Abstract
In this review, we provide a detailed overview of studies on the elusive sex determination (SD) and gonad differentiation mechanisms of zebrafish (Danio rerio). We show that the data obtained from most studies are compatible with polygenic sex determination (PSD), where the decision is made by the allelic combinations of several loci. These loci are typically dispersed throughout the genome, but in some teleost species a few of them might be located on a preferential pair of (sex) chromosomes. The PSD system has a much higher level of variation of SD genotypes both at the level of gametes and the sexual genotype of individuals, than that of the chromosomal sex determination systems. The early sexual development of zebrafish males is a complicated process, as they first develop a ‘juvenile ovary’, that later undergoes a transformation to give way to a testis. To date, three major developmental pathways were shown to be involved with gonad differentiation through the modulation of programmed cell death. In our opinion, there are more pathways participating in the regulation of zebrafish gonad differentiation/transformation. Introduction of additional powerful large-scale genomic approaches into the analysis of zebrafish reproduction will result in further deepening of our knowledge as well as identification of additional pathways and genes associated with these processes in the near future.
Keywords: polygenic sex determination, sex chromosome, gonad differentiation, teleost, fish, Danio rerio
ZEBRAFISH SEX: LOTS OF QUESTIONS AND ONLY A FEW ANSWERS
Teleosts (ray-finned fishes) form the largest group of extant vertebrates with more species than the rest of the other vertebrates combined [1]. The rich diversity of teleosts is observed not only in their phenotypes and behavior (review: [2]), but also in the varieties of their reproductive processes that seem to utilize all known sex determination (SD) mechanisms described for other vertebrates (see reviews [3, 4]). It is believed that the common ancestor of teleosts diverged from that of land vertebrates about 410 million years ago (Mya), well before the split of placental mammals from the latter lineage (ca. 180 Mya) [5]. Therefore, it is not surprising that the evolution of sex determination systems between teleosts and the well-studied mammals show substantial differences. The diversity of reproductive systems in teleosts allows them to contribute to comparative studies on the evolution of SD mechanisms.
One of the most popular model organisms among teleosts is the zebrafish (Danio rerio). Natural habitats of the zebrafish stretch from South Asia (e.g. Pakistan, Nepal and India) to Southeast Asia (e.g. Myanmar) [6]. This small-bodied freshwater species is most commonly found in slow or stagnant waters, such as rivers, ponds and paddy fields [6, 7]. The ambient water temperature at these natural habitats typically ranges from 26–32°C [7].
Among the reasons that make zebrafish a popular laboratory model is its short generation time. In the laboratory, zebrafish reach reproductive maturity at around 3–4 months. Despite its relatively small size, the species is quite fecund: mature females kept under ideal conditions often produce 200–300 eggs regularly on a weekly basis. However, offspring sex ratios can vary over a wide range, including extreme male or female bias. Experiments that require a certain sex ratio necessitate the identification of specific breeding pairs that have been tested earlier and were shown to produce the desired proportion of males/females in their offspring. The identification, maintenance and regular breeding of these pairs could be a time-consuming task.
Although many reviews have been published on SD and gonad differentiation of teleosts (see, e.g. [3, 4, 8–19]), those focusing on zebrafish sex have been few and far between until very recently [11, 20–22]. Over the past few decades, there have been conflicting reports on the mode of SD in zebrafish; some of them predicted genetic mechanisms [23–26], whereas others pointed toward environmental clues [27, 28]. According to our knowledge, ours is the first review that provides a detailed overview on studies spanning nearly four decades in order to improve our knowledge on the elusive SD and gonad differentiation mechanism of zebrafish.
ZEBRAFISH SEX IS DETERMINED PRIMARILY BY GENETIC FACTORS AND NOT ENVIRONMENTAL ONES
In vertebrates, sex is determined either by genetic mechanisms (genetic sex determination or GSD; reviews: [3, 18, 29]) or by the environment (environmental sex determination or ESD; reviews: [3, 4, 8, 12, 15, 30]). Although SD has only been analyzed in a small subset of the 32 000 fish species (reviews: [3, 15]), there are plenty of examples for species with GSD (e.g. Japanese medaka [31], threespine stickleback [32] and Patagonian pejerrey [33]) and ESD (e.g. American eel [34] and bluehead wrasse [35]). In some teleosts, SD (or gonad differentiation) can be overridden by environmental effects, most often temperature (thermal effect (TE) on GSD; review: [36]).
In GSD, the sex of an individual is determined primarily by genes/chromosomes inherited from the parents. There are two major forms of GSD: (i) SD by a single sex chromosomal pair or chromosomal sex determination (CSD) and (ii) SD by several (i.e. more than one) genetic factors or polygenic (multigenic) sex determination (PSD). In our opinion, the latter includes those with multiple sex chromosome types (e.g. several Lake Malawi cichlid species [37]), weak sex chromosomes easily and often overridden by autosomal modifiers (e.g. X/Y+A or ZW+A according to Devlin and Nagahama [3]) and those regulated by several autosomal loci without any sign of sex chromosomes (e.g. European seabass [38]).
To find out whether zebrafish uses GSD or not, we conducted several experiments [26]. Repeated mating of the same breeding pairs yielded offspring groups with very similar sex ratios, even when they were reared in uncontrolled environmental conditions (i.e. variations were expected in rearing density, amount of feed and ambient water temperature [26]). We also performed a selective breeding experiment, whereby factorial crosses were made with four to six siblings in every generation and brooders for the new generation were chosen from the family that produced the highest bias toward the required direction. With this approach, we generated families with severely biased sex ratios within a short period of time (three to four generations), especially toward male excess [26].
Strong influence of parental genotype on the sex of zebrafish offspring was also described by Abozaid et al. [39, 40]. They observed that offspring from gynogenetic males showed stronger effect of heat-induced masculinization than progenies sired by wild-type males. In addition, they also found that progeny sex ratios from the same breeding pairs were highly reproducible [39].
As indicated by its name, ESD is dependent on a signal from the surrounding environment to determine the gonadal fate (i.e. testis or ovary) of an organism (review: [41]). Species utilizing ESD do not have sexual dimorphism at the genomic level. The most common type of ESD mechanism studied is temperature-dependent SD, in which sex is a function of the ambient temperature during early development. This mode of SD is quite common among reptiles [42, 43], but it has only been shown to exist in a few teleost species (review: [36]). Although several environmental factors, including dissolved oxygen [44] and growth rate [27] have been found to influence sex ratio of zebrafish, the effect of temperature on sex was studied most extensively [39, 40, 45, 46]. Studies on various teleost species, including zebrafish, showed that elevated water temperature affected different gonadal processes, including apoptosis, estrogen biosynthesis and germ cell meiosis [45, 47, 48]. When zebrafish juveniles were subjected to high water temperatures during the period of gonad transformation and differentiation, oocyte apoptosis and decreased activity of gonadal aromatase were observed, resulting in increased male bias at adulthood [45]. In European seabass (Dicentrarchus labrax), high temperature caused increased DNA methylation of the aromatase promoter, resulting in decreased expression and male-biased sex ratio [47]. Apparently, masculinizing effect of high temperature is not restricted to the gonad differentiation period in zebrafish, as families exposed to increased temperature during larval stage (5–42 h postfertilization) also produced significantly more males [39]. Interestingly, in the latter experiment some families tended to be more sensitive to heat-induced masculinization than others [40]. This showed that interaction between genotype and the environment exists in zebrafish.
All in all, the above studies showed that zebrafish sexual development could be affected by various environmental factors. However, none of the environmental factors exerted a strong effect on sex within their natural range, and some of them seemed to be largely dependent on the fish genotype. These data argue against zebrafish being a true ESD species, further supporting the earlier conclusion by Piferrer’s group that this is a TE on GSD species [36].
ZEBRAFISH SEX IS DETERMINED BY MULTIPLE LOCI (WITH OR WITHOUT SEX CHROMOSOMES)
In the best known form of SD that is found in mammals and avians as well as many other vertebrate species, including some teleosts, there is a so-called ‘master switch’ that typically resides on a special chromosomal pair, the well-differentiated sex chromosomes [9, 49, 50]. If the ‘master switch’ is on the ‘heterogametic’ member of the sex chromosomal pair that is present in only one of the two sexes (male-specific Y or female-specific W), then it will initiate the biological programs in representatives of that sex only. On the other hand, if the ‘master switch’ is located on the ‘homogametic sex chromosome’ that is present in both sexes, then the outcome will be determined by a dosage-based mechanism (e.g. Drosophila melanogaster [51]). In addition to the classical, well-differentiated sex chromosomes, there are vertebrate species with undifferentiated sex chromosomal pairs, where both the so-called proto-X and proto-Y might contain the master switch, but in a different allelic form (e.g. Takifugu rubripes [52]). In this review, we will summarize the results of over two dozen studies—performed with several different methods— that have been conducted for the identification of sex chromosomes in zebrafish.
The traditional method for identification of sex chromosomes is detection of a heteromorphic chromosomal pair in one of the two sexes through karyotyping. Highly differentiated sex chromosomes (e.g. most mammalian ones) are easily distinguished cytogenetically due to their distinct size difference (heteromorphism) caused by suppression of recombination and degeneration of the heterogametic sex chromosome (reviews: [53–55]). Search for a heteromorphic chromosomal pair in zebrafish started nearly half a century ago when its karyotype was described [56]. Since then there were a number of publications on zebrafish karyotypes, the majority of them did not observe any size dimorphic chromosomal pair [56–65], but see potential indications for a ZW/ZZ system [66, 67]. It must be noted here that pairing of the zebrafish chromosomes on karyotypes is difficult due to their monomorphic nature, whereas the usage of different stains resulted in different chromosome banding patterns [63], making cross-validation of the results from different experiments challenging. Arkhipchuk [68] found differentiated sex chromosomes in about 10% of the ca. 2000 teleost karyotypes he reviewed. As species with undifferentiated sex chromosomes containing a short nonrecombining region (i.e. proto-X/proto-Y) is not cytogenetically recognizable, the exact proportion of teleost species with sex chromosomes might be larger.
Comparative analysis of meiotic recombination rates in males versus females can also be used to detect sex chromosomal systems. Species with heterogametic sex chromosomal pairs usually display different recombination rate that is sex-specific due to suppression of recombination between the pair of sex chromosomes [69]. Using genetic mapping approach, the Postlethwait’s lab compared the recombination rate between a double-haploid zebrafish male [70] and female [71, 72]. They reported that the male androgenetic map had lower recombination rate relative to the female gynogenetic map [70]. Later, they also confirmed these results in normal (wild-type) zebrafish [25]. However, it was also suggested that the lower recombination rate observed in male zebrafish is most likely due to lesser numbers of human mutL homolog 1 (Mlh1) foci in male genome and not because of recombination suppression [73]. The latter observation was supported by data obtained through synaptonemal complex karyotyping [74, 75], which did not find any difference between the two sexes. These results led to the conclusion that zebrafish is unlikely to have a pair of highly differentiated sex chromosomes.
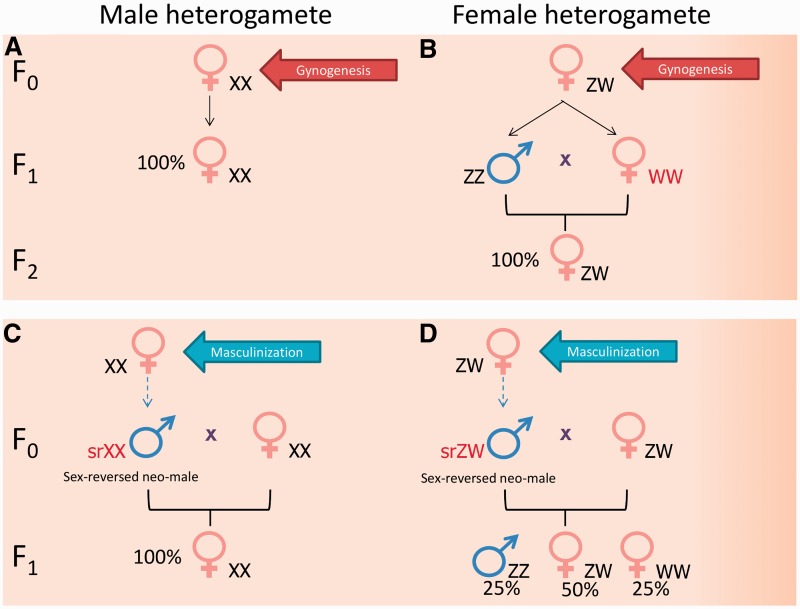
Whole genome manipulation (or induced parthenogenesis) techniques are also used for identification of species with CSD (reviews: [76–80]). The approach is based on the exclusion of the paternal or maternal genome from the inheritance by irradiation of sperm or egg, followed by inhibition of first cell division. This will result in duplication of the remaining haploid genome, hence the name double haploids. Double haploids generated by the exclusion of paternal genome are called gynogenotes, whereas those generated by the elimination of maternal genome are named androgenotes. In the presence of a pair of differentiated sex chromosomes, double haploids have equal chances to inherit either of them. For species that are male heterogametic (XY), the expected sex ratio of F1 double haploids from gynogenesis would be 100% females (Figure 1A). On the other hand, for female heterogametic species (ZW), F1 gynogenotes are expected to have a 1:1 male to female phenotypic sex ratio (Figure 1B), assuming full survival of F1 with WW sex chromosomal set that does not occur in nature. Crossing of WW female with a normal male (ZZ) should produce an all-female progeny (Figure 1B). The reverse is true for androgenesis, whereby F1 double haploids from male heterogametic species should have an equal proportion of males and females (again, assuming full survival of YY males), while female heterogametic species should produce 100% male androgenotes. For this review, we collated data from four sets of zebrafish gynogenesis experiments [81–84] and one androgenesis experiment [28]. The number of double haploids obtained per experiment ranged from 5 to 146 and sex ratio showed high variation from 0% to 95% females (summary of data available in Table 1). Some of these data seem to be incompatible with the XX/XY sex chromosomal system, others with the ZZ/ZW system, whereas the rest do not seem to fit either of the two options. Taken together, they seem to argue against the presence of a strong sex chromosomal system with primary effect on SD in zebrafish.
Figure 1:
Inferring the type of sex chromosomal system by progeny sex ratio as a result of genome manipulation or artificial sex reversal. (A) Performing gynogenesis on male heterogametic species will produce an all-female (XX) F1 offspring. (B) For the female heterogametic species (assuming full viability of all genotypes), gynogenesis will produce F1 progeny with about 1:1 sex ratio and all F1 females will have WW sex chromosomes. [In case of WW lethality, an all-male (ZZ) F1 offspring will be produced, whereas partial survival of WWs will yield intermediate results]. When F1 females (WW) are crossed with normal males (ZZ), their F2 offspring will be all female (ZW). (C) When hormone- or temperature-based masculinization is performed in a male heterogametic species, crossing of sex-reversed neo-males (srXX) with normal females will produce all-female F1 offspring. (D) For female heterogametic species (assuming full viability of all genotypes), the expected phenotypic sex ratio in the F1 progeny, from crossing the sex-reversed neo-male (srZW) with normal female (ZW), will be 25% males and 75% females. Among the F1 females, one-third of them will have WW sex chromosomal pair that does not normally occur in nature and yield an all-female offspring when crossed with a normal male (ZZ). (If WWs showed lethality, then 33% ZZ males and 67% ZW females will be expected, whereas their partial survival will yield intermediate results).
Table 1:
Data from induced parthenogenesis experiments on zebrafish yield sex ratios incompatible with one or both sex chromosomal systems
| Double haploid | Strain | Number of double haploid | Observed sex ratio (percent female) | Type of sex chromosome excluded | Remark | Reference |
|---|---|---|---|---|---|---|
| Gynogenotes | Wild | 8 | 0 | XX/XY | If ZW/ZZ, then WWs are inviable | [81] |
| Gynogenotes | Golden | 8 | 0 | XX/XY | If ZW/ZZ, then WWs are inviable | [81] |
| Gynogenotes | gol-mix | 15 | <20 | both | Per family sex ratios cover the range from <20% to >80% | [82] |
| Gynogenotes | gol-mix | 146 | <20 | both | Per family sex ratios cover the range from <20% to >80% | [82] |
| Gynogenotes | gol-FL1 | 28 | 92 | both | Per family sex ratios cover the range from <20% to >80% | [82] |
| Gynogenotes | C29 | 85 | 95 | ZW/ZZ | If XX/XY, then YY viability is high | [83] |
| Gynogenotes | Unknown | Unknown | 6 | XX/XY | If ZW/ZZ, the viability of WWs is very low | [84] |
| Androgenotes | AB | 5 | 0 | both | Sex ratio of progeny is highly skewed in some families | [28] |
The third experimental technique used for detecting the presence of sex chromosomes is based on analyzing sex ratios of offspring produced by crossing sex-reversed individuals with wild-type partners (review: [3, 85]). In an XX/XY species, when sex-reversed (srXX) neo males produced by masculinization of genetic females are crossed with wild-type females, a monosex female offspring is expected (Figure 1C). However, in ZW/ZZ species, the proportion of females in the progeny of srZW × ZW cross is expected to be between 75% and 50% (Figure 1D), depending on the relative survival rate of the WW individuals compared with ZW. When neo females generated by feminization of genetic males are crossed with wild-type males, the expected sex ratios are: 25–33% females for XX/XY (depending on the survival of the YYs) and 100% males for ZZ/ZW.
Over the years, numerous experiments were performed to achieve hormonal sex reversal of zebrafish (a set of examples available in Table 1 of [21]). However, according to our knowledge, most of them did not report on the offspring sex ratio produced by sex-reversed neo males or females. We only managed to find a single study that successfully masculinized zebrafish by hormone treatment and identified ‘super females’ among the resulting F1 fish [23]. When these ‘super females’ were crossed with normal males, severely female-biased (90–98%) offspring was produced. This led them to suggest that zebrafish has a ‘female dominance chromosomal-based SD mechanism’. In our view, this proposed mechanism is identical to the female heterogametic system (ZW/ZZ) that has been found to be incompatible with zebrafish SD based on data obtained earlier by several genome manipulation experiments (Table 1). Unfortunately, there is no indication in the publication whether the authors have attempted to identify the proposed sex chromosomes (or any of their markers).
As described earlier, masculinization of zebrafish can also be achieved by high temperature applied during the gonadal transformation window ([39, 40, 45]; review: [36]). In an experiment conducted by Abozaid et al. [40], F1 neo males produced by heat treatment were backcrossed to their mother (normal female) for offspring sex ratio analysis. The F1 generation had an average of 16% males, which had a notable difference from the expected sex ratio for both a male heterogametic (expected: no males) and female heterogametic (expected: 50% males) systems. Surprisingly, the F2 progenies had sex ratios very similar to the expected value of a female heterogametic sex system. However, as the F1 progeny sex ratio could not be explained by either of the two sex chromosomal systems, the authors concluded that at least two or more autosomal factors might be involved in zebrafish SD further strengthening indications against the compatibility of zebrafish SD with a simple sex chromosomal system.
The most direct way to search for sex chromosomes is to look for genomic differences between the male and female genomes. Polymerase chain reaction (PCR)-based methods such as random amplified polymorphic DNA (RAPD) [86] and amplified fragment length polymorphism (AFLP) [87] are the most popular molecular methods for isolation of sex markers, but other procedures have also been used [88]. Information obtained from sex markers can be used to identify the type of GSD mechanism the organism is utilizing (review: [89]). We screened through the zebrafish genome using our improved PCR-based assay method, called fluorescent Motif Enhanced Polymorphism (fluoMEP), which combined the simplicity of RAPD and the higher throughput of AFLP [90]. Using fluoMEP, we managed to isolate male-specific sex markers from two fish species known to have sex chromosomes, i.e. guppy (Poecilia reticulata) and rosy barb (Puntius conchonius) [26, 91–94]. However, we could not find any universal sex marker after detailed comparative analysis of the zebrafish males and females genomes in two strains (AB and wild-type) using the same method [26]. This is likely due to the fact that there are no universal major structural differences between the chromosomal sets of the zebrafish sexes in these two strains.
The availability of the whole zebrafish genomic sequence [95, 96] has enabled the use of higher resolution genomic tools to search for sex chromosomes. One such technique is array comparative genomic hybridization (aCGH), which screens the whole genome at regular intervals for copy number variable regions (CNVRs) between two different samples [97, 98] and was recently used to study genetic diversity of different zebrafish strains [99]. In a well-differentiated sex chromosomal system, the SD region should be unique to the heterogametic sex chromosome. As a consequence, it should show different copy numbers between the two sexes; hence, it is possible to search for such sex chromosomes using aCGH. To screen through the zebrafish genome at higher resolution than fluoMEP, we custom-designed an aCGH oligonucleotide microarray that can detect CNVRs longer than 50 kb. Two strains of zebrafish (AB and Singapore-based wild-type) were screened using the custom-designed aCGH array. No universal sex-linked CNVR was detected from the two pairs of zebrafish families tested [26]. These data and the fluoMEP results further strengthen the notion that there are no substantial differences between the zebrafish sexes that would be uniform for every strain.
Genome-wide linkage analysis is another molecular technique that takes advantage of whole genome sequences. DNA markers are used to search for loci that show significant association with the trait of interest. For zebrafish, three labs had performed single-nucleotide polymorphism (SNP)-based genetic linkage mapping on F2 generation for which the grandparents were derived from two different zebrafish strains, followed by full-sib crosses of the F1 hybrids [24, 25, 95]. Altogether, five different regions on four chromosomes (i.e. chromosome 3, 4, 5 and 16) were identified (Table 2). Although all crosses involved an AB strain grandparent, none of the putative sex-associated regions were consistent between any two labs. The detection of five different sex-associated regions from three independent genome-wide studies indicates that zebrafish SD is a complex trait, where multiple autosomal factors and not just one pair of sex chromosome, if any, will instruct individuals to assume one of the two sexes. These data also indicate that there might be differences among the various zebrafish strains with regard to the gene sets that play a role in the SD mechanism.
Table 2:
At least five sex-associated regions were identified in zebrafish strains by genome-wide studies
| Cross-typea | Method | Marker densityb (kb) | Chr | Chr location (Zv9) | Genetic interval | References |
|---|---|---|---|---|---|---|
| IN × AB | SNP typing | 3.80 | 5 | 44 453 011–46 626 084 | 1.7 cM | [24] |
| IN × AB | SNP typing | 3.80 | 16 | 12 952 287–16 952 809 | 3.5 cM | [24] |
| AB# × NA | SNP typing | 357.38 | 4 | 60 837 953–62 094 675 | >1 cM | [25] |
| NA × AB# | SNP typing | 295.82 | 4 | 60 837 953–62 094 676 | >1 cM | [25] |
| NA × AB# | SNP typing | 295.82 | 3 | 15 234 176–29 193 306 | 5.8 cM | [25] |
| AB × Tu | SNP typing | 13.62 | 16 | ∼19–22 Mb | N/P | [95] |
| AB × AB; Toh × Toh | aCGH | 15.74 | N/A | N/A | N/A | [26] |
Note: aFor the first three studies [24, 25, 95], brooders from two different varieties were crossed (males shown first), then F1 full-sib cross was performed to produce the F2 mapping family. For the fourth study [26], two pairs of brooders each from two strains were crossed. The parents and four of their offspring from each cross were analyzed.
bBased on 1.714 Gb genome size (Gregory TR. Animal Genome Size Database. http://www.genomesize.com). IN, wild-type from Northeast India; AB, AB stock; AB#, AB stocks derived from Streisinger's original AB line; NA, Nadia stocks originated from Indian wild-type from the Kolkata region; Tu, Tubingen line; Toh, a farm-derived strain from Singapore; N/P, not provided; N/A, not applicable.
The pathways involved in gonad differentiation are highly conserved among the vertebrates, with many of the genes having the same functions [15, 22, 100–104]. Candidate gene approach as well as large-scale comparative transcriptomic analysis (such as microarray), were used to identify potential master sex determining (MSD) genes. Such a gene is typically only carried by one of the sexes and the chromosome with MSD gene is the sex chromosome. Thus, mapping of genes with sexually dimorphic expression level onto the genome provides a means to detect sex chromosomes. Using this approach the MSD gene for Patagonian pejerrey (Odontesthes hatcheri) and rainbow trout (Oncorhynchus mykiss), amhy [33] and sdY [105], respectively, were identified and mapped to the Y chromosome. Zebrafish also shared many of the common candidate genes involved in gonad differentiation (refer to Table 2 of [21]). There were also several large-scale comparative transcriptomic studies performed between the zebrafish sexes and they identified many genes showing sex dimorphic expression level [106–110]. However, no zebrafish chromosome has significant clustering of these genes and to date no primary SD switch with preferential role was identified. The identification of a MSD gene works most efficiently when the genetic sex of the fish species can be determined at larval stage. This is because the MSD gene might be expressed only during early developmental stages, like the expression of amhy in male Patagonian pejerrey is observed only up to 20 weeks after hatching [33]. Therefore, for an organism like the zebrafish that does not have distinct sexual dimorphism during the larvae stage, ‘blind analysis’ of a large number of samples collected at multiple time points from families with known sex ratios must be performed, making these experiments tedious and expensive.
ZEBRAFISH SEX DETERMINATION IS POLYGENIC
The data reviewed so far on sex chromosome searches in zebrafish shows the following: (i) there is no cytogenetic evidence of heteromorphic chromosomal pair; (ii) the observed progeny sex ratios from various genome manipulation studies did not concur with those expected from a simple sex chromosome model; (iii) lack of substantial, universal differences between genomes of the two sexes in multiple strains and (iv) genome-wide linkage analysis identified multiple regions on different chromosomes associated with SD. All these results convincingly indicate: (i) high level of variability of the SD process among different zebrafish strains and (ii) the likely absence of strong, highly differentiated sex chromosomes in all zebrafish varieties. At the same time, the reviewed data also point to strong influence of parental genotypes on progeny sex ratio. In such cases the most probable SD mechanism is PSD (reviews: [111–113]). (In our opinion, a weak sex chromosomal pair that is not present in every strain and unable to control sexual development of vast majority of individuals due to the action of multiple autosomal modifiers would not exclude zebrafish from this category). Due to the complexity of the mechanism, there are only a few publications on this system and in addition to zebrafish, currently only few fish species, including several Lake Malawi cichlid species [37], several tilapia species [114, 115], European seabass [38] and green swordtail [113], were reported to utilize this mode of SD.
According to Bull [111], the criteria for PSD systems are as follows: (i) wide range of progeny sex ratio among different families; (ii) strong influence from parental genotype and (iii) sex ratio response to selection. Our and other’s data on zebrafish fulfilled all these three criteria. First, we observed huge differences in the progeny sex ratio (4.8–97.3% males) among 62 zebrafish families of various strains [26]. Second, at least two independent studies found strong parental genotype influence on the progeny sex ratio by performing repeated crossings of the same breeding pair [26] and crossing of gynogenetic males with different normal females [39, 40]. Finally, we were able to prove sex ratio response to selection by maintaining sex-biased zebrafish lines and decreased progeny sex ratio variation among families from the same line through selective breeding [26].
HOW DOES PSD WORK AND WHICH GENES ARE INVOLVED?
In a polygenic system, the sex of an individual is not determined by the presence or absence of a special chromosome. Instead, decision is made by the allelic combinations of loci, whose products are involved in the SD process (Figure 2). These loci are typically dispersed throughout the genome, but in some species a few of them might be located on a preferential pair of chromosomes. In other words, the presence of sex chromosomes, per se, should not exclude PSD.
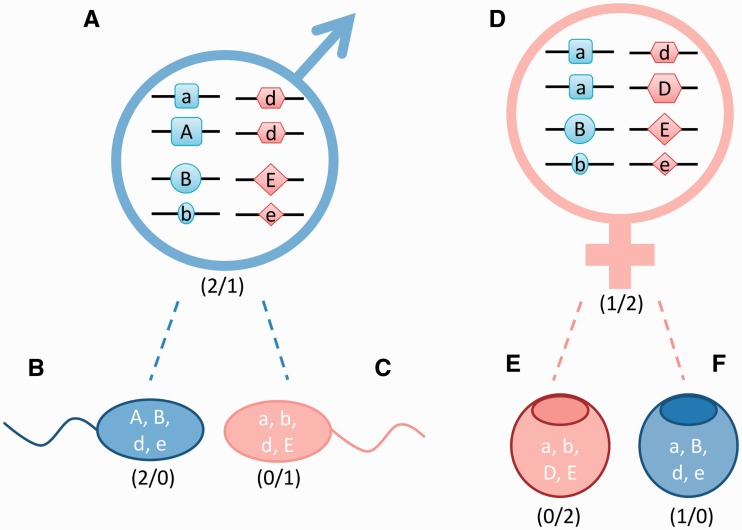
Figure 2:
A simplified mechanistic model for a PSD system based on the involvement of four autosomal genes. In this theoretical PSD system, protein products of four genomic loci determine sex. Two of the protein products perform a function that pushes the gonad toward femaleness D and E loci, while the remaining two are proteins with pro-male function A and B loci. For simplicity, it is assumed that (i) for every locus there is a strong (larger shape and upper case letter) and weak (smaller shape and lower case letter) effect allele; (ii) the effect of the four strong alleles are equal and the same is true for the four weak alleles at a lower level and (iii) the four products do not exert any direct effect on the functions of each other. If we assign a binary code to the alleles (strong—1 and weak—0), then the outcome could be predicted by simply comparing the sum of male and female alleles (assuming that in case of a tie the individual would continue to develop into a female). In this system, a weak male that has two strong male alleles and one strong female allele (A) could produce sperm cells of different sexual genotypes [see (B) and (C) for examples], among them some that would have excess of pro-female alleles [see (C)]. Similarly, a weak female with one strong male and two strong female alleles (D) could also produce oocytes with excess strong female alleles (E) or more strong male alleles (F). Symbols with different shapes label genes located on four different autosomes. Ratios of strong male to strong female alleles are indicated in brackets.
The complexity of PSD system is the result of different allelic combinations with variable effect on gonad differentiation. Protein products of several genes located on different chromosomes are involved in SD with some having pro-female function while others promoting maleness. Different alleles of the same gene can have strong or weak effect. If we assigned a binary code to the allele effect (strong = 1 and weak = 0), then the sex of this PSD species could be predicted by simply comparing the sum of male to female alleles. Gonad differentiation would be directed to the sex with the highest proportion of strong alleles (more detailed explanation is available in Figure 2).
Needless to say, the maximum number of variations (i.e. allelic combinations) in the germ cells would drastically increase in parallel with increasing number of loci involved; for eight loci it would be 128 (27), whereas for a dozen it would be 2056 (211). These calculations exclude those females that are heterozygous for all loci, otherwise the numbers would be twice higher. This is in sharp contrast with CSD systems, where the total number of genetic varieties for the SD loci among the gametes is two.
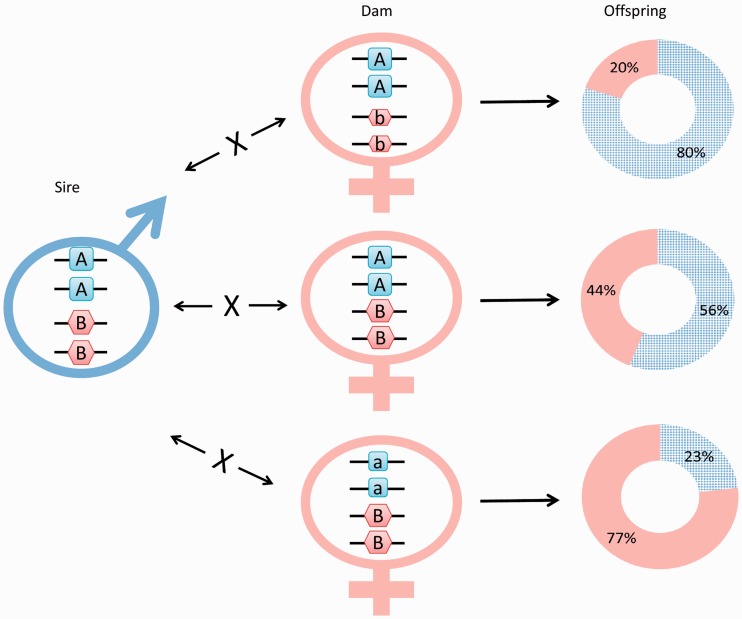
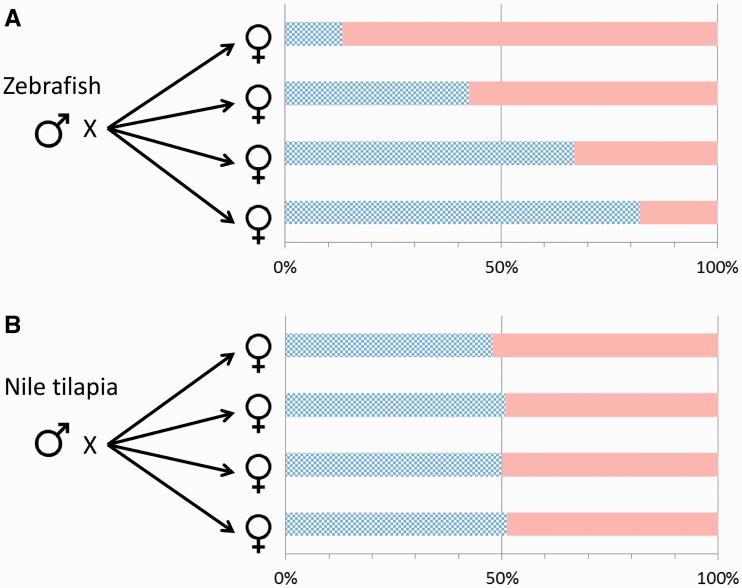
One characteristic of the PSD system is that representatives of the two sexes show a much higher level of variation than that in CSD systems. This variation is then passed over to the next generation in the form of highly variable offspring sex ratios among different combinations of parental pairs. When the same zebrafish male is crossed with three different females, sex ratios of the resulting families spread over a surprisingly wide range (Figure 3), providing additional indication that only part of the complexity due to allelic diversity is observed here. Such a wide variation in the sex ratio of PSD species (Figure 4A) differs markedly from the narrow range observed in the four families produced by a CSD species (Figure 4B).
Figure 3:
The combination of parental ‘SD allele sets’ have a profound effect on the sex ratio of the offspring. Using bigenic genotyping data from the offspring groups analyzed by Bradley et al. (Supplementary Table 4 of [24]), we have created imaginary crosses using one male genotype with three different female genotypes. The parental genotypes were chosen in such a way that they could only produce a single genotype per cross that was among those detected by the authors. Wide range of sex ratios obtained and two opposite sexes with the same SD genotype both indicate a higher level of complexity than what could be explained by participation of just two loci in the SD process. On the pie charts, checkered background indicates males, and solid background indicates females.
Figure 4:
Offspring sex ratios show much higher level of variability for zebrafish (PSD) than Nile tilapia (CSD). (A) Crossing a single zebrafish male with four different female partners yielded offspring groups with very different sex ratios (13.1–81.8% males; data are from [26]). (B) Crossing a Nile tilapia male (XY) with four different female partners (XX) yielded offspring groups with very similar sex ratios (data are from [116]). Checkered bars indicate males, and solid bars indicate females.
Unfortunately, it is still not known: (i) how many genes regulate PSD in zebrafish; (ii) what are these genes and (iii) how much, if any, is the overlap among these gene sets and those of other PSD species in vertebrates. Data obtained from the two genome-wide linkage studies mentioned above [24, 25] provided first set of indications toward the genomic regions that are likely to contain subset of these genes in zebrafish. They found genes with roles in gonad differentiation (dmrt1 and fancg) and sex hormone biosynthesis (cyp21a2 and hsd17b1) to be closely located to the proposed SD regions [24, 25]. Another feature of PSD is the cumulative effect of multiple genes involved in SD. Different allelic combinations exerting variable effects further complicate the process. Experimental evidence for the cumulative effect of the allelic combinations was observed on sex-linked loci reported by Bradley and colleagues (see Supplementary Table 4 of [24]). Given the fact, that the two SD regions identified by them accounted for only 16% of the sex variance trait [24], we propose that there could be a large number of genes involved in zebrafish SD.
MOLECULAR REGULATION OF GONAD DIFFERENTIATION IN ZEBRAFISH IS A COMPLEX PROCESS
The complexity of zebrafish gonad differentiation process was first reported by Takahashi [117], who pointed out that every individual starts to form an ovary (later it was named ‘juvenile ovary’) and future males later have to undergo a transformation before they can start to produce a testis. The ‘juvenile ovary’ is made up of mostly oogonia and primary oocytes [118]. First histological sign of ‘juvenile ovary’ phase can be observed from zebrafish as young as 2.5 weeks post-fertilization (wpf) to around 4 wpf [117, 118]. Likewise, the extent of ‘juvenile ovary’ phase was found to vary among individuals from a few days to more than a week [119]. Following the ‘juvenile ovary’ phase, the gonad either continues to develop into a functional ovary or undergoes a drastic transformation to form a testis (review: [21]).
The hallmark of this ‘juvenile ovary to testis’ transformation process is a rapid degeneration and eventual elimination of oocytes by an apoptotic (or programmed cell death) wave that was first described by Uchida and colleagues [84]. Although the primary signal that triggers the wave is still unknown, it is likely that it acts through a change of hormonal environment of the oocytes that result in increased ratio of male to female hormones. The number of primordial germ cells also seems to play a role in the transformation process: their complete or partial depletion leads to testis development [120, 121]. In addition, germ cells are also important for maintenance of ovarian function in adults [122]. The development of transgenic zebrafish lines that showed differential reporter signals between the developing female (strong fluorescence) and male (weak fluorescence) gonads [123–125] has opened up new possibilities to analyze the underlying processes. The overall nature of gonad differentiation and transformation processes were described [21], the developmental time limits were determined [117, 118] and expression profiles of a few genes (e.g. cyp19a1a, amh and cyp11c1) have been analyzed [22, 126, 127].
The improvement of platforms, especially the broadening application of expression microarrays and characterization of mutants with relevant phenotypes (e.g. fanclnkhg10aEt/nkhg10aEt [128], nanos3fh49/fh49 [129]), has allowed researchers to start looking for large-scale changes and study the involvement of major developmental pathways. Through the analysis of a zebrafish line with mutation in the Fanconi anemia complementation group L (fancl; NM_212982) gene, the Postlethwait’s lab has obtained new information on the regulation of apoptotic processes during gonad transformation [130]. According to their results, tumor protein 53 (Tp53; [131]) activates the apoptotic processes in the male gonads resulting in the removal of oocytes and thereby tilting the balance toward testicular differentiation [128].
The involvement of NF-кB (nuclear factor of kappa light polypeptide gene enhancer in B cells; [132]) pathway in zebrafish gonad differentiation was discovered recently. Juvenile zebrafish subjected to heat-killed bacteria showed a sharp increase in female bias at adulthood: transcriptomic analysis of the underlying processes identified the upregulation of NF-кB pathway through induced inflammation as the cause [133]. When the pathway was chemically inhibited, the resulting adult sex ratio shifted toward increased proportion of males [133].
We have performed a microarray-based transcriptomic analysis of developing gonads using the Tg(vasa:vasa-EGFP) line developed by Olsen lab [123]. The analysis of differentially expressed genes indicated potential involvement of canonical Wnt (or Wnt/beta-catenin) signaling pathway [134] that was shown earlier to play an important role in mammalian ovary formation and maintenance (reviews: [135, 136]). Subsequent downregulation of the pathway through transgenic inhibition has resulted in a significant decrease of the proportion of males, providing functional proof for its importance in ovary formation [and possibly maintenance (Sreenivasan and Jiang, personal communication)].
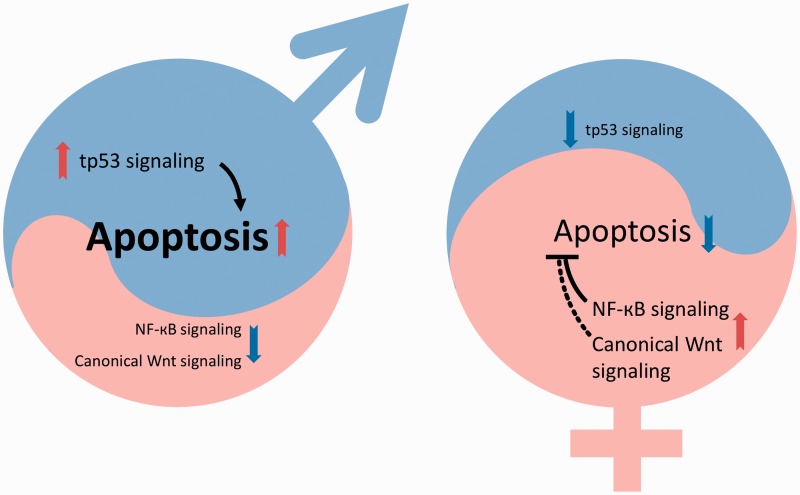
To summarize the current status: one pro-male (Tp53-activated apoptosis) and two pro-female (NF-кB and canonical Wnt) pathways were found to be involved with gonad transformation and differentiation in zebrafish. Two of them (possibly all three) have an effect on apoptotic processes that control the fate of cell types in developing gonad. The shift of balance of these three pathways into one or the other direction will tilt the hormonal balance through the steroidogenic pathway and eventually push the development toward one of the two gonad types (Figure 5). As the involvement of additional pathways seems very likely, further studies will be needed to unravel all the details of this beautiful and complex process. We have preliminary data that seem to indicate that some environmental factors affecting zebrafish sex are also likely to act through altering some of these processes into one or the other direction.
Figure 5:
Shift in the balance of pro-male and pro-female pathways will determine direction of gonad differentiation in zebrafish. Three major developmental pathways (Tp53-apoptosis, NF-кB and canonical Wnt) have been shown to participate in the process. The number of germ cells also has a profound effect on the final outcome (data not shown). In the males (left), numbers of primary oocytes originating from the ‘juvenile ovary’ are low, tp53 is upregulated, apoptotic processes are fully active and the NF-кB as well as canonical Wnt signaling pathways are both downregulated. The hormonal balance is shifted toward maleness. In the females (right), the number of primary oocytes originating from the ‘juvenile ovary’ is high, the NF-кB pathway and canonical Wnt signaling pathways are both upregulated, whereas tp53 is downregulated. Apoptosis is inhibited and the hormonal balance is shifted toward femaleness. The timing and causative effects of these processes are not fully understood.
CONCLUDING REMARKS AND AN OUTLOOK TO THE FUTURE
Despite having a PSD system, the downstream cascade of sex differentiation in zebrafish is still likely to involve several pathways and genes that were shown earlier to play a role in other vertebrates (review: [20]). On the other hand, their role and position in the cascade might vary across the different vertebrate groups [137, 138].
A molecular event, which involves multiple autosomal genes, such as PSD, will require a complex network of regulations to ensure proper development. Additional regulatory mechanisms, involving DNA methylation, histone modification and noncoding RNAs, are likely to contribute to these processes. Recent data showed that epigenetic regulatory mechanisms contribute to SD and reproductive organ development in plants and animals (review: [139]). With advances in next generation sequencing and the improved zebrafish genome assembly [95, 96] concerted analysis of the transcriptome, genome, miRNome and gonadal methylome will soon become a routine tool helping us to uncover additional genes and pathways as well as their interactions. All this will lead to a more detailed understanding of these hitherto elusive ‘complicated affairs’.
Key Points.
Zebrafish SD is genetic; environmental factors exert only secondary effects onto the sexual development of this species.
Most data indicate that SD in zebrafish is not based primarily on the action of sex chromosomes.
The zebrafish has PSD system, i.e. more than one autosomal genes have an effect on the direction of gonad differentiation (with or without contribution from sex chromosomes).
The main characteristics of PSD observed in zebrafish include: (i) cumulative effect of multiple genes on SD; (ii) wide ranging familial sex ratios (including severely biased lines) that are stable in repeated crosses; (iii) strong influence from parental genotypes and (iv) a large number of expected sexual genotypes among the gametes, not just two as in CSD species.
There are at least three developmental pathways involved with gonad transformation and differentiation, the contribution of several others is expected.
Acknowledgements
The authors are grateful to past and current members of the Orbán lab, especially Richárd Bártfai, Junhui Jiang, Yang Li, Rajini Sreenivasan and Xingang Wang, for data, discussions, suggestions and knowledge that have contributed to ideas described in the review. Arguments and discussions with Matt Harris, Per-Erik Olsson, Francesc Piferrer, Laia Ribas, Reade Roberts and Kellee Siegfried have also added depth to the story. The authors also thank two anonymous reviewers for their suggestions and criticisms that helped to improve an earlier version as well as Shubha Vij and Natascha May Thevasagayam for proofreading the manuscript. The work at Orbán lab is supported by the National Research Foundation of Singapore, the Agri-Food and Veterinary Authority of Singapore and the Strategic Research Program of Temasek Life Sciences Laboratory.
Biographies
Woei Chang Liew is a PhD candidate at the Orbán group at TLL. His research interest is in sexual development of teleosts and other vertebrates with special focus on the area of sex determination.
László Orbán has been educated in Hungary and in the USA. He has been studying (zebra)fish reproduction for over 20 years. Currently, he is a group leader at Temasek Life Sciences Laboratory (TLL, Singapore).
FUNDING
This study was supported by the Strategic Research Program of Temasek Life Sciences Laboratory and the Agri-Food and Veterinary Authority of Singapore.
References
- 1.Nelson JS. Fishes of the World. 4th. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2007. [Google Scholar]
- 2.Lévêque C, Oberdorff T, Paugy D, et al. Global diversity of fish (Pisces) in freshwater. Hydrobiologia. 2008;595:545–67. [Google Scholar]
- 3.Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- 4.Penman DJ, Piferrer F. Fish gonadogenesis. Part I: Genetic and environmental mechanisms of sex determination. Rev Fish Sci. 2008;16:16–34. [Google Scholar]
- 5.Graves JAM. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet. 2008;42:565–86. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- 6.Spence R, Gerlach G, Lawrence C, et al. The behaviour and ecology of the zebrafish, Danio rerio. Biol Rev Camb Philos Soc. 2008;83:13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- 7.Engeszer RE, Patterson LB, Rao AA, et al. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish. 2007;4:21–40. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- 8.Baroiller JF, D'Cotta H. Environment and sex determination in farmed fish. Comp Biochem Physiol C Toxicol Pharmacol. 2001;130:399–409. doi: 10.1016/s1532-0456(01)00267-8. [DOI] [PubMed] [Google Scholar]
- 9.Charlesworth B. Sex determination: primitive Y chromosomes in fish. Curr Biol. 2004;14:R745–7. doi: 10.1016/j.cub.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Nagahama Y. Molecular mechanisms of sex determination and gonadal sex differentiation in fish. Fish Physiol Biochem. 2005;31:105–9. doi: 10.1007/s10695-006-7590-2. [DOI] [PubMed] [Google Scholar]
- 11.Volff JN, Nanda I, Schmid M, et al. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sex Dev. 2007;1:85–99. doi: 10.1159/000100030. [DOI] [PubMed] [Google Scholar]
- 12.Baroiller JF, D'Cotta H, Saillant E. Environmental effects on fish sex determination and differentiation. Sex Dev. 2009;3:118–35. doi: 10.1159/000223077. [DOI] [PubMed] [Google Scholar]
- 13.Herpin A, Schartl M. Molecular mechanisms of sex determination and evolution of the Y-chromosome: Insights from the medakafish (Oryzias latipes) Mol Cell Endocrinol. 2009;306:51–8. doi: 10.1016/j.mce.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Mank JE, Avise JC. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev. 2009;3:60–7. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- 15.Sandra GE, Norma MM. Sexual determination and differentiation in teleost fish. Rev Fish Biol Fish. 2010;20:101–21. [Google Scholar]
- 16.Chen J, Hu W, Zhu Z. Progress in studies of fish reproductive development regulation. Chin Sci Bull. 2012;58:7–16. [Google Scholar]
- 17.Kikuchi K, Hamaguchi S. Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn. 2013;242:339–53. doi: 10.1002/dvdy.23927. [DOI] [PubMed] [Google Scholar]
- 18.Hattori RS, Strussmann CA, Fernandino JI, et al. Genotypic sex determination in teleosts: Insights from the testis-determining amhy gene. Gen Comp Endocrinol. 2013;192:55–9. doi: 10.1016/j.ygcen.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 19.Piferrer F, Guiguen Y. Fish gonadogenesis. Part II: Molecular biology and genomics of sex differentiation. Rev Fish Sci. 2008;16:35–55. [Google Scholar]
- 20.von Hofsten J, Olsson PE. Zebrafish sex determination and differentiation: involvement of FTZ-F1 genes. Reprod Biol Endocrinol. 2005;3:63. doi: 10.1186/1477-7827-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orban L, Sreenivasan R, Olsson PE. Long and winding roads: testis differentiation in zebrafish. Mol Cell Endocrinol. 2009;312:35–41. doi: 10.1016/j.mce.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Siegfried KR. In search of determinants: gene expression during gonadal sex differentiation. J Fish Biol. 2010;76:1879–1902. doi: 10.1111/j.1095-8649.2010.02594.x. [DOI] [PubMed] [Google Scholar]
- 23.Tong SK, Hsu HJ, Chung BC. Zebrafish monosex population reveals female dominance in sex determination and earliest events of gonad differentiation. Dev Biol. 2010;344:849–56. doi: 10.1016/j.ydbio.2010.05.515. [DOI] [PubMed] [Google Scholar]
- 24.Bradley KM, Breyer JP, Melville DB, et al. An SNP-based linkage map for zebrafish reveals sex determination loci. G3. 2011;1:3–9. doi: 10.1534/g3.111.000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson JL, Rodriguez Mari A, Braasch I, et al. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One. 2012;7:e40701. doi: 10.1371/journal.pone.0040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liew WC, Bartfai R, Lim Z, et al. Polygenic sex determination system in zebrafish. PLoS One. 2012;7:e34397. doi: 10.1371/journal.pone.0034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence C, Ebersole JP, Kesseli RV. Rapid growth and out-crossing promote female development in zebrafish (Danio rerio) Environ Biol Fishes. 2008;81:239–46. [Google Scholar]
- 28.Corley-Smith GE, Lim CJ, Brandhorst BP. Production of androgenetic zebrafish (Danio rerio) Genetics. 1996;142:1265–76. doi: 10.1093/genetics/142.4.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manolakou P, Lavranos G, Angelopoulou R. Molecular patterns of sex determination in the animal kingdom: a comparative study of the biology of reproduction. Reprod Biol Endocrinol. 2006;4:59. doi: 10.1186/1477-7827-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piferrer F, Ribas L, Diaz N. Genomic approaches to study genetic and environmental influences on fish sex determination and differentiation. Mar Biotechnol. 2012;14:591–604. doi: 10.1007/s10126-012-9445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda M, Nagahama Y, Shinomiya A, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–63. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 32.Peichel CL, Ross JA, Matson CK, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–24. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 33.Hattori RS, Murai Y, Oura M, et al. A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci USA. 2012;109:2955–9. doi: 10.1073/pnas.1018392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krueger WH, Oliveira K. Evidence for environmental sex determination in the American eel, Anguilla rostrata. Environ Biol Fishes. 1999;55:381–9. [Google Scholar]
- 35.Warner RR, Swearer SE. Social-control of sex-change in the bluehead wrasse, Thalassoma-bifasciatum (Pisces, Labridae) Biol Bull. 1991;181:199–204. doi: 10.2307/1542090. [DOI] [PubMed] [Google Scholar]
- 36.Ospina-Alvarez N, Piferrer F. Temperature-dependent sex determination in fish revisited: prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS One. 2008;3:e2837. doi: 10.1371/journal.pone.0002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ser JR, Roberts RB, Kocher TD. Multiple interacting loci control sex determination in Lake Malawi cichlid fish. Evolution. 2010;64:486–501. doi: 10.1111/j.1558-5646.2009.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandeputte M, Dupont-Nivet M, Chavanne H, et al. A polygenic hypothesis for sex determination in the European sea bass Dicentrarchus labrax. Genetics. 2007;176:1049–57. doi: 10.1534/genetics.107.072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abozaid H, Wessels S, Horstgen-Schwark G. Effect of rearing temperatures during embryonic development on the phenotypic sex in zebrafish (Danio rerio) Sex Dev. 2011;5:259–65. doi: 10.1159/000330120. [DOI] [PubMed] [Google Scholar]
- 40.Abozaid H, Wessels S, Horstgen-Schwark G. Elevated temperature applied during gonadal transformation leads to male bias in zebrafish (Danio rerio) Sex Dev. 2012;6:201–9. doi: 10.1159/000336297. [DOI] [PubMed] [Google Scholar]
- 41.Strüssmann C, Nakamura M. Morphology, endocrinology, and environmental modulation of gonadal sex differentiation in teleost fishes. Fish Physiol Biochem. 2002;26:13–29. [Google Scholar]
- 42.Pieau C, Dorizzi M, Richard-Mercier N. Temperature-dependent sex determination and gonadal differentiation in reptiles. Cell Mol Life Sci. 1999;55:887–900. doi: 10.1007/s000180050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modi WS, Crews D. Sex chromosomes and sex determination in reptiles. Curr Opin Genet Dev. 2005;15:660–5. doi: 10.1016/j.gde.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Shang EH, Yu RM, Wu RS. Hypoxia affects sex differentiation and development, leading to a male-dominated population in zebrafish (Danio rerio) Environ Sci Technol. 2006;40:3118–22. doi: 10.1021/es0522579. [DOI] [PubMed] [Google Scholar]
- 45.Uchida D, Yamashita M, Kitano T, et al. An aromatase inhibitor or high water temperature induce oocyte apoptosis and depletion of P450 aromatase activity in the gonads of genetic female zebrafish during sex-reversal. Comp Biochem Physiol A Mol Integr Physiol. 2004;137:11–20. doi: 10.1016/s1095-6433(03)00178-8. [DOI] [PubMed] [Google Scholar]
- 46.Villamizar N, Ribas L, Piferrer F, et al. Impact of daily thermocycles on hatching rhythms, larval performance and sex differentiation of zebrafish. PLoS One. 2012;7:e52153. doi: 10.1371/journal.pone.0052153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro-Martin L, Vinas J, Ribas L, et al. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 2011;7:e1002447. doi: 10.1371/journal.pgen.1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi T, Kitano T. High temperature induces cyp26b1 mRNA expression and delays meiotic initiation of germ cells by increasing cortisol levels during gonadal sex differentiation in Japanese flounder. Biochem Biophys Res Commun. 2012;419:287–92. doi: 10.1016/j.bbrc.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–14. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 50.Gschwend AR, Weingartner LA, Moore RC, et al. The sex-specific region of sex chromosomes in animals and plants. Chromosome Res. 2012;20:57–69. doi: 10.1007/s10577-011-9255-y. [DOI] [PubMed] [Google Scholar]
- 51.Cline TW. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 1993;9:385–90. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- 52.Kamiya T, Kai W, Tasumi S, et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (Fugu) PLoS Genet. 2012;8:e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delbridge M, Graves J. Mammalian Y chromosome evolution and the male-specific functions of Y chromosome-borne genes. Rev Reprod. 1999;4:101–9. doi: 10.1530/ror.0.0040101. [DOI] [PubMed] [Google Scholar]
- 54.Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet. 2011;12:157–66. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- 55.Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes–an evolving understanding. Bioessays. 1995;17:311–20. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 56.Post A. Vergleichende untersuchungen der chromosomenzahlen bei susswassernteleosteern. Z Zool Syst Evol. 1964;3:47–93. [Google Scholar]
- 57.Endo A, Ingalls TH. Chromosomes of the zebra fish. A model for cytogenetic, embryologic, and ecologic study. J Hered. 1968;59:382–4. doi: 10.1093/oxfordjournals.jhered.a107755. [DOI] [PubMed] [Google Scholar]
- 58.Rishi KK. Karyotypic studies on four species of fishes. Nucleus. 1976;19:95–8. [Google Scholar]
- 59.Schreeb K, Groth G, Sachsse W, et al. The karyotype of the zebrafish (Brachydanio rerio) J Exp Anim Sci. 1993;36:27–31. [PubMed] [Google Scholar]
- 60.Pijnacker LP, Ferwerda MA. Zebrafish chromosome banding. Genome. 1995;38:1052–5. doi: 10.1139/g95-140. [DOI] [PubMed] [Google Scholar]
- 61.Daga R, Thode G, Amores A. Chromosome complement, C-banding, Ag-NOR and replication banding in the zebrafish Danio rerio. Chromosome Res. 1996;4:29–32. doi: 10.1007/BF02254941. [DOI] [PubMed] [Google Scholar]
- 62.Gornung E, Gabrielli I, Cataudella S, et al. CMA3-banding pattern and fluorescence in situ hybridization with 18S rRNA genes in zebrafish chromosomes. Chromosome Res. 1997;5:40–46. doi: 10.1023/a:1018441402370. [DOI] [PubMed] [Google Scholar]
- 63.Amores A, Postlethwait JH. The zebrafish: Genetics and genomics. In: Detrich III HW, Westerfield M, Zon LI, editors. Method. Cell Biol. San Diego, CA, USA: Academic Press; 1998. pp. 323–38. [Google Scholar]
- 64.Ueda T, Naoi H. BrdU-4Na-EDTA-Giemsa band karyotypes of 3 small freshwater fish, Danio rerio, Oryzias latipes, and Rhodeus ocellatus. Genome. 1999;42:531. [Google Scholar]
- 65.Phillips RB, Reed KM. Localization of repetitive DNAs to zebrafish (Danio rerio) chromosomes by fluorescence in situ hybridization (FISH) Chromosome Res. 2000;8:27–35. doi: 10.1023/a:1009271017998. [DOI] [PubMed] [Google Scholar]
- 66.Fontana F, Chiarelli B, Rossi AC. Il cariotipo di alcune specie di Cyprinidae, Centrarchidae, Characidae studiate mediante culture in vivo. Caryologia. 1970;23:549–64. [Google Scholar]
- 67.Sharma K, Sharma O, Tripathi M. Female heterogamety in Danio rerio (Cypriniformes: Cyprinidae) Proc Ind Natl Sci Acad. 1998;68:123–6. [Google Scholar]
- 68.Arkhipchuk VV. Role of chromosomal and genome mutations in the evolution of bony fishes. Hydrobiol J. 1995;31:55–65. [Google Scholar]
- 69.Tease C, Hulten MA. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet Genome Res. 2004;107:208–15. doi: 10.1159/000080599. [DOI] [PubMed] [Google Scholar]
- 70.Singer A, Perlman H, Yan Y, et al. Sex-specific recombination rates in zebrafish (Danio rerio) Genetics. 2002;160:649–57. doi: 10.1093/genetics/160.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Postlethwait JH, Johnson SL, Midson CN, et al. A genetic linkage map for the zebrafish. Science. 1994;264:699–703. doi: 10.1126/science.8171321. [DOI] [PubMed] [Google Scholar]
- 72.Postlethwait JH, Yan YL, Gates MA, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–9. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 73.Kochakpour N, Moens PB. Sex-specific crossover patterns in zebrafish (Danio rerio) Heredity. 2008;100:489–95. doi: 10.1038/sj.hdy.6801091. [DOI] [PubMed] [Google Scholar]
- 74.Traut W, Winking H. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res. 2001;9:659–72. doi: 10.1023/a:1012956324417. [DOI] [PubMed] [Google Scholar]
- 75.Wallace BMN, Wallace H. Synaptonemal complex karyotype of zebrafish. Heredity. 2003;90:136–40. doi: 10.1038/sj.hdy.6800184. [DOI] [PubMed] [Google Scholar]
- 76.Purdom CE. Radiation-induced gynogenesis and androgenesis in fish. Heredity. 1969;24:431–44. doi: 10.1038/hdy.1969.59. [DOI] [PubMed] [Google Scholar]
- 77.Parsons JE, Thorgaard GH. Production of androgenetic diploid rainbow trout. J Hered. 1985;76:177–81. doi: 10.1093/oxfordjournals.jhered.a110060. [DOI] [PubMed] [Google Scholar]
- 78.Ihssen PE, Mckay LR, Mcmillan I, et al. Ploidy manipulation and gynogenesis in fishes - cytogenetic and fisheries applications. T Am Fish Soc. 1990;119:698–717. [Google Scholar]
- 79.Komen H, Thorgaard GH. Androgenesis, gynogenesis and the production of clones in fishes: A review. Aquaculture. 2007;269:150–73. [Google Scholar]
- 80.Horváth L, Orbán L. Genome and gene manipulation in the common carp. Aquaculture. 1995;129:157–81. [Google Scholar]
- 81.Hörstgen-Schwark G. Production of homozygous diploid zebra fish (Brachydanio rerio) Aquaculture. 1993;112:25–37. [Google Scholar]
- 82.Pelegri F, Schulte-Merker S. The zebrafish: Genetics and genomics. In: Detrich III HW, Westerfield M, Zon LI, editors. Method. Cell Biol. San Diego, CA, USA: Academic Press; 1999. pp. 1–20. [Google Scholar]
- 83.Streisinger G, Walker C, Dower N, et al. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–6. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 84.Uchida D, Yamashita M, Kitano T, et al. Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J Exp Biol. 2002;205:711–8. doi: 10.1242/jeb.205.6.711. [DOI] [PubMed] [Google Scholar]
- 85.Pandian TJ, Sheela SG. Hormonal induction of sex reversal in fish. Aquaculture. 1995;138:1–22. [Google Scholar]
- 86.Williams JGK, Kubelik AR, Livak KJ, et al. DNA polymorphisms amplified by arbitrary primers are useful as genetic-markers. Nucleic Acids Res. 1990;18:6531–5. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–14. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen JJ, Du QY, Yue YY, et al. Screening and identification of male-specific DNA fragments in common carps Cyprinus carpio using suppression subtractive hybridization. J Fish Biol. 2010;77:403–13. doi: 10.1111/j.1095-8649.2010.02700.x. [DOI] [PubMed] [Google Scholar]
- 89.Charlesworth D, Mank JE. The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics. 2010;186:9–31. doi: 10.1534/genetics.110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang A, Liew WC, Chuah A, et al. FluoMEP: a new genotyping method combining the advantages of randomly amplified polymorphic DNA and amplified fragment length polymorphism. Electrophoresis. 2007;28:525–34. doi: 10.1002/elps.200600715. [DOI] [PubMed] [Google Scholar]
- 91.Kirankumar S, Anathy V, Pandian TJ. Hormonal induction of supermale golden rosy barb and isolation of Y-chromosome specific markers. Gen Comp Endocrinol. 2003;134:62–71. doi: 10.1016/s0016-6480(03)00218-1. [DOI] [PubMed] [Google Scholar]
- 92.Kirankumar S, Pandian TJ. Production and progeny testing of androgenetic rosy barb Puntius conchonius. J Exp Zool Part A. 2004;301A:938–51. doi: 10.1002/jez.a.117. [DOI] [PubMed] [Google Scholar]
- 93.Nanda I, Feichtinger W, Schmid M, et al. Simple repetitive sequences are associated with differentiation of the sex-chromosomes in the guppy fish. J Mol Evol. 1990;30:456–62. [Google Scholar]
- 94.Tripathi N, Hoffmann M, Willing EM, et al. Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc Biol Sci. 2009;276:2195–208. doi: 10.1098/rspb.2008.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patowary A, Purkanti R, Singh M, et al. A sequence-based variation map of zebrafish. Zebrafish. 2013;10:15–20. doi: 10.1089/zeb.2012.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 98.Pinto D, Darvishi K, Shi X, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29:512–20. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown KH, Dobrinski KP, Lee AS, et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci USA. 2012;109:529–34. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wilkins AS. Moving up the hierarchy: A hypothesis on the evolution of a genetic sex determination pathway. Bioessays. 1995;17:71–77. doi: 10.1002/bies.950170113. [DOI] [PubMed] [Google Scholar]
- 101.Morrish BC, Sinclair AH. Vertebrate sex determination: many means to an end. Reproduction. 2002;124:447–57. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- 102.Graham P, Penn JK, Schedl P. Masters change, slaves remain. Bioessays. 2003;25:1–4. doi: 10.1002/bies.10207. [DOI] [PubMed] [Google Scholar]
- 103.Bagheri-Fam S, Sinclair AH, Koopman P, et al. Conserved regulatory modules in the Sox9 testis-specific enhancer predict roles for SOX, TCF/LEF, Forkhead, DMRT, and GATA proteins in vertebrate sex determination. Int J Biochem Cell B. 2010;42:472–7. doi: 10.1016/j.biocel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 104.Graves JAM, Peichel CL. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010;11:205. doi: 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yano A, Guyomard R, Nicol B, et al. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr Biol. 2012;22:1423–8. doi: 10.1016/j.cub.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 106.Sreenivasan R, Cai M, Bartfai R, et al. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS One. 2008;3:e1791. doi: 10.1371/journal.pone.0001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wen C, Zhang Z, Ma W, et al. Genome-wide identification of female-enriched genes in zebrafish. Dev Dyn. 2005;232:171–9. doi: 10.1002/dvdy.20210. [DOI] [PubMed] [Google Scholar]
- 108.Small CM, Carney GE, Mo Q, et al. A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics. 2009;10:579. doi: 10.1186/1471-2164-10-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zheng W, Xu H, Lam SH, et al. Transcriptomic analyses of sexual dimorphism of the zebrafish liver and the effect of sex hormones. PLoS One. 2013;8:e53562. doi: 10.1371/journal.pone.0053562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Santos EM, Kille P, Workman VL, et al. Sexually dimorphic gene expression in the brains of mature zebrafish. Comp Biochem Physiol A Mol Integr Physiol. 2008;149:314–24. doi: 10.1016/j.cbpa.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 111.Bull JJ. Evolution of Sex Determining Mechanisms. Melon Park, CA, USA: Benjamin-Cummings Publishing Company; 1983. [Google Scholar]
- 112.Moore EC, Roberts RB. Polygenic sex determination. Curr Biol. 2013;23:R510–2. doi: 10.1016/j.cub.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 113.Kosswig C. Polygenic sex determination. Experientia. 1964;20:190–9. doi: 10.1007/BF02135395. [DOI] [PubMed] [Google Scholar]
- 114.Desprez D, Briand C, Hoareau MC, et al. Study of sex ratio in progeny of a complex Oreochromis hybrid, the Florida red tilapia. Aquaculture. 2006;251:231–7. [Google Scholar]
- 115.Cnaani A, Lee BY, Zilberman N, et al. Genetics of sex determination in tilapiine species. Sex Dev. 2008;2:43–54. doi: 10.1159/000117718. [DOI] [PubMed] [Google Scholar]
- 116.Tessema M, Müller-Belecke A, Hörstgen-Schwark G. Effect of rearing temperatures on the sex ratios of Oreochromis niloticus populations. Aquaculture. 2006;258:270–77. [Google Scholar]
- 117.Takahashi H. Juvenile hermaphroditism in the zebrafish, Brachydanio rerio. Bull Fac Fish Hokkaido Univ. 1977;28:57–65. [Google Scholar]
- 118.Maack G, Segner H. Morphological development of the gonads in zebrafish. J Fish Biol. 2003;62:895–906. [Google Scholar]
- 119.Wang XG, Bartfai R, Sleptsova-Freidrich I, et al. The timing and extent of ‘juvenile ovary' phase are highly variable during zebrafish testis differentiation. J Fish Biol. 2007;70:33–44. [Google Scholar]
- 120.Siegfried KR, Nusslein-Volhard C. Germ line control of female sex determination in zebrafish. Dev Biol. 2008;324:277–87. doi: 10.1016/j.ydbio.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 121.Slanchev K, Stebler J, de la Cueva-Méndez G, et al. Development without germ cells: The role of the germ line in zebrafish sex differentiation. Proc Natl Acad Sci USA. 2005;102:4074–9. doi: 10.1073/pnas.0407475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dranow DB, Tucker RP, Draper BW. Germ cells are required to maintain a stable sexual phenotype in adult zebrafish. Dev Biol. 2013;376:43–50. doi: 10.1016/j.ydbio.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 123.Krøvel AV, Olsen LC. Expression of a vas::EGFP transgene in primordial germ cells of the zebrafish. Mech Develop. 2002;116:141–50. doi: 10.1016/s0925-4773(02)00154-5. [DOI] [PubMed] [Google Scholar]
- 124.Hsiao CD, Tsai HJ. Transgenic zebrafish with fluorescent germ cell: a useful tool to visualize germ cell proliferation and juvenile hermaphroditism in vivo. Dev Biol. 2003;262:313–23. doi: 10.1016/s0012-1606(03)00402-0. [DOI] [PubMed] [Google Scholar]
- 125.Onichtchouk D, Aduroja K, Belting HG, et al. Transgene driving GFP expression from the promoter of the zona pellucida gene zpc is expressed in oocytes and provides an early marker for gonad differentiation in zebrafish. Dev Dyn. 2003;228:393–404. doi: 10.1002/dvdy.10392. [DOI] [PubMed] [Google Scholar]
- 126.Rodríguez-Marí A, Yan Y-L, BreMiller RA, et al. Characterization and expression pattern of zebrafish anti-Müllerian hormone (amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr Patterns. 2005;5:655–67. doi: 10.1016/j.modgep.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 127.Wang XG, Orban L. Anti-Mullerian hormone and 11 beta-hydroxylase show reciprocal expression to that of aromatase in the transforming gonad of zebrafish males. Dev Dyn. 2007;236:1329–38. doi: 10.1002/dvdy.21129. [DOI] [PubMed] [Google Scholar]
- 128.Rodríguez-Marí A, Cañestro C, BreMiller RA, et al. Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS Genet. 2010;6:e1001034. doi: 10.1371/journal.pgen.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Beer RL, Draper BW. nanos3 maintains germline stem cells and expression of the conserved germline stem cell gene nanos2 in the zebrafish ovary. Dev Biol. 2013;374:308–18. doi: 10.1016/j.ydbio.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 130.Rodríguez-Marí A, Postlethwait JH. The zebrafish: Disease models and chemical screens. In: Detrich III HW, Westerfield M, Zon LI, editors. Method Cell Biol. San Diego, CA, USA: Academic Press; 2011. pp. 461–90. [Google Scholar]
- 131.Lane DP, Crawford LV. T antigen is bound to a host protein in SY40-transformed cells. Nature. 1979;278:261–3. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 132.Hayden MS, Ghosh S. Shared principles in NF-kB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 133.Pradhan A, Khalaf H, Ochsner SA, et al. Activation of NF-kappaB protein prevents the transition from juvenile ovary to testis and promotes ovarian development in zebrafish. J Biol Chem. 2012;287:37926–38. doi: 10.1074/jbc.M112.386284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sreenivasan R, Wang X, Bartfai R, et al. Global expression profiling in zebrafish reveals genes with potential roles in sexual differentiation. Biol Reprod. 2009;78:116. [Google Scholar]
- 135.Bernard P, Harley VR. Wnt4 action in gonadal development and sex determination. Int J Biochem Cell B. 2007;39:31–43. doi: 10.1016/j.biocel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 136.Nef S, Vassalli JD. Complementary pathways in mammalian female sex determination. J Biol. 2009;8:74. doi: 10.1186/jbiol173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cutting A, Chue J, Smith CA. Just how conserved is vertebrate sex determination? Dev Dyn. 2013;242:380–7. doi: 10.1002/dvdy.23944. [DOI] [PubMed] [Google Scholar]
- 138.Herpin A, Adolfi MC, Nicol B, et al. Divergent expression regulation of gonad development genes in medaka shows incomplete conservation of the downstream regulatory network of vertebrate sex determination. Mol Biol Evol. 2013;30:2328–46. doi: 10.1093/molbev/mst130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Piferrer F. Epigenetics of sex determination and gonadogenesis. Dev Dyn. 2013;242:360–70. doi: 10.1002/dvdy.23924. [DOI] [PubMed] [Google Scholar]