Abstract
Wolf–Hirschhorn syndrome (WHS) is a complex genetic disorder caused by the loss of genomic material from the short arm of chromosome 4. Genotype–phenotype correlation studies indicated that the loss of genes within 4p16.3 is necessary for expression of the core features of the phenotype. Within this region, haploinsufficiency of the genes WHSC1 and LETM1 is thought to be a major contributor to the pathogenesis of WHS. We present clinical findings for three patients with relatively small (<400 kb) de novo interstitial deletions that overlap WHSC1 and LETM1. 3D facial analysis was performed for two of these patients. Based on our findings, we propose that hemizygosity of WHSC1 and LETM1 is associated with a clinical phenotype characterized by growth deficiency, feeding difficulties, and motor and speech delays. The deletion of additional genes nearby WHSC1 and LETM1 does not result in a marked increase in the severity of clinical features, arguing against their haploinsufficiency. The absence of seizures and typical WHS craniofacial findings in our cohort suggest that deletion of distinct or additional 4p16.3 genes is necessary for expression of these features. Altogether, these results show that although loss-of-function for WHSC1 and/or LETM1 contributes to some of the features of WHS, deletion of additional genes is required for the full expression of the phenotype, providing further support that WHS is a contiguous gene deletion disorder.
Keywords: 4p16.3 deletion, Wolf–Hirschhorn syndrome, WHSC1, LETM1, 3D facial analysis
Introduction
Wolf–Hirschhorn syndrome (WHS; OMIM #194190) is a complex genetic disorder caused by deletions in the short arm of chromosome 4 (4p) and occurring in 1 in 50 000 to 1 in 20 000 births.1 The phenotype of WHS consists of a broad range of clinical findings. The core features include a typical craniofacial appearance, growth deficiency, developmental delays, intellectual disability of variable degree and seizures (or EEG anomalies) in most patients. Variable clinical findings include feeding difficulties, congenital malformations including orofacial cleft, cardiac, renal and urogenital malformations, skeletal and dental anomalies, hearing loss, recurrent infections and other complications.2, 3, 4
The broad clinical spectrum of WHS can be attributed to variability in both the mechanism and genomic content of 4p deletions. WHS is most often caused by terminal deletions, however, interstitial deletions, unbalanced translocations and complex genomic rearrangements account for a significant proportion of cases.2, 5 The majority of 4p deletions are non-recurrent. Genotype–phenotype correlation studies have shown a relationship between deletion size and severity of clinical presentation.6, 7, 8 Of patients meeting the minimal diagnostic criteria,9 the smallest terminal deletion identified was ∼1.9 Mb,1, 9, 10 suggesting that genes within this interval of 4p16.3 are responsible for the core features of WHS.8, 11 Beyond this region, the loss of additional critical genes appears to be responsible for variably present features, such as congenital malformations or hearing loss.1, 5, 8
The identification of patients with atypical 4p deletions has provided key insight into which regions of 4p16.3 may (or may not) contribute to the pathogenesis of WHS. For example, small terminal deletions (up to 400 kb) have been inherited from phenotypically normal individuals;12, 13, 14 indicating that monosomy of this region is likely benign.11 Between ∼1.8 and 2.0 Mb from the 4p terminus, two adjacent critical regions were proposed based on the smallest region of overlap (SRO) among the deletions of individuals with or without the core features of WHS. The more proximal critical region (WHSCR) was delineated first15 and mapping within this 165-kb interval identified two genes, WHSC1 and WHSC2.16, 17 The identification of two WHS patients with more distal 4p16.3 terminal deletion breakpoints9, 10 shifted and expanded the critical region (WHSCR-2) to a 300–600-kb region overlapping the 5′ end of WHSC1 and encompassing LETM1, a candidate gene for seizures.18 Although there is evidence to support a contribution of WHSC1 and LETM1 to the core WHS phenotype,19, 20 focal deletions or mutations of these genes have not been identified in WHS patients.1, 8, 16 Furthermore, not all patients with deletions overlapping both WHSC1 and LETM1 express the full WHS phenotype1, 12, 13 and deletions elsewhere in 4p16.3 have been identified in patients with features overlapping WHS.1, 21, 22, 23, 24 Altogether, these findings suggest that multiple loci within the ∼0.4–2.0 Mb terminal region of 4p16.3 contribute to WHS.
To provide additional insight into the genetic etiology of WHS, we present clinical findings for three patients with relatively small interstitial deletions that overlap WHSCR-2. These patients do not express all of the core features of WHS; they lack the typical craniofacial gestalt and do not exhibit seizures, but do share certain aspects of the WHS phenotype including growth delays, feeding difficulties, and developmental delays in motor abilities and speech, although of a milder degree compared with WHS patients. Our findings support the hypothesis that haploinsufficiency of WHSC1 and/or LETM1 contributes to WHS, but that loss of distinct and/or additional genes in 4p16.3 is necessary for the expression of the core WHS phenotype.
Materials and methods
Clinical and molecular cytogenetic studies
Patients 1–3 were referred to ARUP Laboratories (Salt Lake City, UT, USA) through JCC and/or their respective medical genetics providers. Patients 1 and 3 were ascertained by DLE at Seattle Children's Hospital (Seattle, WA, USA). Patient 2 was ascertained by DC and colleagues at Boston Children's Hospital (Boston, MA, USA). Cytogenomic microarray analysis (CMA) and FISH testing were performed through regular clinical services in CLIA-certified laboratories. CMA for patient 3 and his family was performed as a research study at ARUP Laboratories. Genomic coordinates for CMA results are reported in relation to the current human reference sequence (hg19/GRCh37). Metaphase FISH analysis was performed using a commercially available probe set for WHSCR (WHSC1 and D4Z1; Vysis, Inc., Abbott Laboratories, Green Oaks, IL, USA). Details are provided in the Results section. Permission to publish clinical descriptions, results of genetic testing and patient photographs/images was obtained with written parental consent through the University of Utah Clinical Genetics Research Program and with approval from the University of Utah Institutional Review Board. CMA results were not submitted to a public database.
3D facial analysis
3D facial analysis using dense surface modeling was performed for patients 1 and 3 (patient 2 was not available for this analysis). 3D face images of patients 1 and 3 were collected at age 2 years 10 months and 2 years 1 month, respectively, using a commercial photogrammetric device at Seattle Children's Hospital. Analysis was performed by PH using methods described previously12, 23 and software developed by MS and previous lab members. Comparative image data sets using age- and sex-matched controls and individuals with WHS were used for each patient.
Clinical reports
Patient 1
Patient 1 (Figures 1a and b) is a 3-year-old female who presented with postnatal onset growth delays, feeding difficulties and developmental delays. She was born to healthy parents, a 34-year-old G4P1-2 SAB2 mother and 37-year-old father, at 41 weeks gestation after an uncomplicated pregnancy. All growth parameters at birth were 25th centile, weight was 3.2 kg, length was 48 cm and OFC was 34 cm. At 15 months, the patient was referred to Genetics for evaluation and testing because of concerns for her linear growth and development. Her height and weight Z-scores were <–2 and OFC was 25th centile. She had a delayed bone age, delayed dental eruption and a mild degree of hypotonia. Growth hormone therapy (Omnitrope) was started at age 19 months and resulted in gains in linear growth; at 2 years 9 months her height, weight and OFC measurements were 5th–10th, 23rd and 50th centile, respectively. She had fed well as an infant and transitioned to solids at 6 months without difficulty; however, feeding issues arose at 12 months, and by 15 months, she was refusing most solid foods. Her limited intake was considered to be secondary to oral aversion and dysphagia and was accompanied by reflux, vomiting and constipation. Sleep interruptions were noted, and linked to her feeding routine. The patient had frequent infections of the upper respiratory tract and ears, for which tubes were placed. An immunologic workup that included quantitative serum immunoglobulins (IgA, IgG and IgM), B-cell and T-cell counts, specific antibody response and mitogen studies was essentially normal. An echocardiogram at 19 months revealed a left aortic arch and retroesophogeal course of the subclavian artery. These findings were considered to be unrelated to her feeding issues and required no intervention. Awake and drowsy EEG studies at 17 months were normal, and at 3 years 3 months she had no recognized seizure activity. Renal ultrasound and audiologic testing at 18 months were normal. The patient was found to have a mild degree of bilateral esotropia, which improved without intervention. Developmental testing at age 2 years 9 months showed cognitive skills at the 75th centile, social/emotional skills at the 16th centile and adaptive skills at the 21st centile (Bayley); fine motor skills at the 12th centile and gross motor at the 10th centile (PDMS-2); auditory comprehension was at the 68th centile and expressive communication at the 66th centile (PLS-4); and articulation was 41st centile (GFTA-2). The patient underwent behavioral intervention for feedings, as well as occupational and speech therapy.
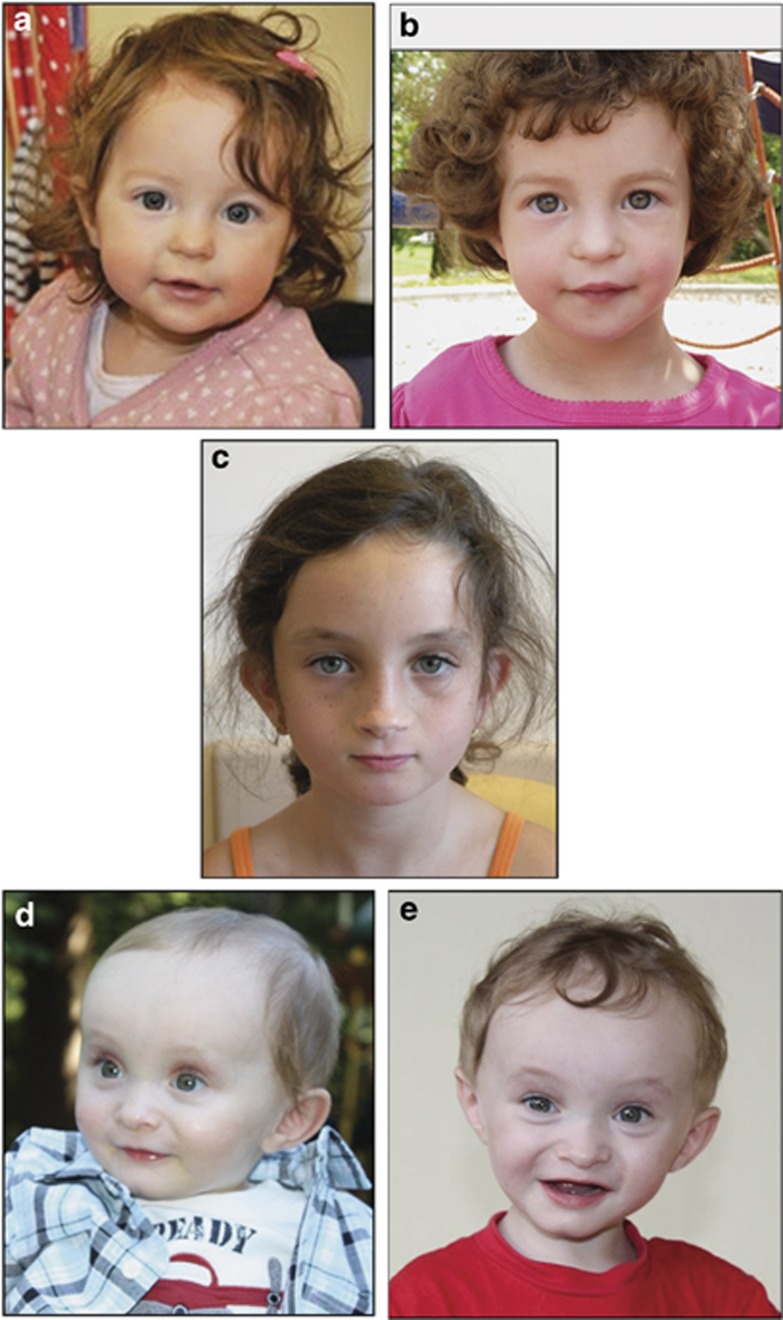
Figure 1.
Photographs of patients 1–3. (a, b) Patient 1 at age 15 months (a) and 2 years 9 months (b). (c) Patient 2 at age 7. (d, e) Patient 3 at age 12 months (d) and 18 months (e).
Patient 2
Patient 2 (Figure 1c) is an 11-year-old female who presented with prenatal onset growth delays, feeding difficulties, developmental delays, and behavioral and learning disabilities. She was born at 41 weeks gestation weighing 2.5 kg (<5th centile) to a healthy 26-year-old G3P1 mother after a pregnancy complicated by IUGR. The proposita was the only child to her parents, who were noted to have normal height and intelligence. During infancy, the patient had failure to thrive and a mild degree of hypotonia that were compounded by poor eating and oral feeds aversion. A gastronomy tube was placed because of feeding issues that included recurrent vomiting and GERD. Vesicoureteral reflux was identified during early infancy and the patient was treated with a prophylactic antibody regimen. Echocardiogram, ECG and renal ultrasound were normal. At age 7 years, the patient's growth parameters were all within the 10th–25th centile. On physical examination, craniofacial findings included a small, triangular face and flat profile, a high nasal bridge, mildly cupped ears, and a small mouth. At 11 years, seizure activity had not been observed. A developmental evaluation at 3 years identified delayed fine and gross motor skills and language development and the patient was diagnosed with a communication disorder (NOS; delayed language acquisition). Subsequently, she was also diagnosed with ADHD/combined type. At age 7 years, the proposita had been receiving services in regular class and had repeated kindergarten. Formal IQ testing revealed a low to normal range score (∼85, full scale – WASI). She was also diagnosed with a developmental coordination disorder and was noted to be at risk for a learning disability.
Patient 3
Patient 3 (Figures 1d and e) is a 2-year-old male with prenatal onset growth deficiency, feeding difficulties, and mild-to-moderate developmental delays. He was born to healthy parents, a 33-year-old G3P2 mother and 40-year-old father, at 38 weeks following a pregnancy complicated by reduced fetal movements and IUGR identified at 30 weeks. At birth, his weight was 2.0 kg (<3rd centile), length was 48 cm (<3rd centile) and head circumference was 34 cm (10–25th centile). Apgar scores were normal. He also passed his newborn hearing screen. At 10 months, the patient was referred to Genetics for evaluation and testing for failure to thrive and developmental delays. His length and weight z-scores were <–2 and OFC was 10th–25th centile. A skeletal study was normal. The patient had delayed dental eruption, with only upper and lower central incisors at 20 months. Feeding difficulties included a limited oral intake and oral aversion. He had no history of gastroesophageal reflux or constipation. A swallowing study at 18 months revealed oropharyngeal phase dysphagia and delayed oral skills. Interventions included a nasogastric tube placement and feedings from 11 to 20 months and occupational and feeding therapy. His weight gain plateaued following the discontinuation of his nasogastric tube feedings. At 2 years 2 months, his length and weight z-scores were −2.05 and −3.82 and his OFC was 10–25th centile. An undescended right testis and distal ventral chordee identified in infancy were corrected at 14–15 months. A renal ultrasound at 15 months identified a mild left pelvocaliectasis with slight dilation of the left proximal ureter, which subsequently resolved. The patient had chronic rhinitis, but had not had immunologic labs. Frequent ear infections were denied. Results of an echocardiogram and dilated ophthalmologic exam were normal. Awake and drowsy EEG studies at 14–15 months were normal, and at 2 years 5 months he had no recognized seizure activity. Sleep problems were denied. Craniofacial features were considered normal and include a high and broad forehead and mild micrognathia. The patient was noted to have flammeus nevus to his occiput, but no other birthmarks. Developmental delays were identified in the areas of motor skills and language acquisition; the patient crawled at 9–10 months and at 20 months, he was walking, using a few single words and had started to babble in a conversational manner. The patient received physical therapy for motor delays.
Results
Molecular cytogenetic studies
CMA for patient 1 performed at Seattle Children's Hospital on a Roche NimbleGen (Madison, WI, USA) 135K oligonucleotide array platform identified a single copy loss of 377-kb (arr[hg19] 4p16.3(1 743 630–2 120 247)). The minimally deleted region encompasses 10 genes: 8 OMIM genes (3/16 3′ exons of TACC3, FGFR3, LETM1, WHSC1, WHSC2, C4orf48, NAT8L and 7/24 3′ exons of POLN) and 2 noncoding genes (SCARNA22 and MIR943). Parental testing by metaphase FISH was normal (data not shown), indicating a de novo deletion. No other clinically relevant copy number variants were identified.
CMA for patient 2 performed at Boston Children's Hospital on a custom 244K ArrayCGH (Agilent Technologies, Santa Clara, CA, USA) detected a single copy loss of 170-kb (arr[hg19] 4p16.3(1 827 029–1 997 169)). The minimally deleted region includes LETM1 (7/14 5′ exons), WHSC1, SCARNA22, MIR943 and WHSC2 (10/11 3′ exons). No additional copy number alterations were reported. Parental arrays were normal, indicating a de novo deletion. Parental FISH analysis ruled out the possibility of a balanced rearrangement (data not shown).
Patient 3 was initially studied by CMA at Seattle Children's Hospital, where he was found to have a 60-kb deletion involving the LETM1 and WHSC1 genes (data not shown). Upon referral to ARUP Laboratories, CMA testing was performed for both the patient and his parents on an Affymetrix CytoScan HD (Affymetrix, Santa Clara, CA, USA) platform. A 67-kb de novo single copy loss (arr[hg19] 4p16.3(1 828 866–1 895 728)) was detected, which results in the partial deletion of the genes LETM1 (6/14 5′ exons) and WHSC1 (minimum 3/25 and maximum 5/25 5′ exons). Owing to limitations in the resolution of the array, the involvement of WHSC1 exons 4 and 5 could not be determined by this analysis. No additional clinically concerning copy number alterations were identified.
3D facial analysis
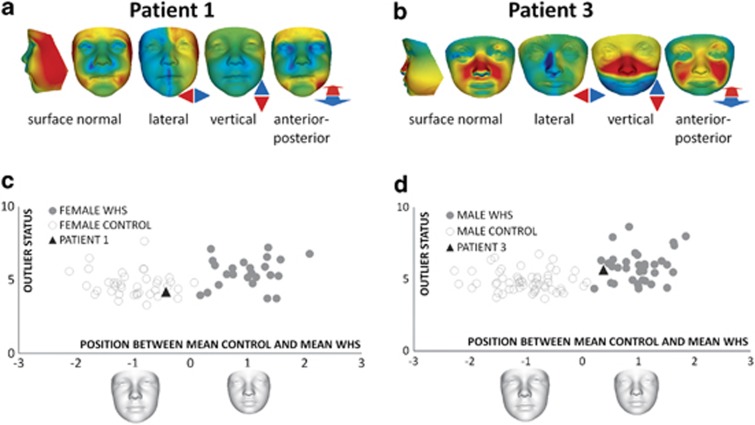
Patients 1 and 3 were further studied by 3D facial analysis. Animated comparison of the face of patient 1 with an age- and sex-matched control mean showed a few differences: lateral narrowing, slight flattening of the forehead and mildly low set ears (Supplementary Video 1). Heat-mapped face signatures showed the significance of the reduced face width in surface normal and lateral comparisons (Figure 2a). Patient 1's vertical face length was normalized, perhaps by a year of hGH treatment, as similar findings have been observed in Prader–Willi syndrome patients treated with hGH.25 The only other significant difference from controls was in forward displacement of the inner and outer canthi, shown in the anterior–posterior signature. The bilateral red regions adjacent to the lower lip in the anterior–posterior comparison are likely due to a non-neutral pose.
Figure 2.
3D facial analysis of patient 1 (a, c) and patient 3 (b, d). (a, b) Heat-mapped face signatures showing normalized face shape differences (displacement of surface points orthogonal to face surface and parallel to three axes) compared with matched control mean. The red/green/blue coloring reflects displacement orthogonal to the surface in the first signature (red/green/blue=contraction/coincidence/expansion) and in the three axial signatures parallel to the colored arrows with green=no displacement and maximal red-blue hues reflecting 2 SDs or more. (c, d) Comparison of subject profiles with WHS patients and controls by blinded closest mean classification. X axis—similarity to average faces of sex-matched control (−1) and WHS (+1) subgroups. Y axis – outlier status (distance from control-WHS mean axis). For patient 1: n(WHS)=24 and n(control)= 38. For patient 3: n(WHS)=35 and n(control)= 56. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Patient 3 showed some dysmorphic facial features, including a long nose, elongated midface with malar flattening, depressed nasal bridge, tall orbits, flattening and downward displacement of the maxillary region, and a small mandible (Supplementary Video 2). Heat-mapped signatures clearly indicate these differences (Figure 2b).
To determine whether patients 1 and 3 have facial characteristics more similar to controls or to WHS patients, a blinded closest mean classification was conducted comparing the patients with the average faces of age- and sex-matched control and WHS subgroups selected from an existing data set.12 In both Figures 2c and d the control and WHS average faces are normalized to the positions −1 and +1, respectively, on the X axis of the scatter plots. Patient 1 was classified to the periphery of the control group, suggesting no overlapping WHS facial characteristics, whereas patient 3 was classified to the periphery of the WHS group, suggesting some overlap with the typical WHS facial gestalt, but of a mild nature.
Discussion
We describe the findings for three previously uncharacterized and unrelated patients with relatively small deletions that overlap the critical region for WHS (WHSCR-2). All three patients exhibited clinical findings observed in WHS (summarized in Table 1); however, based on the minimal diagnostic criteria,9 these patients do not meet a WHS diagnosis. Shared findings included growth deficiency, feeding difficulties, and developmental delays in motor function and speech. Variably present features in at least two patients included IUGR, delayed dental eruption and recurrent infections. Patients 2 and 3 both had anomalies of the genitourinary system, although they were different abnormalities. Photographs of the subjects show clearly that these patients do not have the typical WHS gestalt (Figure 1).
Table 1. Summary of molecular cytogenetic and clinical findings.
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| Age at most recent observation | 2 years 5 months | 11 years | 20 months |
| Molecular cytogenetic findings | |||
| Deletion size | 377 kb | 170 kb | 67 kb |
| Deletion location on chr. 4 (GRCh37/hg19) | 1 743 630–2 120 247 | 1 827 029–1 997 169 | 1 828 867–1 895 728 |
| Inheritance | De novo | De novo | De novo |
| Clinical findings | |||
| IUGR | − | + | + |
| Postnatal growth delay | + | + | + |
| Feeding difficulties | + | + | + |
| Hypotonia | + | + | + |
| Motor delays | + | + | + |
| Speech delays | + | + | + |
| Intellectual disability | NA | Mild | NA |
| Behavioral concerns | − | + | − |
| Seizures/EEG anomalies | −/− | −/NA | −/− |
| Microcephaly | − | − | − |
| WHS craniofacial profile | − | − | − |
| Major congenital malformations | − | − | − |
| Echocardiogram findings | + | − | − |
| Renal ultrasound findings | − | − | + |
| Genitourinary anomalies | − | + | + |
| Delayed bone age | + | NA | − |
| Delayed dental eruption | + | − | + |
| Recurrent infections | + | − | + |
| Hearing deficiency | − | − | − |
| Sleep problems | + | − | − |
(+), feature present; (−), feature absent; NA, feature not assessed.
3D facial analysis provides an objective method for the evaluation of craniofacial features, especially in cases where a mild degree of overlap with a recognizable syndrome such as WHS is suspected. This analysis classified patient 1 within the control group while patient 3 showed peripheral overlap with the WHS patients (Figure 2, Supplementary Videos 1 and 2). These findings were somewhat unexpected based on deletion size comparison. Evidence from a recent study of patients with Prader–Willi syndrome suggests that dysmorphic features persist even if face length is normalized by growth hormone therapy,25 arguing against a misclassification of patient 1.
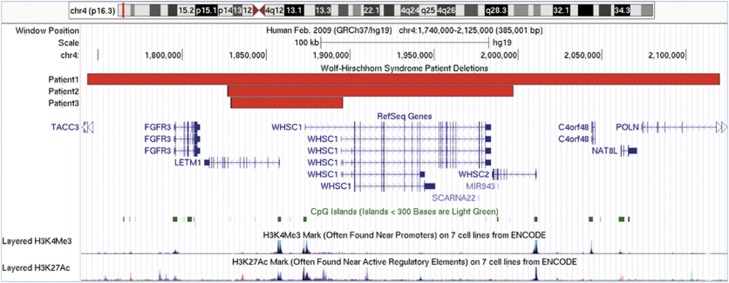
The SRO is the deleted region of patient 3 (Figure 3). This 67-kb segment overlaps the 5′ ends of LETM1 and WHSC1 and includes the putative promoters and regulatory elements of these genes. Deletion of this region is presumed to result in their loss-of-function; although for WHSC1, transcriptional complexity and uncertain involvement of exons 4 and 5 raises the possibility of retained function through expression from an alternative promoter. Similar clinical findings among our cohort argue against this hypothesis.
Figure 3.
Genomic region of 4p16.3 involved in the deletions of patients 1–3. Snapshot of genomic region 1.740–2.125 Mb of chr. 4 displayed using the UCSC genome browser (GRCh37/hg19) showing the location of deletions of patients 1–3. The RefSeq Genes track shows the mapped location and exon/intron structure of gene transcripts from the NCBI RNA reference sequences collection of known protein-coding (dark-colored) and non-protein-coding (light-colored) genes. Non-coding exons are displayed as half the height of coding exons and the direction of transcription with arrowheads. The CpG islands track shows the location of predicted clusters of methylated CG dinucleotides, which are common near transcription start sites and may be associated with promoter regions. The ENCODE layered H3K4Me3 and H3K27Ac tracks show the location and levels of enrichment of H3K4Me3 and H3K27Ac histone marks, which are molecular correlates of promoters and enhancers, respectively.
WHSC1 encodes a nuclear-localized, SET domain-containing transcriptional regulatory protein with histone methyltransferase (HMT) activity.26, 27 Histone modification is essential for the establishment and maintenance of transcriptional programs during development and gene dosage alterations of histone modifiers are a known cause of intellectual disability syndromes.28 The HMT function of WHSC1 has been implicated in diverse biological processes, including early development,29 cytokine signaling,30 the DNA damage response31, 32 and class switch recombination.33 In addition to its functional diversity, WHSC1 has a complex pattern of expression, with the potential to regulate multiple processes during development.16 In mice, deletion of the SET domain results in a phenotype characterized by severe growth restriction, craniofacial malformations and midline fusion defects,29 consistent with phenotypes observed in WHS. Furthermore, dosage sensitivity of WHSC1 (also known as MMSET/NSD2) is supported by its overexpression in a variety of cancer types.34, 35 These findings suggest WHSC1 haploinsufficiency may contribute to the growth and developmental delays of patients 1–3, and perhaps also to variably present minor malformations, immunologic findings and mild dysmorphic features.
LETM1 encodes a ubiquitously expressed leucine zipper-EF-hand-containing transmembrane 1 protein18 with intracellular localization to mitochondria36, 37 and the ER.38 Its cellular function as an ion exchanger37, 38, 39, 40, 41 suggests potential roles in cell signaling and energy production. In flies and worms, LETM1 loss-of-function causes severe growth restriction and decreased viability.40, 42 Conditional knockdown studies of DmLETM1 revealed a requirement for LETM1 in a variety of tissues, including the nervous system.42 These studies support a role for LETM1 in the developmental phenotypes of patients 1–3, yet the haploinsufficiency of LETM1 remains uncertain. Investigations into the function of LETM1 in human fibroblast cells from patients with WHS did not recapitulate mitochondrial phenotypes observed in model systems;39, 40 although a cell-type-specific effect could explain this discrepancy. Furthermore, an intragenic LETM1 deletion identified in a patient with developmental abnormalities different those observed in our patients and patients with WHS43 calls into question the pathogenicity of focal LETM1 alteration. Additional studies will be necessary to elucidate the functional consequence of LETM1 deletion.
The observation that patients 1 and 2 did not have a more severe clinical presentation compared with patient 3 argues against the pathogenicity of deleted genes outside of the SRO. WHSC2 is a ubiquitously expressed gene,17 encoding the protein NELF-A, a subunit of the negative elongation factor complex involved in multiple aspects of mRNA processing and the cell cycle.44, 45 Defects in these processes were observed in a study of lymphoblast cells isolated from patients with non-focal 4p16.3 deletions,46 supporting a role for WHSC2 in the more global aspects of WHS, although WHSC1 and LETM1 appear to have a similar broad pathogenic potential. Additional evidence will be necessary to determine whether or not WHSC2 is haploinsufficient. For other deleted genes in this region, clinical consequence of hemizygosity has not been identified.
Distinctive craniofacial findings are a key component of the clinical diagnosis of WHS. In the literature, there are several reports of patients with atypical 4p16.3 deletions and a facial profile similar to WHS,12, 19, 21, 23, 24 suggesting that multiple genes contribute to the WHS gestalt. In addition to WHSC1, the gene fibroblast growth factor receptor-like 1 (FGFRL1) has also been associated with craniofacial development phenotypes in the mouse.29, 47 Our study provides additional evidence for a multigenic etiology of the WHS gestalt.
Seizures are among the most challenging of clinical management issues for patients with WHS.4, 48 In the past, LETM1 has been a favored candidate for the manifestation of seizures, yet both patients with LETM1 deletions and without seizures1, 12, 13, 43 as well as patients with seizures and with 4p16.3 deletions exclusive of LETM11, 22, 24 have been described. Thus far, none of the patients in this study have exhibited seizure activity and patients 1 and 3 had normal EEG findings. These findings suggest that hemizygous deletion of LETM1 is not sufficient to cause seizures. We propose that additional or distinct loci in 4p16.3 are responsible for this aspect of the syndrome.
The growing body of data suggests that WHS is truly a contiguous gene deletion syndrome and that haploinsufficiency of multiple genes is responsible for the core features of the syndrome. Our study provides further evidence in support of this notion and suggests that deletions involving the SRO may result in a phenotype characterized by growth deficiency, feeding difficulties, motor and speech delays, and perhaps other clinical findings described herein. Determining the individual phenotypic contributions of 4p genes will rely on the detailed characterization of individuals with more focal deletions such as those described here, in addition to studying their function. Altogether, our results suggest haploinsufficiency of one or both of the genes within the SRO is required, but is not sufficient for full expression of the WHS phenotype.
Acknowledgments
We sincerely thank the patients and their families for their participation in this study. We thank Maria Sederberg for technical assistance in the workup of patient 3 and his family and Jeff Swensen and Attila Kumanovics for helpful discussions concerning this study. The project described was supported by the Clinical Genetics Research Program at the University of Utah, Department of Pediatrics and the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant UL1RR025764. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Maas NM, Van Buggenhout G, Hannes F, et al. Genotype-phenotype correlation in 21 patients with Wolf-Hirschhorn syndrome using high resolution array comparative genome hybridisation (CGH) J Med Genet. 2008;45:71–80. doi: 10.1136/jmg.2007.052910. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Carey JC, South ST, Wright TJ.Wolf-Hirschhorn syndromein: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, (eds): GeneReviews Seattle, WA, USA: University of Washington, Seattle; 1993 [Google Scholar]
- Battaglia A, Carey JC, Wright TJ. Wolf-Hirschhorn (4p-) syndrome. Adv Pediatr. 2001;48:75–113. [PubMed] [Google Scholar]
- Battaglia A, Filippi T, Carey JC. Update on the clinical features and natural history of Wolf-Hirschhorn (4p-) syndrome: experience with 87 patients and recommendations for routine health supervision. Am J Med Genet C Sem Med Genet. 2008;148C:246–251. doi: 10.1002/ajmg.c.30187. [DOI] [PubMed] [Google Scholar]
- South ST, Whitby H, Battaglia A, Carey JC, Brothman AR. Comprehensive analysis of Wolf-Hirschhorn syndrome using array CGH indicates a high prevalence of translocations. Eur J Hum Genet. 2008;16:45–52. doi: 10.1038/sj.ejhg.5201915. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, et al. Effect of the size of the deletion and clinical manifestation in Wolf-Hirschhorn syndrome: analysis of 13 patients with a de novo deletion. Eur J Hum Genet. 2000;8:519–526. doi: 10.1038/sj.ejhg.5200498. [DOI] [PubMed] [Google Scholar]
- Zollino M, Di Stefano C, Zampino G, et al. Genotype-phenotype correlations and clinical diagnostic criteria in Wolf-Hirschhorn syndrome. Am J Med Genet. 2000;94:254–261. doi: 10.1002/1096-8628(20000918)94:3<254::aid-ajmg13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Zollino M, Murdolo M, Marangi G, et al. On the nosology and pathogenesis of Wolf-Hirschhorn syndrome: genotype-phenotype correlation analysis of 80 patients and literature review. Am J Med Genet C Sem Med Genet. 2008;148C:257–269. doi: 10.1002/ajmg.c.30190. [DOI] [PubMed] [Google Scholar]
- Zollino M, Lecce R, Fischetto R, et al. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet. 2003;72:590–597. doi: 10.1086/367925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L, Zollino M, Climent S, et al. The new Wolf-Hirschhorn syndrome critical region (WHSCR-2): a description of a second case. Am J Med Genet A. 2005;136:175–178. doi: 10.1002/ajmg.a.30775. [DOI] [PubMed] [Google Scholar]
- South ST, Hannes F, Fisch GS, Vermeesch JR, Zollino M. Pathogenic significance of deletions distal to the currently described Wolf-Hirschhorn syndrome critical regions on 4p16.3. Am J Med Genet C Sem Med Genet. 2008;148C:270–274. doi: 10.1002/ajmg.c.30188. [DOI] [PubMed] [Google Scholar]
- Hammond P, Hannes F, Suttie M, et al. Fine-grained facial phenotype-genotype analysis in Wolf-Hirschhorn syndrome. Eur J Hum Genet. 2012;20:33–40. doi: 10.1038/ejhg.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buggenhout G, Melotte C, Dutta B, et al. Mild Wolf-Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet. 2004;41:691–698. doi: 10.1136/jmg.2003.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnan JB, Tepperberg JH, Papenhausen P, et al. Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J Med Genet. 2006;43:478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TJ, Ricke DO, Denison K, et al. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6:317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen GJ, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Costa JL, Naranjo C, Francis-West P, Altherr MR. Comparative analysis of a novel gene from the Wolf-Hirschhorn/Pitt-Rogers-Danks syndrome critical region. Genomics. 1999;59:203–212. doi: 10.1006/geno.1999.5871. [DOI] [PubMed] [Google Scholar]
- Endele S, Fuhry M, Pak SJ, Zabel BU, Winterpacht A. LETM1, a novel gene encoding a putative EF-hand Ca(2+)-binding protein, flanks the Wolf-Hirschhorn syndrome (WHS) critical region and is deleted in most WHS patients. Genomics. 1999;60:218–225. doi: 10.1006/geno.1999.5881. [DOI] [PubMed] [Google Scholar]
- Rauch A, Schellmoser S, Kraus C, et al. First known microdeletion within the Wolf-Hirschhorn syndrome critical region refines genotype-phenotype correlation. Am J Med Genet. 2001;99:338–342. doi: 10.1002/ajmg.1203. [DOI] [PubMed] [Google Scholar]
- Izumi K, Okuno H, Maeyama K, et al. Interstitial microdeletion of 4p16.3: contribution of WHSC1 haploinsufficiency to the pathogenesis of developmental delay in Wolf-Hirschhorn syndrome. Am J Med Genet A. 2010;152A:1028–1032. doi: 10.1002/ajmg.a.33121. [DOI] [PubMed] [Google Scholar]
- Engbers H, van der Smagt JJ, van ‘t Slot R, Vermeesch JR, Hochstenbach R, Poot M. Wolf-Hirschhorn syndrome facial dysmorphic features in a patient with a terminal 4p16.3 deletion telomeric to the WHSCR and WHSCR 2 regions. Eur J Hum Genet. 2009;17:129–132. doi: 10.1038/ejhg.2008.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faravelli F, Murdolo M, Marangi G, Bricarelli FD, Di Rocco M, Zollino M. Mother to son amplification of a small subtelomeric deletion: a new mechanism of familial recurrence in microdeletion syndromes. Am J Med Genet A. 2007;143A:1169–1173. doi: 10.1002/ajmg.a.31723. [DOI] [PubMed] [Google Scholar]
- Hannes F, Hammond P, Quarrell O, Fryns JP, Devriendt K, Vermeesch JR. A microdeletion proximal of the critical deletion region is associated with mild Wolf-Hirschhorn syndrome. Am J Med Genet A. 2012;158A:996–1004. doi: 10.1002/ajmg.a.35299. [DOI] [PubMed] [Google Scholar]
- Misceo D, Baroy T, Helle JR, Braaten O, Fannemel M, Frengen E. 1.5Mb deletion of chromosome 4p16.3 associated with postnatal growth delay, psychomotor impairment, epilepsy, impulsive behavior and asynchronous skeletal development. Gene. 2012;507:85–91. doi: 10.1016/j.gene.2012.07.021. [DOI] [PubMed] [Google Scholar]
- de Souza MA, McAllister C, Suttie M, et al. Growth hormone, gender and face shape in prader-willi syndrome Am J Med Genet A 2013;. epub ahead of print 5 August 2013; doi: 10.1002/ajmg.a.36100 [DOI] [PubMed]
- Kim JY, Kee HJ, Choe NW, et al. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol Cell Biol. 2008;28:2023–2034. doi: 10.1128/MCB.02130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marango J, Shimoyama M, Nishio H, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- Garlisi CG, Uss AS, Xiao H, et al. A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription. Am J Respiratory Cell Mol Biol. 2001;24:90–98. doi: 10.1165/ajrcmb.24.1.4224. [DOI] [PubMed] [Google Scholar]
- Hajdu I, Ciccia A, Lewis SM, Elledge SJ. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci USA. 2011;108:13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Wu X, Liu T, Yu K, Jelinek DF, Lou Z. The histone methyltransferase MMSET regulates class switch recombination. J Immunol. 2013;190:756–763. doi: 10.4049/jimmunol.1201811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. [PubMed] [Google Scholar]
- Hudlebusch HR, Santoni-Rugiu E, Simon R, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17:2919–2933. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- Schlickum S, Moghekar A, Simpson JC, et al. LETM1, a gene deleted in Wolf-Hirschhorn syndrome, encodes an evolutionarily conserved mitochondrial protein. Genomics. 2004;83:254–261. doi: 10.1016/j.ygeno.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Nowikovsky K, Froschauer EM, Zsurka G, et al. The LETM1/YOL027 gene family encodes a factor of the mitochondrial K+ homeostasis with a potential role in the Wolf-Hirschhorn syndrome. J Biol Chem. 2004;279:30307–30315. doi: 10.1074/jbc.M403607200. [DOI] [PubMed] [Google Scholar]
- Kuum M, Veksler V, Liiv J, Ventura-Clapier R, Kaasik A. Endoplasmic reticulum potassium-hydrogen exchanger and small conductance calcium-activated potassium channel activities are essential for ER calcium uptake in neurons and cardiomyocytes. J Cell Sci. 2012;125:625–633. doi: 10.1242/jcs.090126. [DOI] [PubMed] [Google Scholar]
- Dimmer KS, Navoni F, Casarin A, et al. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum Mol Genet. 2008;17:201–214. doi: 10.1093/hmg/ddm297. [DOI] [PubMed] [Google Scholar]
- Hasegawa A, van der Bliek AM. Inverse correlation between expression of the Wolfs Hirschhorn candidate gene Letm1 and mitochondrial volume in C. elegans and in mammalian cells. Hum Mol Genet. 2007;16:2061–2071. doi: 10.1093/hmg/ddm154. [DOI] [PubMed] [Google Scholar]
- Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326:144–147. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban AG, Joza N, Megighian A, et al. A Drosophila mutant of LETM1, a candidate gene for seizures in Wolf-Hirschhorn syndrome. Hum Mol Genet. 2010;19:987–1000. doi: 10.1093/hmg/ddp563. [DOI] [PubMed] [Google Scholar]
- Luo Y, Hermetz KE, Jackson JM, et al. Diverse mutational mechanisms cause pathogenic subtelomeric rearrangements. Hum Mol Genet. 2011;20:3769–3778. doi: 10.1093/hmg/ddr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, et al. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J Biol Chem. 2010;285:6443–6452. doi: 10.1074/jbc.M109.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzendorfer C, Hannes F, Colnaghi R, et al. Characterizing the functional consequences of haploinsufficiency of NELF-A (WHSC2) and SLBP identifies novel cellular phenotypes in Wolf-Hirschhorn syndrome. Hum Mol Genet. 2012;21:2181–2193. doi: 10.1093/hmg/dds033. [DOI] [PubMed] [Google Scholar]
- Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te Welscher P. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Model Mech. 2009;2:283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia A, Carey JC. Seizure and EEG patterns in Wolf-Hirschhorn (4p-) syndrome. Brain Dev. 2005;27:362–364. doi: 10.1016/j.braindev.2004.02.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.