes | ||||||
| Response to clonazepam | Y | U | Y | Y | U | Y |
| Additional information | Attacks have become more severe with age | |||||
| EA during pregnancy | Marked improvement | No pregnancies | ||||
| Interictal examination findings | N/A | N/A | N/A | |||
| Nystagmus | N | N | N | |||
| Gait ataxia | N | N | Y | |||
| Myokymia | N | N | N | |||
| Dysarthria | N | |||||
| Tremor | Y | N | Y | |||
| Additional interictal symptoms | N | N | N | N | N | N |
Abbreviations: N, no; N/A, data not available; U unknown; Y, yes.
All patients developed symptoms of EA in early infancy. Many of the individuals suffer at least one attack per day, however the frequency and severity of attacks lessens with increasing age. No individual responded to acetazolamide but the attacks are eased by clonazepam.
Patient II-6 was 45 years old when first seen and described attacks since the age of 2 years. Attacks were usually triggered by anxiety, stress, cold weather or physical fatigue, and each attack would last between 10 min and 12 hours. An attack would be preceded by a ‘warning' feeling, which she found difficult to describe. She would then develop generalized weakness associated with slurred speech and imbalance, which would prevent her walking. She also described twitching around the eyes and mouth, which occurred during an attack but could also occur independently of an attack. Attacks occurred up to twice daily. In between attacks, she felt normal. She also described migrainous headaches that occurred on a weekly basis. On examination, there was eyelid myokymia, mild dysarthria and intention tremor of both upper limbs. Tandem gait was mildly impaired. Power, sensation and reflexes were normal.
Although there are a number of similarities between the features in this family to the other EA subtypes (EA1–EA7), some differences also exist. The onset of symptoms, usually by age 2 years, is much earlier than that in EA 1–7. In comparison with EA1, attacks are much longer, and there is no muscle hypertrophy. In addition, there is no kinesigenic provocation or limb or neck jerking during attacks. This family does not report vertigo and tinnitus, features of EA3. Although some affected individuals have minimal gaze-evoked nystagmus, prominent eye movement abnormalities such as those seen in EA2 and EA4 are not seen. Acetazolamide is not effective in these patients; however, they demonstrate a clear response to clonazepam.
SNP genotyping and linkage analysis
DNA from the 13 family members identified above was available for molecular analysis. Genomic DNA was extracted from peripheral blood and genotyped for over 300 000 markers on the HumanCytoSNP-12 microarray using the recommended protocol. Samples were processed on an Illumina BeadStation 500GX platform (Illumina, San Diego, CA, USA) using default settings. Analysis and genotype calling were performed using the IlluminaBeadStudio software v3.3.7 (Illumina). Experimental quality control was assessed using the sample independent and sample dependent Illumina internal controls.
Data cleaning was performed using Plink v1.06.9 Samples were excluded if the call rate was <95%. SNPs were excluded based on missingness (100% call rate required), Hardy–Weinberg equilibrium (<0.001), minor allele frequency (<10%) and the presence of Mendel errors. Following this, the data was thinned to 20%. Pairwise linkage analysis was undertaken using Merlin 1.1.2.10 An autosomal dominant model with high penetrance (0.99) and low disease allele frequency (0.0001) was assumed.
Exome sequencing and data analysis
DNA from six family members (I-2, II-3, II-6, II-7, III-6 and III-12) was selected for whole-exome sequencing, which was performed at Atlas Biolabs (http://www.atlas-biolabs.de). The exome of each individual was enriched and sequenced using the Human EZ Exome Enrichment kit (Roche NimbleGen, Madison, WI, USA) and HiSeq platform (Illumina). Single-sequence reads were aligned to the hg18 reference genome using the Burrows–Wheeler Alignment tool version 0.5.7.11 PCR duplicates and low-quality sequences (Q<20) were excluded. GATK12 was used to recalibrate the quality scores for the aligned reads. Fivefold coverage was required for inclusion in analysis. Variants and indels were identified by SAMtools.13 Variants were annotated using the software ANNOVAR.14 All chromosome locations are based on the March 2006 (NCBI36/hg18) assembly. For regions with low read depth in linkage peaks, ‘consensus' affected reads and ‘consensus' unaffected reads were constructed where individual reads (ie, all affected or all unaffected individuals) were combined into a single read.
As this is an extremely rare form of a highly infrequent dominant disorder, it is very unlikely that any variants reported in other individuals are disease-causing mutations. Therefore, only novel variants were included. Missense, nonsense, frameshift and indels were identified and included for further analysis. Variants that were present in a heterozygous state in all four affected family members but absent in the two unaffected family members were identified in the first stage of analysis. Variants were further restricted to those present in loci identified in the linkage analysis and excluded if present in our own database of 38 Irish exomes. Variants present in two additional control data sets were excluded. These were (1) the 1000 Genomes Project (http://www.1000genomes.org/) and (2) the NHLBI GO Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/). Later analysis included visual inspection of linkage region variants present in three out of four affected individuals to ascertain whether the exclusion was a result of low coverage of the region (ie, to identify potential false negative findings). This was undertaken using the Generic Genome Browser version 2.4.3 (part of the Generic Model Organism suite of genome analysis software tools, http://www.gmod.org/wiki/Main_Page). Variants were also filtered in the absence of the dbSNP database in order to ensure potential disease-causing variants were not excluded because of their presence in this database. A minor allele frequency of 0.1% was selected as the threshold for inclusion/exclusion.
The possible effect of the two candidate mutations was assessed through the use of bioinformatics programmes (Polyphen-2,15 SIFT,16 SNAP17 and PMUT18). For an average-effect calculation, the values for Polyphen-2 were converted to values from 0 to 1 by dividing all estimates by the max reported value (2.257). For the average calculation, the SIFT values were subtracted from 1 to ensure that the higher scores were associated with the most damaging effect.
Results
Linkage results
The average call rate per sample was 97.96%. Following data cleaning and filtering of SNP data, 30 233 SNPs remained for linkage analysis. The maximum LOD score for this family study was calculated to be 3.3065. Pairwise linkage analysis ruled out all loci previously reported for EA. The highest linkage values for the 2 cM region spanning each EA gene were as follows: KCNA1=−14.3, CACNA1A=−6.9, CACNB4=−10.6, SLC1A3=−14.6 and PRRT2=−1.3.
One novel locus (LOD=3.29) was identified on chromosome 1p36.13-p34.3 (18.5 Mb, rs2743201-rs215791) and spanned 283 genes and 10 miRNAs (see Supplementary Table 1 for more information). Linkage maps for all additional chromosomal regions are present in Supplementary Information.
Sequence results
Exome sequencing resulted in an average of 10.88 Gigabases of mappable sequence data per individual, with ∼35.6% mapping to the targeted exome. The median targeted exome coverage was 117 fold, and the average percentage of the exome sequenced to 10-fold was 96.8%. Regions with low or zero read depth are detailed in Supplementary Table 2. Following data cleaning, the number of variants identified per individual ranged from 119 037 to 150 980 (mean=13 8748, SD=10 667).
Data were cleaned, filtered and analyzed according to the criteria described in the methods section. Filtering the data according to these criteria and presence within the linkage peaks resulted in a dramatic reduction in the number of candidate disease-causing variants (see Table 3). Initially three candidate variants were identified in the FCN3, HSPG2 and UBR4 genes. However, literature reviews have identified that the FCN3 variant (1637delC; rs28357092) has been reported previously.19 The FCN3 gene produces a ficolin protein that has a role in the innate immune system. Homozygosity of this variation has been reported in a single individual with immunodeficiency and recurrent infections. Heterozygosity of this variant does not produce any phenotypic effects reminiscent of an ataxic disorder. Therefore, this variant was excluded as a candidate gene for EA.
Table 3. Summary of variant analysis and segregation with EA phenotype.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Control 1 | Control 2 | All patients | Not in control | |
|---|---|---|---|---|---|---|---|---|
| Q20 and 5x coverage | 150980 | 141952 | 141305 | 119037 | 137102 | 142110 | 52560 | 1375 |
| +Missense, nonsense, frameshift | 10326 | 10391 | 10373 | 10132 | 10352 | 10320 | 5229 | 116 |
| +Not in dbSNP (version 130) | 1269 | 1319 | 1304 | 1258 | 1283 | 1319 | 289 | 20 |
| +In candidate linkage regions | 14 | 12 | 5 | 7 | 6 | 6 | 4 | 4 |
| +Not in Irish control exomes | 10 | 8 | 3 | 6 | 4 | 5 | 3 | 3 |
| +Not in 1000 genomes or ESP | 6 | 6 | 3 | 5 | 3 | 3 | 3 | 3 |
Exome sequencing of six family members, four with EA and two without EA identified multiple variants. Data were cleaned and filtered to include variants that were likely to alter protein function and structure, and not previously reported in dbSNP130. To identify candidate disease-causing variants, exome data were analyzed in conjunction with previous linkage analysis. Four variants within the linkage peaks were common to all patients with EA but absent in the unaffected family members. Three of these variants (in HSPG2, UBR4 and FCN3) were also absent in individuals in the Irish exome database. The variant in FCN3 has been previously reported and is not considered a disease-causing candidate variation. The data prioritization method supports the remaining variants as candidate variants for EA.
Two candidate variants remained. The UBR4 variant (chr1:19277109-19277109) involves a C to T transition that results in an arginine to histidine substitution at amino-acid position 5091. The HSPG2 variant (chr1:22050646-22050646) involves a T to G transversion leading to a serine to arginine substitution at amino-acid position 2380. The data for variants present in linkage peaks in three out of four affected individuals, while absent in the unaffected individuals, were also analyzed. No additional candidate variants were identified. Filtering of the data based solely on the presence/absence and minor allele frequency using the ESP and 1000 genomes data resulted in the identification of the same two variants (see Supplementary Table 3).
The use of ‘consensus' affected and unaffected reads allowed the identification of a single additional variant at chr1:19511144-19511144. This variant (rs148182255) was found to be present with a maf of 0.02 in the 1000 Genomes data. It was excluded from further analysis.
The potential effects of the HSPG2 and UBR4 variants were assessed. The UBR4 variant (average score =0.812 compared with 0.607 for HSPG2) was predicted to be more damaging by each of the prediction programs (see Supplementary Table 4 for more details). All programs reported a non-neutral effect. The HSPG2 variant was predicted to have no effect using SIFT and PMut. The UBR4 variant was validated by Sanger sequencing and shown to segregate with the disorder.
Discussion
Seven different EA subtypes have been described to date, with genes and disease loci reported for many (see Table 1 for more details). In this paper, we report a novel form of dominant EA in a large non-consanguineous Irish family. The phenotype is relatively uniform in all affected individuals. The response to clonazepam but not acetazolamide is consistent within affected individuals. Most forms of EA reported to date respond to acetazolamide, possibly because of its effect on carbonic anhydrase causing metabolic acidosis or because of effects on ion channels.20, 21 Clonazepam is not typically used in EA; however, it has been used successfully in vestibular migraine, a condition suspected to be due to channelopathy in some cases.22 It increases the frequency of chloride ion channel opening, enhancing the inhibitory effect of GABA at the GABAa receptor;23 however, it has multiple other central effects. Benzodiazepines have also been shown to have an inhibitory effect on voltage-dependant calcium channels,24 which may be relevant in the setting of the UBR4 genetic variant that we have described in this family.
Exome sequencing provided the most economic and efficient method for variant screening because of the large number of candidate genes in the single disease locus identified. As expected, exome sequencing confirmed that there were no causative variants in any of the previously identified EA genes. It has been reported that deletions within the CACNA1A gene can cause the development of EA.25 If a deletion within CACNA1A or indeed any of the previously reported EA genes, a haplotype common to all of the affected family members would surround the gene and the deletion. These haplotypes would be detected by linkage analysis. Linkage scores surrounding the known EA genes (1Mb centromeric and telomeric) were not significant. Variants that were novel, within the linkage peaks and only present in the affected individuals, were limited to just two genes, HSPG2 and UBR4.
In the absence of functional work, it remains possible that the variants reported here are not disease causing but instead are exonic variants of no functional consequence that happen to be present in the linkage peaks identified in the family. A disease-causing variant within an intron or outside the gene may be the true-disease causing one. Expanded repeats are common causes of ataxic disorders, for example, SCA1 and SCA2. However, almost all EA mutations identified to date have been missense or truncating variants. An expanded repeat in the CACNA1A gene has been reported in a Korean family in which some affected individuals presented with a phenotype similar to spinocerebellar ataxia 6, others with a phenotype similar to EA2, while other members of the family also had expanded repeats but exhibited no clinical symptoms of either disorder.26 Triplet-repeat disorders often lead to the phenomena of anticipation. No anticipation with regard to age of onset or severity of disorder has been observed in this three-generation family.
The protein produced by HSPG2 is a heparan sulfate-containing proteo-glycan commonly known as perlecan. It is a large complex protein in the extracelluar matrix and has a crucial role in maintaining the structure and function of the neuromuscular junction and vascular system. In humans, mutations in this gene have been reported in two recessive disorders, Schwartz–Jampel syndrome type 1 (SJS, OMIM 255800)27, 28 and Silver–Handmaker type of dyssegmental dysplasia (DDSH, OMIM 22410).29 Parents, despite being obligate carriers for a single copy of a mutation, have no symptoms consistent with an EA phenotype or indeed any neurological defects (personal communication with Prof L Al Gazali, United Arab Emirates University, UAE). Further, the HSPG variant lies within the Ig-like C2-type 9 domain. No DDSH or SJS variants have been reported to be located within this domain.
Although HSPG is an unlikely candidate, its role in the development of EA cannot be absolutely excluded. One possible hypothesis for perlecan may involve binding of immunoglobulin-like (Ig-like) repeats in perlecan (n=22) and those in ion channel proteins. The HSPG2 variant in this family is located in domain IV of the protein, which contains 22 Ig-like domains. Such interactions have already been proposed (Kirn-Safran et al30). The voltage-gated sodium channels in rats are composed of a one alpha and two beta subunits. The beta subunits are both predicted to form Ig folds in their extracellular domains.31 The deletion of the extracellular domain of the beta-1 prevents sodium channel modulation.32 Beta-1 knockout mice develop an ataxic gait and frequent generalized seizures that are followed by periods of immobility.33 Mutations in sodium channel alpha subunit genes have already been reported to cause complex neurological phenotypes that include ataxia including SCN2A34 and SCN8A.35
The UBR4 gene encodes an ubiquitin-protein ligase that interacts in the nucleus with the retinoblastoma-associated protein. In the cytoplasm, it has been localized to and shown to interact with the smooth endoplasmic reticulum in neurons.36 These are known to be a major intracellular calcium store. It has also been shown to interact with calcium bound calmodulin in the cytoplasm,37 highlighting the potential role for UBR4 in calcium signaling. Calmodulin has its role in calcium signaling through calcium-dependent inactivation or calcium-dependent facilitation.38, 39 It has been shown to bind and act as a regulator of neural excitability in KCNQ40 and Cav2.1 channels (see Halling et al for a recent review41). The CACNA1A gene encodes the Cav2.1 channel, and it is mutations in this gene that cause the most common form of EA (EA2). The ataxic mouse model (‘Pogo') has shown to have altered purkinje cell responses and calmodulin content.42 Nakatani et al37 hypothesized that UBR4 may act as a sensor of Ca2+, which is released through ITPR1, an inositol triphosphate receptor. ITPR1 is an intracellular calcium-release channel gated by IP3 (inositol 1,4,5-triphosphate), and purkinje cells have dense concentrations of ITPR1 on their post-synaptic endoplasmic reticulums.43 ITPR1 is a major determinant of motor coordination in mammals and may well be the ‘centre of the storm' with regard to the development of ataxic disorders in addition to other neurological conditions (see Schorge et al44 for a recent review). Null and missense mutations of ITPR1 can result in SCA15.45 Reduced ITPR1 expression was reported in post-mortem study of the cerebellums of two patients with ataxia.46 In addition, ITPR1 has been reported to be a binding protein for CA8. Mutations in the gene encoding this protein can lead to congenital cerebellar ataxia and mild mental retardation (OMIM 613227). A mouse model (the ‘waddles' mouse) develops ataxia, appendicular dystonia and abnormal ambulation as a result of a 19bp deletion in exon 8.47 It is possible that atypical binding of UBR4 with calmodulin and/or ITPR1 may result in an abnormal calcium sensor system within the neuron and hence the development of ataxia. These reported findings highlight an interesting possible role of UBR 4 in the development of ataxia.
In summary, we present the clinical details of a family presenting with a novel form of episodic ataxia. Utilizing linkage analysis, we report a previously unreported locus on chromosome 1p36.13-p34.3. Furthermore, next-generation sequencing has identified two potential disease-causing variants. The first lies in the HSPG2 gene (S2380R). The second variant lies in the UBR4 gene (R5091H). This gene is known to interact with calmodulin, a calcium sensor protein, which has been previously reported to be a regulator of the Cav2.1 channel. These findings highlight a novel candidate gene for EA.
Acknowledgments
The authors of this paper would like to thank the family members who participated in the research. We would also like to thank Ataxia Ireland for funding this project. We also thank L Al Gazali for providing additional clinical data related to her patients.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Brandt T, Strupp M. Episodic ataxia type 1 and 2 (familial periodic ataxia/vertigo) Audiol Neurootol. 1997;2:373–383. doi: 10.1159/000259262. [DOI] [PubMed] [Google Scholar]
- Jen J, Kim GW, Baloh RW. Clinical spectrum of episodic ataxia type 2. Neurology. 2004;62:17–22. doi: 10.1212/01.wnl.0000101675.61074.50. [DOI] [PubMed] [Google Scholar]
- Escayg A, De Waard M, Lee DD, et al. Coding and noncoding variation of the human calcium-channel beta4-subunit gene CACNB4 in patients with idiopathic generalized epilepsy and episodic ataxia. Am J Hum Genet. 2000;66:1531–1539. doi: 10.1086/302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen JC, Wan J, Palos TP, Howard BD, Baloh RW. Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology. 2005;65:529–534. doi: 10.1212/01.wnl.0000172638.58172.5a. [DOI] [PubMed] [Google Scholar]
- Gardiner AR, Bhatia KP, Stamelou M, et al. PRRT2 gene mutations: from paroxysmal dyskinesia to episodic ataxia and hemiplegic migraine. Neurology. 2012;79:2115–2121. doi: 10.1212/WNL.0b013e3182752c5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labate A, Tarantino P, Viri M, et al. Homozygous c.649dupC mutation in PRRT2 worsens the BFIS/PKD phenotype with mental retardation, episodic ataxia, and absences. Epilepsia. 2012;53:e196–e199. doi: 10.1111/epi.12009. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Cader MZ, Steckley JL, Dyment DA, McLachlan RS, Ebers GC. A genome-wide screen and linkage mapping for a large pedigree with episodic ataxia. Neurology. 2005;65:156–158. doi: 10.1212/01.wnl.0000167186.05465.7c. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- Munthe-Fog L, Hummelshoj T, Ma YJ, et al. Characterization of a polymorphism in the coding sequence of FCN3 resulting in a Ficolin-3 (Hakata antigen) deficiency state. Mol Immunol. 2008;45:2660–2666. doi: 10.1016/j.molimm.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Eguchi H, Tsujino A, Kaibara M, et al. Acetazolamide acts directly on the human skeletal muscle chloride channel. Muscle Nerve. 2006;34:292–297. doi: 10.1002/mus.20585. [DOI] [PubMed] [Google Scholar]
- Gordon N. Episodic ataxia and channelopathies. Brain Dev. 1998;20:9–13. doi: 10.1016/s0387-7604(97)00086-7. [DOI] [PubMed] [Google Scholar]
- Bisdorff AR. Management of vestibular migraine. Ther Adv Neurol Disord. 2011;4:183–191. doi: 10.1177/1756285611401647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riss J, Cloyd J, Gates J, Collins S. Benzodiazepines in epilepsy: pharmacology and pharmacokinetics. Acta Neurol Scand. 2008;118:69–86. doi: 10.1111/j.1600-0404.2008.01004.x. [DOI] [PubMed] [Google Scholar]
- Taft WC, DeLorenzo RJ. Micromolar-affinity benzodiazepine receptors regulate voltage-sensitive calcium channels in nerve terminal preparations. Proc Natl Acad Sci USA. 1984;81:3118–3122. doi: 10.1073/pnas.81.10.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Mamsa H, Johnston JL, et al. Large Genomic Deletions in CACNA1A Cause Episodic Ataxia Type 2. Front Neurol. 2011;2:51. doi: 10.3389/fneur.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh SH, Kim HT, Kim SH, Lee GY, Kim J, Kim MH. Spinocerebellar ataxia type 6 and episodic ataxia type 2 in a Korean family. J Korean Med Sci. 2001;16:809–813. doi: 10.3346/jkms.2001.16.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicole S, Davoine CS, Topaloglu H, et al. Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz-Jampel syndrome (chondrodystrophic myotonia) Nat Genet. 2000;26:480–483. doi: 10.1038/82638. [DOI] [PubMed] [Google Scholar]
- Stum M, Davoine CS, Vicart S, et al. Spectrum of HSPG2 (Perlecan) mutations in patients with Schwartz-Jampel syndrome. Hum Mutat. 2006;27:1082–1091. doi: 10.1002/humu.20388. [DOI] [PubMed] [Google Scholar]
- Arikawa-Hirasawa E, Wilcox WR, Le AH, et al. Dyssegmental dysplasia, Silverman-Handmaker type, is caused by functional null mutations of the perlecan gene. Nat Genet. 2001;27:431–434. doi: 10.1038/86941. [DOI] [PubMed] [Google Scholar]
- Kirn-Safran C, Farach-Carson MC, Carson DD. Multifunctionality of extracellular and cell surface heparan sulfate proteoglycans. Cell Mol Life Sci. 2009;66:3421–3434. doi: 10.1007/s00018-009-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom LL, Ragsdale DS, De Jongh KS, et al. Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- McCormick KA, Isom LL, Ragsdale D, Smith D, Scheuer T, Catterall WA. Molecular determinants of Na+ channel function in the extracellular domain of the beta1 subunit. J Biol Chem. 1998;273:3954–3962. doi: 10.1074/jbc.273.7.3954. [DOI] [PubMed] [Google Scholar]
- Chen C, Westenbroek RE, Xu X, et al. Mice lacking sodium channel beta1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J Neurosci. 2004;24:4030–4042. doi: 10.1523/JNEUROSCI.4139-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Anttonen AK, Liukkonen E, et al. SCN2A mutation associated with neonatal epilepsy, late-onset episodic ataxia, myoclonus, and pain. Neurology. 2010;75:1454–1458. doi: 10.1212/WNL.0b013e3181f8812e. [DOI] [PubMed] [Google Scholar]
- Trudeau MM, Dalton JC, Day JW, Ranum LP, Meisler MH. Heterozygosity for a protein truncation mutation of sodium channel SCN8A in a patient with cerebellar atrophy, ataxia, and mental retardation. J Med Genet. 2006;43:527–530. doi: 10.1136/jmg.2005.035667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SY, Wang J, Asada N, et al. Protein 600 is a microtubule/endoplasmic reticulum-associated protein in CNS neurons. J Neurosci. 2008;28:3604–3614. doi: 10.1523/JNEUROSCI.5278-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Konishi H, Vassilev A, et al. p600, a unique protein required for membrane morphogenesis and cell survival. Proc Natl Acad Sci USA. 2005;102:15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaria CD, Soong TW, Alseikhan BA, Alvania RS, Yue DT. Calmodulin bifurcates the local Ca2+ signal that modulates P/Q-type Ca2+ channels. Nature. 2001;411:484–489. doi: 10.1038/35078091. [DOI] [PubMed] [Google Scholar]
- Liang H, DeMaria CD, Erickson MG, Mori MX, Alseikhan BA, Yue DT. Unified mechanisms of Ca2+ regulation across the Ca2+ channel family. Neuron. 2003;39:951–960. doi: 10.1016/s0896-6273(03)00560-9. [DOI] [PubMed] [Google Scholar]
- Shahidullah M, Santarelli LC, Wen H, Levitan IB. Expression of a calmodulin-binding KCNQ2 potassium channel fragment modulates neuronal M-current and membrane excitability. Proc Natl Acad Sci USA. 2005;102:16454–16459. doi: 10.1073/pnas.0503966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling DB, Aracena-Parks P, Hamilton SL. Regulation of voltage-gated Ca2+ channels by calmodulin. Sci STKE. 2005. p. re15. [DOI] [PubMed]
- Lee KY, Kim JS, Kim SH, et al. Altered Purkinje cell responses and calmodulin expression in the spontaneously ataxic mouse, Pogo. Eur J Neurosci. 2011;33:1493–1503. doi: 10.1111/j.1460-9568.2011.07641.x. [DOI] [PubMed] [Google Scholar]
- Bardo S, Cavazzini MG, Emptage N. The role of the endoplasmic reticulum Ca2+ store in the plasticity of central neurons. Trends Pharmacol Sci. 2006;27:78–84. doi: 10.1016/j.tips.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Schorge S, van de Leemput J, Singleton A, Houlden H, Hardy J. Human ataxias: a genetic dissection of inositol triphosphate receptor (ITPR1)-dependent signaling. Trends Neurosci. 2010;33:211–219. doi: 10.1016/j.tins.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Leemput J, Chandran J, Knight MA, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic N, Milosevic A, Ehrlich BE. Calcium signaling molecules in human cerebellum at midgestation and in ataxia. Early Hum Dev. 1999;54:103–116. doi: 10.1016/s0378-3782(98)00090-5. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Yan J, Zhao Y, et al. Carbonic anhydrase-related protein VIII deficiency is associated with a distinctive lifelong gait disorder in waddles mice. Genetics. 2005;171:1239–1246. doi: 10.1534/genetics.105.044487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.