Abstract
The zebrafish (Danio rerio) is an important model organism for studying development and human disease. The zebrafish has an excellent reference genome and the functions of hundreds of genes have been tested using both forward and reverse genetic approaches. Recent years have seen an increasing number of large-scale mutagenesis projects and the number of mutants or gene knockouts in zebrafish has increased rapidly, including for the first time conditional knockout technologies. In addition, targeted mutagenesis techniques such as zinc finger nucleases, transcription activator-like effector nucleases and clustered regularly interspaced short sequences (CRISPR) or CRISPR-associated (Cas), have all been shown to effectively target zebrafish genes as well as the first reported germline homologous recombination, further expanding the utility and power of zebrafish genetics. Given this explosion of mutagenesis resources, it is now possible to perform systematic, high-throughput phenotype analysis of all zebrafish gene knockouts.
Keywords: zebrafish, mutagenesis, phenotyping, resources, knockouts
INTRODUCTION
In the age of the sequenced human genome, diseases and phenotypes can be rapidly mapped by genome-wide association studies (GWAS) to potential candidate genes [1] (http://www.genome.gov/gwastudies/) and candidates are increasingly identified by exome sequencing [2], but in both cases these merely represent correlations with diseases and cannot prove disease causation alone. A key issue still remains in determining genetic causes of disease: the functions of the vast majority of human genes have only been predicted computationally and have never been tested or verified in vivo. It is essential that functional testing of every gene be carried out so that better predictions for candidate disease genes from GWAS or exome/genome sequencing can be made. For decades, Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster have been favorite model systems for geneticists to carry out functional genetic studies, these models have contributed immensely to our understanding of signaling pathways, metabolism, the cell cycle, embryonic patterning, aging, homeostasis and many other areas. Their utility was enhanced even more when their genomes were sequenced, opening new avenues for systematic testing of gene function. Although these nonvertebrates are excellent model systems to study conserved developmental pathways, many aspects of vertebrate embryonic development such as tissue patterning and morphogenesis have features unique to the vertebrate lineage. Mouse (Mus Musculus) is the most commonly used vertebrate model organism with a high-quality reference genome, with nearly all genes having been identified. There are many powerful genetic tools available (e.g. targeted conditional knockouts) to study gene function in mouse, however, the maintenance of large mouse colonies is expensive, making it difficult for large-scale genetic screens and phenotyping studies. Three decades ago, George Streisinger and colleagues [3] introduced the small, freshwater teleost fish the zebrafish (Danio rerio) as a model organism to geneticists. Since then, zebrafish has gained significant momentum as a model for studying vertebrate development and modeling human disease. The zebrafish genome is only the third vertebrate genome to be ‘finished’ [4] recently joining human [5–7] and mouse [8] in having a high quality reference genome sequence. Annotations show that zebrafish has the largest number of genes (26 000) of any sequenced vertebrate [4]. Comparison of the zebrafish genome to the human genome revealed that 70% of all human genes have at least one zebrafish ortholog. Comparing the 3176 potential human diseases genes listed in Online Mendelian Inheritance in Man (OMIM) database to the list of zebrafish genes shows that there are 2601 (82%) present in zebrafish [4], and it reaffirms the potential for zebrafish in studying vertebrate development, biological pathways and human disease. Many human diseases such as cancer, infections, congenital and hereditary malformations, immunological diseases, heart defects, muscle degeneration, neurological problems, ocular degeneration and kidney disease have been effectively modeled in zebrafish [9, 10]. Zebrafish offers many advantages over mouse for studying early development as the embryos are externally fertilized and develop outside the mother, the embryos are transparent, the fish have high fecundity and they are relatively inexpensive for large-scale screens. The completed genome sequencing project in zebrafish identified the majority of the functional genes allowing researchers the opportunity to study the associated phenotypes systematically after mutagenesis. Therefore, high-throughput reverse genetic approaches are a potentially important tool for studying human genes by first testing their function in zebrafish. The last 10 years have seen an explosion in the number of tools and techniques available to manipulate the zebrafish genome (Table 1), and the number of novel genotypes in the ZFIN database has increased from 5645 in 2007 to 15 464 in 2012 [11]. Recently, significant progress has been made in large-scale mutagenesis projects that have generated the second largest collection of mutants in a vertebrate model, and these efforts will continue generating more alleles in the coming years [12–14]. Here, we review different mutagenesis strategies in zebrafish including insertional mutagenesis, chemical-mediated random mutagenesis and targeted mutagenesis as well as large-scale phenotyping aimed at studying zebrafish genes in a systematic fashion.
Table 1:
Mutagenesis resources and databases for zebrafish research
| Tools | Description | Website |
|---|---|---|
| Zebrafish Insertion Collection (ZInC) | Database of retroviral insertional mutants. | http://research.nhgri.nih.gov/zinc |
| Zebrafish Mutagenesis Project (ZMP) | Resource for loss-of-function mutations and phenotypes. | http://www.sanger.ac.uk/Projects/D_rerio/zmp/ |
| Zebrafish TILLING project | Database of loss-of-function mutations from TILLING project. | https://webapps.fhcrc.org/science/tilling/index.php |
| zfishbook | The International Protein trap consortium. | http://zfishbook.org/ |
| Digital Fish | Database of conditional Flip Trap lines | http://www.fliptrap.org/static/index_new.html |
| zCre | A database of Cre/lox based tools. | http://zcre.org.uk/Overview.html |
| CreZoo | Database of CreERT2 driver lines. | http://crezoo.crt-dresden.de/crezoo/ |
| zTrap | Database of gene traps and enhancer traps | http://kawakami.lab.nig.ac.jp/ztrap/ |
| ZIFIT | Tools to design ZFN,TALEN and CRISPR. | http://zifit.partners.org/ZiFiT/ |
| MOJO Hand | TALEN design tool | http://talendesign.com/mojohand_main.php |
| MegaMapper | A computational pipeline for positional cloning of mutations by whole-genome sequencing. | https://wiki.med.harvard.edu/SysBio/Megason/MegaMapper |
| MMAPPR | Analysis pipeline for mapping mutations using RNA-seq. | http://yost.genetics.utah.edu/software.php |
RANDOM MUTAGENESIS APPROACHES
Chemical-mediated mutagenesis
ENU (N-ethyl-N-nitrosourea) is the most commonly used chemical mutagen in zebrafish and was used for the two largest forward genetic screens that identified thousands of mutants with embryonic developmental phenotypes [15, 16]. The identification of mutated genes by positional cloning is still laborious, although the positional cloning methods have simplified over the years [17]. Recent advances in genomic technologies and next-generation sequencing further improved these methods and the mutated genes can now often be identified by whole-genome sequencing at low coverage (3–8X). Voz et al. developed a fast mapping method using the whole-genome sequencing (8-fold coverage); in this method the affected locus can be identified by the analysis of single-nucleotide polymorphism (SNP) homozygosity. Compared with traditional positional cloning, this method requires many fewer mutant embryos and can be performed in a few weeks [18]. However, given the high number of variations within or between different strains of zebrafish, it could be challenging to distinguish a homozygous mutation-causing variant with linked but low-frequency SNPs. Bowen et al. documented and developed an extensive SNP database in zebrafish that can be useful in mapping mutations with low-coverage whole-genome sequencing [19].
Two additional cloning strategies have been developed using the new sequencing platforms: bulk segregant-based linkage analysis (BSFseq) and homozygosity mapping (HMFseq). In BSFseq, a carrier of the mutant is out-crossed to a different wild-type strain, and the resulting F1 hybrid pairs are repeatedly crossed to generate a few hundred progeny, which are pooled and sequenced. The HMFseq is faster as it does not involve a mapping cross but relies on the inherently high SNP rate present in most of the lab zebrafish lines. Multiple carrier pairs are crossed and mutant larvae are collected, pooled and sequenced. Both strategies use sequences from a pool of mutants and analysis of whole-genome sequence for causative mutations using a similar bioinformatics pipeline. An open source tool MegaMapper (https://wikis.utexas.edu/display/bioiteam/MegaMapper) is also available for the analysis of both the HMFseq and BSFseq approaches [20, 21].
Two approaches based on transcriptome sequencing have been developed: Mutation Mapping Analysis Pipeline for Pooled RNA-seq (MMAPPR) and RNA-seq-based bulk segregant analysis (BSAseq), [21, 22]. As the size of the transcriptome is smaller than the whole genome, these approaches could be significantly cheaper if multiplexing is desired. MMAPPR (http://yost.genetics.utah.edu/software.php) can identify mutations without sequencing the parental strain or utilizing a SNP database. The library of pooled mutants and their phenotypically wild-type siblings are pooled and sequenced by RNA sequencing (RNA-seq). There are typically enough SNPs in the exonic sequences, that loss of heterozygosity can be identified from the sequenced complementary DNAs (cDNAs).
ENU-based mutagenesis has also been used in reverse genetics approaches in which the site of mutation is detected first and then the associated phenotypes measured (i.e. reverse genetics). One major method of identifying mutations in this fashion is called TILLING (targeting induced lesions in genomes) and has been applied to variety of plants and animals [23–27], including zebrafish [28–30]. The initial TILLING method used the Cel1 enzyme that cleaves heteroduplex DNA at a mismatched basepair site and the resulting fragments subsequently analyzed by polyacrylamide gel to detect mutations. Because of a high false negative rate and the labor-intensive nature of the technique, this method was superseded by different sequencing methods in which target genes are sequenced from a large number of mutagenized fish [31]. To leverage the cost-effectiveness of large-scale sequencing efforts, a public genome-wide Zebrafish Mutagenesis Project (ZMP), is being carried out at the Wellcome Trust Sanger Institute. This project aims to generate knockout alleles in every protein-coding gene in the zebrafish genome. The mutations are identified by whole exome enrichment followed by next generation sequencing on the Illumina platform. The sequencing throughput is further increased by pooling up to eight barcoded F1 genomic libraries into a single sequencing lane. The exomes of more than 1600 mutagenized F1 individuals have been sequenced, and as of this review, this project has mutated 11 892 genes and 24 000 alleles have been identified. There are mutations in the orthologs of 3188 of the 5494 genes identified in genome-wide association studies, and at least 1 allele in 2505 of the 3176 human diseases genes in the OMIM database [12]. A similar public effort is being carried out by a consortium of researchers from the Fred Hutchinson Cancer Research Center, Vanderbilt University and the University of Oregon. This project has generated a cryopreserved library of 8640 ENU-mutagenized zebrafish and identified loss-of-function mutations in over 140 genes. Researchers can submit the request for the sequencing of their genes of interest at http://www.zfishtilling.org/zfish/. Although ENU is an incredibly effective mutagenesis approach, it is primarily limited to missense or nonsense mutations and does not have a high likelihood of generating conditional alleles. Other techniques are required to generate conditional alleles, or gene fusions with reporter proteins or other functional moieties.
Retroviral-mediated mutagenesis
Insertional mutagenesis in zebrafish was introduced as an alternative approach to chemicals where exogenous DNA (initially) from retroviruses was used as mutagen and inserted into the genome. When an insertion occurs in any given gene, it can create a loss of function. Transposons, which were easier to use and had more flexible design capabilities were developed, significantly expanding the utility of the insertional approach. Retroviruses and transposons have different insertion site preferences, and depending on the site of insertion and the nature of the inserted element, they can either produce hypomorphic or null alleles, or may not cause loss of function at all. The advantage of insertional mutagenesis over chemical mutagenesis is the rapid identification of the mutated gene and newer technologies based on high-throughput sequencing for the identification of insertional elements make this process even more robust and cost-effective [14, 32, 33]. The disadvantage is that there has not been a transposon or retroviral vector reported that is as efficient in mutagenizing the genome as ENU.
Initially, the most efficient method of insertional mutagenesis in zebrafish was retroviral-mediated mutagenesis. In the early 1990s, Lin et al. [34] demonstrated that moloney murine leukemia virus (MoMLV) pseudotyped with the envelope protein from vesicular stomatitis virus (VSV) and injected into zebrafish embryos could infect the embryos, and the integrated DNA would pass through the germline to the F1 generation offspring. A large-scale forward genetic screen was performed to identify genes involved in embryonic development, and ∼500 mutants were recovered in almost 400 loci. Retroviral-mediated mutagenesis is not as easy to perform as chemical mutagenesis but the identification of the affected loci can subsequently be achieved by relatively simple and straightforward linker-mediated polymerase chain reaction (PCR), as evidenced by the fact that 335/400 loci were identified [35–37]. Most of the insertional screens in model organisms used forward genetics approach, i.e. screened for phenotypes first and then identified the affected genes. These screens were effective in identifying mutations with visible phenotypes, but were limited in addressing the issues of gene redundancy or genes that have more subtle phenotypes. As high-quality genome sequencing data became available, the MLV retrovirus became an effective mutagenesis tool for reverse genetics. As a proof-of-principle, Wang et al. developed a reverse genetics approach in which sperm from F1 fish were cryopreserved and at the same time insertions were identified from a genomic DNA sample. The insertional mutagenesis pipeline started with the production of high-titer pseudotyped MoMLV retroviruses [38]; this virus was then injected into blastula stage embryos to generate the founder fish (F0). The founder fish were raised and outcrossed to wild-type fish and sperm were cryopreserved from the male F1 fish. Integrations were mapped by identifying flanking genomic sequences to the proviral insertion, and a simple and rapid protocol utilizing combination of linker-mediated PCR and Sanger sequencing was developed. Approximately 900 insertions were mapped and fish carrying insertions could be recovered by in vitro fertilization [38, 39]. Recently, this technology was modernized and scaled up to generate a large-scale, genome-wide knockout library. A highly multiplexed and high-throughput strategy was developed to map retroviral insertions using the Illumina sequencing platform. In this method each fish was tagged with a unique 6 bp barcode and up to 500 fish were sequenced together in a single lane of a flow cell. Once the insertions were mapped to a genomic locus, they were assigned to their respective fish based on the 6 base barcode. Varshney et al. reported 6144 mutagenized and archived F1 fish. To date, 15 223 proviral integrations were mapped onto the Zv9 assembly of the zebrafish genome, 52% of these insertions were in genes and 12% of all gene hits were in exons, whereas 88% of all gene hits were in introns. Further analysis showed that 40% of the intronic hits were in the first intron. Given insertions in exons generate a truncation at the site of insertion and insertions in the first introns tend to result in hypomorphic alleles; there were 3776 predicted mutations in 3054 genes. The majority of mutated genes were hit only once suggesting this screen has not yet reached saturation [14]. A database called the Zebrafish Insertion Collection (http://research.nhgri.nih.gov/ZInC/) is available to help researchers find insertions in genes of interest [40].
Transposon-mediated mutagenesis
In zebrafish several transposable elements, such as Tol2, Ac/Ds and Sleeping Beauty, isolated from heterologous hosts are being used for transgenesis and mutagenesis [41–43]. Due to their simplicity, ease of use, high insertion efficiencies and their ability to carry complex DNA transgenes, transposons are the preferred choice over retroviral-based vectors for transgenesis. The Tol2 transposon from medaka has been particularly widely used in zebrafish in recent years and has become the primary genetic tool for transgenesis in zebrafish.
Conditional mutants give researchers the power to inactivate gene function temporally or in a tissue-specific manner making them versatile genetic tools in studying gene function in vivo. Recently, several transposon-based gene-trap/protein-trap cassettes have been published. Trinh et al. developed a conditional knockout tool called a ‘FlipTrap’ that offers the functionality of gene traps, a Cre-mediated conditional mutagen and Flp-mediated targeted recombination. When the FlipTrap cassette is inserted into an intron it uses the promoter of the gene and forms a functional fusion protein (citrine tag) thus revealing the native protein expression. This cassette can also be converted to a mutation when exposed to Cre recombinase generating a mCherry-tagged gene trap and a mutant allele. The cassette can also be exchanged with other cassettes by Flp-mediated recombination. There are ∼200 FlipTap lines available to the community and the detailed information can be accessed from http://www.fliptrap.org [44].
The most recent version of a ‘gene-breaking’ transposon included a protein trap that fused a fluorescent reporter to endogenously expressed genes and simultaneously inactivates the gene function. The RP2 cassette contains a splice acceptor that creates a fusion of an endogenous gene to monomeric red fluorescent protein (RFP), which lacks a starting methionine codon (ATG), thus, relying on translation initiation from the endogenous gene. By capturing the endogenous transcript, RP2 efficiently knocked down endogenous gene expression to <1% of normal, making it an efficient mutagen. The RP2 construct also has a 3′ poly-adenylation trap that contains green fluorescent protein (GFP) driven by the β-actin promoter. This drives the expression of GFP regardless of the insertion site by bypassing the endogenous promoter. The RP2 cassette is flanked by two loxP sites that can rescue the trapped gene by Cre recombinase or morpholinos that specifically target the trap’s splice acceptor [45]. This reversibility is a critical feature in proving the integration is causing the observed phenotype, as rescue is the classic test in genetics for proving causation.
Another Tol2-based vector was developed, which employs a highly mutagenic, bi-directional gene trap within an FlEx cassette. In one direction, the construct contains a ‘strong’ splice acceptor with an mCherry reporter, trapping essentially all of the transcripts and causing a mutagenic truncation of the gene. In the opposite direction, there is no functional trap, allowing for normal expression. This allows the entire construct to be inverted in vivo by either the Cre or Flp recombinase. When the gene trap is inserted into the intron of a gene in the same orientation as the gene transcript, it is expected to inactivate the gene function similar in function to the trap in the RP2 cassette. However, the FlEx cassette may be inverted by either expressing Cre or Flp in a tissue-specific manner resulting in tissue-specific rescue or by injecting Cre/Flp mRNA into one cell stage embryos resulting into global rescue. Once the insertion is in the inverted orientation of the tagged gene it will not have any mutagenic effect. As the FlEx casstte contains both FRT and loxP sites, it can be inverted once more to achieve the desired global or conditional gene knockout [46, 47].
In order to maximize the utility of conditional mutant alleles, a wide range of tissue-specific drivers are needed to express Cre or Flp temporally. An effort is being made to generate multiple Cre driver lines using a gene trap with a reporter consisting of an mCherry–Cre fused to the mutated human ligand binding domain of the estrogen receptor (mCherry–CreERT2). This gives the researcher dual functionality for the Cre drivers, not only can you control where the Cre recombinase is expressed, you can control activity temporally by adding tamoxifen at the desired time. Jungke et al. created a database called CreZoo (http://crezoo.crt-dresden.de/crezoo/) to help the community find a specific driver to suit their interests. As of this review, there are 47 Cre driver lines expressing CreERT2 in different spatiotemporal patterns but this is an ongoing effort and more lines are to be added [48].
Similar to P-elements in Drosophila, Tol2 transposons can also be re-mobilized to generate genomic deletions induced by imprecise excision. This can be accomplished by injecting transposase mRNA in the zebrafish’s one cell stage embryo. Using this method, up to 1.3 kb deletions were generated in enhancer trap transgenic lines. Given the large number of Tol2-based transgenic lines available, it could serve as an alternative approach to inactivate gene function in previously characterized transgenic lines. [49].
All of the described techniques to this point rely on randomly mutagenizing the genome and identifying mutations or integrations. The techniques are powerful, but there are inherent limitations to any random approach. Next we will describe how recently engineered DNA-binding proteins are being utilized to make targeted mutations in the zebrafish genome.
TARGETED MUTAGENESIS APPROACHES
Zinc finger nucleases-mediated mutagenesis
Recently, researchers have shown that it is possible to attach a nonspecific DNA nuclease to a DNA-binding protein that recognized a specific DNA sequence. Utilizing such a protein fusion, it becomes possible to target specific genomic loci in the zebrafish genome. The first of these nucleases to come along were the zinc finger nucleases (ZFNs). ZFN technology was developed and promised to be an effective targeted mutagenesis tool in zebrafish, presenting zebrafish researchers with the first opportunity to specifically target genes. ZFNs are chimeric proteins that are composed of two C2H2-type zinc finger arrays (ZFAs). Each ZFA is fused to the cleavage domain of the nonspecific Fok1 endonuclease that only becomes active upon dimerization. Two ZFAs consist of three ‘fingers’ each that recognizes a nine basepair sequence for a total of 18 bp of recognition. The ZFNs generate sequence-specific double-strand breaks (DSBs) on target DNA that can be repaired by error-prone non-homologous end joining (NHEJ) or by homologous recombination with a donor DNA template [50]. The efficacy of ZFNs in generating targeted mutations was first demonstrated in D. melanogaster [51–53], and later used for manipulating genomes in different organisms and cultured cells [54–56] including zebrafish [57, 58]. Doyon et al. used a proprietary technology from Sangamo Bioscience, and Ming et al. used a publicly available technology to target three genes: golden (gol), no tail/Brachyury (ntl), and kdr (kdra) that had well-characterized mutant phenotypes. In zebrafish, the messenger RNA's (mRNAs) encoding ZFNs are injected into one-cell stage embryos, and mosaic founder fish are tested for lesions. Founders are then raised and out-crossed to get an F1 generation and progeny are genotyped. About 5–25% of the progeny will show a mutation in the desired gene if the ZFN has high specificity and activity.
In zebrafish, ZFNs have been used to generate mutations in many genes [57–67]. There are multiple ways to design ZFNs including the proprietary methods developed by Sangamo bioscience and commercialized by Sigma-Aldrich (CompoZr ZFNs). Open source methods such as modular assembly, oligomerized pool engineering (OPEN) and context-dependent assembly (CoDA) were developed to engineer ZFNs more quickly and cheaply. The off-target effect of ZFNs was assessed by Doyon et al. and Ming et al. Doyon et al. analyzed five potential off-target loci for each ntl ZFN pair and detected no off-target lesions [57], whereas Meng et al. reported that ZFN-induced lesions at the heterodimeric sites were limited to a few sites (∼1–5%) and on-target site cleavage were ∼700-fold more likely to occur than on the off-target sites [58]. ZFNs are an effective tool to generate targeted mutations, however, the synthesis of ZFNs with high efficiency is expensive, labor intensive and time consuming and more recently, the development of alternative technologies such as transcription activator-like effector nucleases (TALENs) and CRISPR (Clustered regularly interspaced short sequences)/Cas (CRISPR-associated) have emerged as faster and cheaper alternatives.
TALENs-mediated mutagenesis
The key breakthrough in zebrafish-targeted mutagenesis came when a new class of sequence-specific nucleases, TALENs was shown to direct DSBs to specific DNA sequences. TALENs are created by fusing transcription activator-like effectors (TALEs) to the catalytic domain of the FokI endonuclease. TALEs are naturally occurring DNA-binding proteins produced by the plant pathogenic bacteria Xanthomonas to regulate the host gene expression. Each TALE DNA-binding protein is composed of an N-terminal domain that recognizes a 5′-T, a central domain comprising variable 33–35 amino acids repeats (each repeat will recognize and bind only a single DNA basepair) and a C-terminal sequence from the naturally occurring TALE proteins. The DNA-binding preference of each unit is determined by two critical amino acids at positions 12 and 13 known as ‘repeat variable di-residue’ (RVD), and each RVD is a pair of amino acids that determine the base to which it binds, for example, NI (Asn Ile) to A, NG (Asn Gly) to T, HD (His Asp) to C and NN (Asn Asn)/NK (Asn Lys) to G. TALEN-induced DSBs can be repaired by either NHEJ or homology-directed repair, and has been shown to generate targeted mutations in many organisms [68–74] including zebrafish [75, 76]. Different groups have published methods describing the rapid assembly of custom TALE repeat arrays using publicly available reagents [70, 75–79]. Most of these methods are based on Golden Gate cloning, which uses a type IIS restriction endonuclease that cuts outside of the recognition site and generates a nonpalindromic 4 bp overhang that cannot be re-digested after ligation [70, 77–79]. All these methods are multi-step and thus labor intensive and not amenable to high-throughput production. Two high-throughput and cost-effective methods called fast ligation-based automatable solid-phase high-throughput (FLASH) [80] and iterative capped assembly (ICA) [81] were developed that use solid-phase magnetic beads to digest and ligate multiple TALE repeat units in an iterative process. A method called unit assembly, developed by Huang et al. used single unit vectors as starting materials with each vector corresponding to the single nucleotide target it recognized. TALE repeats are then synthesized using standard restriction digestions and ligations [75]. Another method of TALEN assembly based on restriction enzyme and ligation (REAL) was published which does not require polymerase chain reaction (PCR) amplification or multi-fragment ligations [82]. More recently, a ligation-independent cloning (LIC) technique for generating TALEs has been published, this method could synthesize up to 600 TALENs in a single day [83]. In zebrafish, TALENs have been shown to generate lesions at targeted loci in both somatic [76] and germline cells [75]. Huang et al. designed and synthesized TALENs pairs using unit assembly for tnikb and dip2a genes and reported the germline transmission rate of ∼10–33%, showing the efficiency close to that observed with ZFNs. To test the possible off-target effects of TALENs, the potential off-target sites for tnikb were calculated and nine off-target sites tested did not show any off-target activity [75]. Bedell et al. developed an enhanced TALEN toolkit using the GoldyTALEN modified scaffold and its efficacy was tested in zebrafish. Authors generated nucleases targeting ponzr1, crhr1, moesina, ppp1cab and cdh5 showing the efficient cleavage at each locus with the efficiency as high as 100% at some loci. GoldyTALENs are capable of inducing mutations at high rate in both somatic and germline tissues. The authors further showed biallelic conversion in somatic tissue and phenotypes were observed in injected embryos similar to seen using morpholino-mediated knockdowns [84]. More recently, a simplified 15-RVD GoldyTALEN structure was tested in zebrafish, 14 TALEN pairs were tested and all of them were able to create lesions at somatic efficacy up to 86% and germ line transmission rate from 18% to 100%. It has also been shown that large inheritable deletions up to 18 kb can be generated by co-injecting two GoldyTALEN pairs [85]. Two recent studies used a pair of TALENs to generate large deletions and inversions in zebrafish [86].
TALENs can also be used to ‘knock-in’ large DNA fragments in zebrafish. GoldyTALENs were used to precisely modify sequences or introduce sequences (knock-in) using single-stranded DNA oligonucleotides and a homology-directed repair mechanism [84]. Zu et al. report the use of TALENs for knocking in large DNA fragments by homologous recombination using double-stranded donor DNA and the modified gene was able to transmit through the germ line [87].
Cade et al. showed both the original homodimeric form of Fok1, and an obligate heterodimeric mutant form can be used in TALENs and both were shown to generate heritable targeted mutations. 7/10 homodimeric TALEN pairs induced targeted mutations with frequencies ranging from 2% to 76% in zebrafish. The authors also found that heterodimeric TALENs have higher mutation frequencies (up to 100%) than homodimeric TALENs [88].
A comparison between TALENs and CoDA ZFNs to induce somatic mutation in zebrafish shows higher success rate for TALENs (20%-76.8%) than CoDA ZFNs (1.1–2.3%) [89]. Chen et al. performed large-scale comparison of ZFNs and TALENs mutagenicity in zebrafish. They found 85% of TALEN pairs tested induced somatic indels at rates >1% compared with 25% of ZFNs tested. They also found TALENs generated significantly more mutations than ZFNs [90]. TALENs and ZFNs pairs were also used to generate deletions up to 69 kb at rates of 1–5% germline transmission. Larger deletions of up to 5.5 Mb were also generated in somatic cells at lower frequency (0.7%) [91]. There are online tools such as Mojo Hand [92], TAL effector-nucleotide targeter (TALE-NT) [93] idTALE [94] and ZiFit [95] to assist researchers in identifying TAL-binding sites for use in TALEN design.
CRISPR/Cas system-mediated mutagenesis
Recent studies reported the use of the CRISPR and Cas system from bacteria and archaea as a simple genome-editing tool [96–98]. The CRISPR/CRISPR-associated (Cas) system protects bacteria and archaea from invading viruses and foreign DNA. The CRISPR/Cas system relies on the use of CRISPR RNA (crRNA) and transactivating crRNA (trcrRNA), the crRNAs anneal to trans-activating RNA (tracrRNAs) that efficiently guide a nuclease (cas9) to its target site for site-specific cleavage and silencing of foreign DNA. A single guide RNA chimera harboring the crRNA:trcrRNA has also been shown to guide the cas9 endonuclease to cleave DNA in a sequence-specific manner in vitro [98], in mammalian cells [96, 97], zebrafish [99, 100], bacteria [101], yeast [102] Drosophila [103, 104] and C. elegans [105].
Three independent studies successfully demonstrated that programmable guide RNAs (gRNAs) could direct cas9 endonuclease to induce in vivo gene targeting in zebrafish [99, 100, 106]. The system relies on a human codon-optimized cas9 endonuclease containing a nuclear localization signal and a gRNA that targets N20-NGG sequences [also called protospacer adjacent motif (PAM)]. The gRNA can be synthesized using two complementary oligonucleotides that are subcloned into an expression vector, then the in vitro transcribed gRNA is co-injected with cas9 endonuclease mRNA into zebrafish embryos. Huang et al. successfully targeted 8 out of 10 genes in zebrafish by NHEJ in embryos with variable efficiency of 24–59%, they further showed these mutations are heritable to the next generation and can also be used in conjunction with single-stranded oligonucleotides to introduce precise nucleotide alterations [107]. The CRIPSR/Cas was able to induce biallelic conversion of the etsrp and gata5 genes in somatic cells, with detectable phenotypes in the injected zebrafish embryos [99]. The CRIPSR/Cas was also used to introduce a mutant loxP (mloxP) site via homology-directed repair (HDR) [99].
This versatile CRIPSR/Cas system has been shown to be effective in multiplex gene targeting, a unique feature that has not been shown with TALENs or ZFNs. The simultaneous disruption of five genes and eight alleles (Tet1,Tet2, Tet3 Sry and Uty) was demonstrated in mouse embryonic stem cells. At the same time, biallelic Tet1 and Tet2 double mutants were also generated by co-injecting cas9 mRNA and the gRNA, with an efficiency of 80% [108]. Given the large number of duplicated genes in zebrafish, multiplexed CRIPSR/Cas system could be a powerful tool, allowing researchers to simultaneously inactivate both genes arising from a gene duplication event. Jao et al. developed an improved CRISPR/Cas system using a zebrafish codon-optimized Cas9 protein and were able to simultaneously target four loci (tyr, golden, mitfa and ddx19) with visible phenotypes in the injected embryos (i.e. bi-allelic conversion) showing its efficacy in targeting multiple genes at once [106]. This new, exciting and fast-developing technology is quick, inexpensive and can target any N20-NGG sequence (PAM) in the genome and unlike ZFNs and TALENs, the gRNA can be assembled by a simple and straightforward technique and without significant initial investments, making the CRIPSR/cas technique amenable for both focused, individual and large-scale, genome-wide mutagenesis efforts.
While CRISPR/cas is a powerful and simple tool for targeted mutagenesis, a recent study showed that it can cause off-target mutations in human cells, and can target the sites that are significantly different from the targeted DNA site. It has been shown that mismatches up to five bases were able to cleave an off-target DNA sequence and the rate of off-target cleavage was as high as for the primary target sequence and in some cases even higher than the targeted cleavage. Work is surely to continue to increase the specificity of the technologies which will make this technology a more robust and reliable tool [109].
High-throughput phenotyping approaches
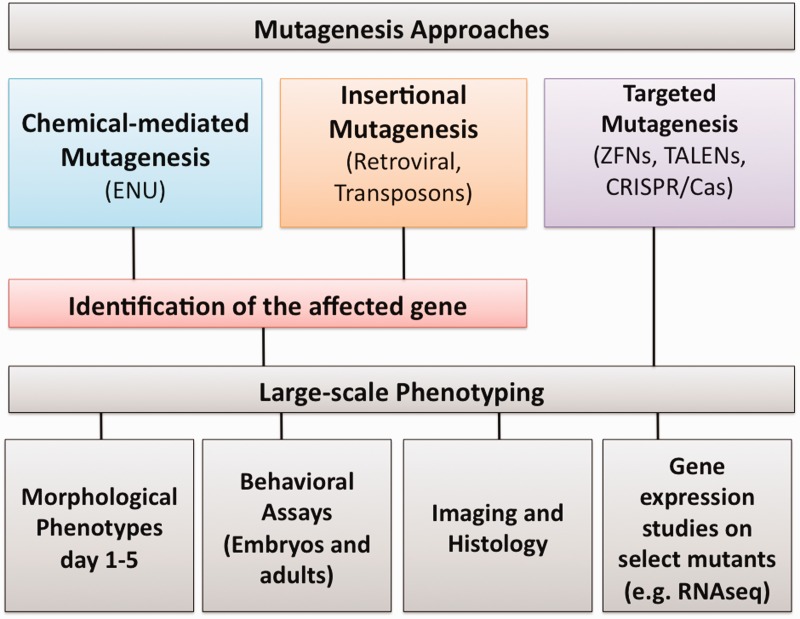
Despite the increasing number of mutagenesis projects and resources (Table1), and a completed genome, the understanding of zebrafish gene function in vivo is limited to a few hundred tested genes. The high degree of gene conservation from fish to humans, high fecundity, transparent embryos and small size make zebrafish an ideal candidate for high-throughput phenotyping. The two largest scale mutagenesis projects together generated more than 15 000 mutants but only a subset of these mutants are currently linked to phenotypes. In March 2009, a meeting of zebrafish researchers from around the world was held at the Sanger Wellcome Trust Institute to form a community opinion on the utility of large-scale systematic phenotyping to generate a zebrafish ‘phenome’ similar to the efforts undertaken by the International Mouse Phenotyping Consortium (http://www.mousephenotype.org/), with a larger, follow-up meeting a year later in Bethesda. A number of assays for detailed phenotyping such as morphology at different stages of development (up to 5 dpf), histology, gene expression profiling and different behavioral assays on both larvae and adults were discussed and prioritized. The final phenotyping strategy is dependent on the mutagenic strategy chosen, but the basic approaches and phenotyping categories are summarized in Figure 1. The forward genetic screens in zebrafish estimated that only ≈10% of zebrafish genes show embryonic phenotypes in the early stage of the development, which means the majority of the genes will either have adult phenotype or no obvious morphological phenotypes by traditional visual inspection. However, if image processing could be automated, the externally fertilized and optically clear embryos present an opportunity for systematic, high-resolution measurements. Recently, an automated imaging approach based on a simple fluidic system was developed that could perform automated optical projection tomography of zebrafish embryos [110]. This system can screen an entire embryo at up to micrometer resolution within seconds and could detect subtle morphological alterations that would not be observed with traditional visual screening, alterations subtle enough that they would not necessarily cause a lethal phenotype.
Figure 1:
High-throughput phenotyping strategies in zebrafish.
Recent efforts have demonstrated an efficient phenotyping pipeline that leverages multiple mutations identified in a mutagenized zebrafish background [12]. By tracking multiple mutations simultaneously in an embryonic cross using fluorescent probes and PCR, it enables significantly higher throughput for basic phenotyping. Used in conjunction with automated imaging and image analysis, ‘first pass’ phenotyping of an entire vertebrate genome becomes a very obtainable goal with tremendous possibilities for both understanding basic biology and for determining the causative mutations in human diseases.
The various protein-, gene- and enhancer-traps can be used for studying tissue-specific phenotypes by first selecting for genes with specific expression in the target tissue. Ding et al. [111] performed such a screen using protein-trapped transgenic lines in which mutated genes expressed in the heart were investigated for heart-related phenotypes such as cardiac hypertrophy. Zebrafish is being used in studying electrophysiology of the heart and tools such as optical mapping can be used for this purpose [112]. Conversely, if the efficacy of CRISPR/Cas or TALEN systems are improved to the point where phenotypes can routinely be assayed directly in the injected embryos, transgenic lines with specific structures fluorescently labeled (e.g. the heart), could be screened for sensitive alterations in structure or morphology by injection of the transgenic fish with mutating nucleases.
Assays are also being developed using systematic histology and micoCT technologies to image older larval or adult tissues. A prototype for automated segmentation and classification of larval eye and gut images has also been developed [113–115]. All of these techniques have the ability to view tissues at high resolution and can convey important information not readily detectable by more cursory phenotyping, but throughput remains a challenge on a genome-wide scale.
Zebrafish has also been used effectively to study different behavioral phenotypes [116] and several behavioral assays are currently being used or developed. A high-throughput method for neurophenotyping of adult zebrafish behavior has been developed which generates temporal and spatial three-dimensional (3D) reconstruction of various fish activities following anxiogenic and anxiolytic responses. A program called ZebraZoom has been developed to automatically track fish movement over a time period and up to 56 larval movements can be tracked simultaneously [117]. Similarly, a novel imaging approach using dark field illumination has the potential for tracking larval movements in a more ‘natural’ setting [118]. As the technique does not rely on bright field illumination, lower frame rates are possible allowing for longer tracking times.
CONCLUSION
Zebrafish is at an important moment in its evolution as a model organism for human disease and general biology. The mutagenic technologies have advanced to the point that it is only a matter of time before all zebrafish genes will be mutated (Table 2). A modestly priced, concerted effort among several labs throughout the world could leverage this mutant zebrafish resource to phenotype all the genes in a vertebrate genome. Having measurable phenotypes for all zebrafish genes would have a profound effect on the field of human genetics and be a major contributor to our understanding of signals identified in GWAS mapping. In addition, a ready-to-use resource of zebrafish mutations will accelerate the pace of discovery for zebrafish researchers looking to quickly test the functioning of their genes of interest.
Key Points.
Completion of the zebrafish reference genome sequence makes systematic mutagenesis possible.
Mutagenesis technologies for making zebrafish gene knockouts are rapidly advancing with targeted and conditional alleles, now possible for the first time.
With mutagenesis of the entire zebrafish genome likely to be done in the next 2 years, an attainable goal would be to characterize the phenotypes of each gene knockout.
Table 2:
Comparison of different mutagenesis approaches
| Mutagenesis | Pros | Cons | Potential mutagenic throughput |
|---|---|---|---|
| Retroviral insertional mutagenesis |
|
|
High throughput. |
| Transposon-based approaches |
|
|
High throughput. |
| ENU Mutagenesis | Highly efficient. |
|
Very high throughput. |
| ZFNs | Targeted. |
|
Low throughput. |
| TALENs | Targeted, every 1–3 bp in zebrasfish genome can be targeted. |
|
Low–moderate throughput. |
| CRISPR-Cas |
|
|
Moderate–high throughput. |
Acknowledgements
Opinions expressed are those of the authors and do not necessarily represent the views of the United States Federal Government.
Biography
Drs Varshney and Burgess are employees of the National Human Genome Research Institute, in the Genome Technology Branch, and are currently working on large-scale mutagenesis in zebrafish.
FUNDING
This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
References
- 1.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Do R, Kathiresan S, Abecasis GR. Exome sequencing and complex disease: practical aspects of rare variant association studies. Hum Mol Genet. 2012;21:R1–9. doi: 10.1093/hmg/dds387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Streisinger G, Walker C, Dower N, et al. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–6. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 4.Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 7.Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 8.Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 9.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–43. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–67. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 11.Howe DG, Bradford YM, Conlin T, et al. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013;41:D854–60. doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kettleborough RN, Busch-Nentwich EM, Harvey SA, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–7. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark KJ, Argue DP, Petzold AM, et al. zfishbook: connecting you to a world of zebrafish revertible mutants. Nucleic Acids Res. 2012;40:D907–11. doi: 10.1093/nar/gkr957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varshney GK, Lu J, Gildea DE, et al. A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res. 2013;23:727–35. doi: 10.1101/gr.151464.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solnica-Krezel L, Schier AF, Driever W. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics. 1994;136:1401–20. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driever W, Solnica-Krezel L, Schier AF, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Zon LI. The zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol. 2011;104:287–309. doi: 10.1016/B978-0-12-374814-0.00016-1. [DOI] [PubMed] [Google Scholar]
- 18.Voz ML, Coppieters W, Manfroid I, et al. Fast homozygosity mapping and identification of a zebrafish ENU-induced mutation by whole-genome sequencing. PLoS One. 2012;7:e34671. doi: 10.1371/journal.pone.0034671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowen ME, Henke K, Siegfried KR, et al. Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics. 2012;190:1017–24. doi: 10.1534/genetics.111.136069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obholzer N, Swinburne IA, Schwab E, et al. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development. 2012;139:4280–90. doi: 10.1242/dev.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill JT, Demarest BL, Bisgrove BW, et al. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013;23:687–97. doi: 10.1101/gr.146936.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller AC, Obholzer ND, Shah AN, et al. RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res. 2013;23:679–86. doi: 10.1101/gr.147322.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilchrist EJ, O'Neil NJ, Rose AM, et al. TILLING is an effective reverse genetics technique for Caenorhabditis elegans. BMC Genomics. 2006;7:262. doi: 10.1186/1471-2164-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taniguchi Y, Takeda S, Furutani-Seiki M, et al. Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol. 2006;7:R116. doi: 10.1186/gb-2006-7-12-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Till BJ, Reynolds SH, Weil C, et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004;4:12. doi: 10.1186/1471-2229-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Till BJ, Zerr T, Bowers E, et al. High-throughput discovery of rare human nucleotide polymorphisms by Ecotilling. Nucleic Acids Res. 2006;34:e99. doi: 10.1093/nar/gkl479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCallum CM, Comai L, Greene EA, et al. Targeted screening for induced mutations. Nature Biotechnol. 2000;18:455–7. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- 28.Draper BW, McCallum CM, Stout JL, et al. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- 29.Wienholds E, Schulte-Merker S, Walderich B, et al. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- 30.Wienholds E, van Eeden F, Kosters M, et al. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–7. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sood R, English MA, Jones M, et al. Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods. 2006;39:220–7. doi: 10.1016/j.ymeth.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Amsterdam A, Varshney GK, Burgess SM. Retroviral-mediated Insertional Mutagenesis in Zebrafish. Methods Cell Biol. 2011;104:59–82. doi: 10.1016/B978-0-12-374814-0.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Stuart L, Ohsumi TK, et al. Transposon activation mutagenesis as a screening tool for identifying resistance to cancer therapeutics. BMC Cancer. 2013;13:93. doi: 10.1186/1471-2407-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S, Gaiano N, Culp P, et al. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science. 1994;265:666–9. doi: 10.1126/science.8036514. [DOI] [PubMed] [Google Scholar]
- 35.Gaiano N, Amsterdam A, Kawakami K, et al. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383:829–32. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 36.Golling G, Amsterdam A, Sun Z, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–40. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 37.Amsterdam A, Burgess S, Golling G, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–24. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jao LE, Burgess SM. Production of pseudotyped retrovirus and the generation of proviral transgenic zebrafish. Methods Mol Biol. 2009;546:13–30. doi: 10.1007/978-1-60327-977-2_2. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Jao LE, Zheng N, et al. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad USA. 2007;104:12428–33. doi: 10.1073/pnas.0705502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varshney GK, Huang H, Zhang S, et al. The Zebrafish Insertion Collection (ZInC): a web based, searchable collection of zebrafish mutations generated by DNA insertion. Nucleic Acids Res. 2013;41:D861–4. doi: 10.1093/nar/gks946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–8. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidson AE, Balciunas D, Mohn D, et al. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Emelyanov A, Gao Y, Naqvi NI, et al. Trans-kingdom transposition of the maize dissociation element. Genetics. 2006;174:1095–104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trinh le A, Hochgreb T, Graham M, et al. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 2011;25:2306–20. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark KJ, Balciunas D, Pogoda HM, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8:506–15. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess SM. The changing conditions of zebrafish mutants. Proc Natl Acad Sci USA. 2012;109:15082–3. doi: 10.1073/pnas.1212832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni TT, Lu J, Zhu M, et al. Conditional control of gene function by an invertible gene trap in zebrafish. Proc Natl Acad Sci USA. 2012;109:15389–94. doi: 10.1073/pnas.1206131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jungke P, Hans S, Brand M. The Zebrafish CreZoo: An Easy-to-Handle Database for Novel CreER-Driver Lines. Zebrafish. 2013;10:259–63. doi: 10.1089/zeb.2012.0834. [DOI] [PubMed] [Google Scholar]
- 49.Huang P, Xu L, Liang W, et al. Genomic deletion induced by Tol2 transposon excision in zebrafish. Nucleic Acids Res. 2013;41:e36. doi: 10.1093/nar/gks1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci. 2009;66:1039–56. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bozas A, Beumer KJ, Trautman JK, et al. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics. 2009;182:641–51. doi: 10.1534/genetics.109.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beumer KJ, Trautman JK, Bozas A, et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:19821–6. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bibikova M, Golic M, Golic KG, et al. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–75. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morton J, Davis MW, Jorgensen EM, et al. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA. 2006;103:16370–5. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santiago Y, Chan E, Liu PQ, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–14. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 57.Doyon Y, McCammon JM, Miller JC, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–8. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng X, Noyes MB, Zhu LJ, et al. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foley JE, Yeh JR, Maeder ML, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta A, Meng X, Zhu LJ, et al. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–92. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta A, Christensen RG, Rayla AL, et al. An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods. 2012;9:588–90. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu C, Smith T, McNulty J, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–64. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xing L, Hoshijima K, Grunwald DJ, et al. Zebrafish foxP2 zinc finger nuclease mutant has normal axon pathfinding. PLoS One. 2012;7:e43968. doi: 10.1371/journal.pone.0043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayes M, Naito M, Daulat A, et al. Ptk7 promotes non-canonical Wnt/PCP-mediated morphogenesis and inhibits Wnt/beta-catenin-dependent cell fate decisions during vertebrate development. Development. 2013;140:1807–18. doi: 10.1242/dev.090183. [DOI] [PubMed] [Google Scholar]
- 65.Liu YJ, Fan HB, Jin Y, et al. Cannabinoid Receptor 2 Suppresses Leukocyte Inflammatory Migration by Modulating the JNK/c-Jun/Alox5 Pathway. J Biol Chem. 2013;288:13551–62. doi: 10.1074/jbc.M113.453811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taibi A, Mandavawala KP, Noel J, et al. Zebrafish churchill regulates developmental gene expression and cell migration. Dev Dyn. 2013;242:614–21. doi: 10.1002/dvdy.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Bebber F, Hruscha A, Willem M, et al. Loss of Bace2 in zebrafish affects melanocyte migration and is distinct from Bace1 knock out phenotypes. J Neurochem. 2013 doi: 10.1111/jnc.12198. doi:10.1111/jnc.12198. [DOI] [PubMed] [Google Scholar]
- 68.Liu J, Li C, Yu Z, et al. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics. 2012;39:209–15. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 69.Li T, Huang S, Zhao X, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–25. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tesson L, Usal C, Menoret S, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–96. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 73.Wood AJ, Lo TW, Zeitler B, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lei Y, Guo X, Liu Y, et al. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci USA. 2012;109:17484–9. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang P, Xiao A, Zhou M, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 76.Sander JD, Cade L, Khayter C, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature Biotechnol. 2011;29:697–8. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang F, Cong L, Lodato S, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–53. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morbitzer R, Elsaesser J, Hausner J, et al. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–9. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weber E, Gruetzner R, Werner S, et al. Assembly of designer TAL effectors by Golden Gate cloning. PLoS One. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reyon D, Tsai SQ, Khayter C, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–5. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Briggs AW, Rios X, Chari R, et al. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reyon D, Khayter C, Regan MR, et al. Engineering designer transcription activator-like effector nucleases (TALENs) by REAL or REAL-Fast assembly. Curr Protoc Mol Biol. 2012 doi: 10.1002/0471142727.mb1215s100. Chapter 12:Unit 12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmid-Burgk JL, Schmidt T, Kaiser V, et al. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–8. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma AC, Lee HB, Clark KJ, et al. High efficiency in vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS One. 2013;8:e65259. doi: 10.1371/journal.pone.0065259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao A, Wang Z, Hu Y, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013;41:e141. doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zu Y, Tong X, Wang Z, et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods. 2013;10:329–31. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 88.Cade L, Reyon D, Hwang WY, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–10. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moore FE, Reyon D, Sander JD, et al. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen S, Oikonomou G, Chiu CN, et al. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41:2769–78. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gupta A, Hall VL, Kok FO, et al. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013;23:1008–17. doi: 10.1101/gr.154070.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neff KL, Argue DP, Ma AC, et al. Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinformatics. 2013;14:1. doi: 10.1186/1471-2105-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doyle EL, Booher NJ, Standage DS, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–22. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li L, Piatek MJ, Atef A, et al. Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification. Plant Mol Biol. 2012;78:407–16. doi: 10.1007/s11103-012-9875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sander JD, Maeder ML, Reyon D, et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–8. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang N, Sun C, Gao L, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–72. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang W, Bikard D, Cox D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–9. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DiCarlo JE, Norville JE, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–43. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gratz SJ, Cummings AM, Nguyen JN, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–35. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bassett AR, Tibbit C, Ponting CP, et al. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–8. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Friedland AE, Tzur YB, Esvelt KM, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–3. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–9. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hwang WY, Fu Y, Reyon D, et al. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang H, Yang H, Shivalila CS, et al. One-Step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pardo-Martin C, Allalou A, Medina J, et al. High-throughput hyperdimensional vertebrate phenotyping. Nat Commun. 2013;4:1467. doi: 10.1038/ncomms2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ding Y, Liu W, Deng Y, et al. Trapping cardiac recessive mutants via expression-based insertional mutagenesis screening. Circ Res. 2013;112:606–17. doi: 10.1161/CIRCRESAHA.112.300603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sabeh MK, Kekhia H, Macrae CA. Optical mapping in the developing zebrafish heart. Pediatr Cardiol. 2012;33:916–22. doi: 10.1007/s00246-012-0300-1. [DOI] [PubMed] [Google Scholar]
- 113.Canada BA, Thomas GK, Cheng KC, et al. SHIRAZ: an automated histology image annotation system for zebrafish phenomics. Multimed Tools Appl. 2011;51:401–40. doi: 10.1007/s11042-010-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cheng KC, Xin X, Clark DP, et al. Whole-animal imaging, gene function, and the Zebrafish Phenome Project. Current Opinion Genet Dev. 2011;21:620–9. doi: 10.1016/j.gde.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sabaliauskas NA, Foutz CA, Mest JR, et al. High-throughput zebrafish histology. Methods. 2006;39:246–54. doi: 10.1016/j.ymeth.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 116.Norton W, Bally-Cuif L. Adult zebrafish as a model organism for behavioural genetics. BMC Neuroscience. 2010;11:90. doi: 10.1186/1471-2202-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mirat O, Sternberg JR, Severi KE, et al. ZebraZoom: an automated program for high-throughput behavioral analysis and categorization. Front Neural Circuits. 2013;7:107. doi: 10.3389/fncir.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martineau PR, Mourrain P. Tracking zebrafish larvae in group - Status and perspectives. Methods. 2013;62:292–303. doi: 10.1016/j.ymeth.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]