Abstract
G protein–coupled receptor 84 (GPR84) is a 7-transmembrane protein expressed on myeloid cells that can bind to medium-chain free fatty acids in vitro. Here, we report the discovery of a 2-bp frameshift deletion in the second exon of the Gpr84 gene in several classical mouse inbred strains. This deletion generates a premature stop codon predicted to result in a truncated protein lacking the transmembrane domains 4-7. We sequenced Gpr84 exon 2 from 58 strains representing different groups in the mouse family tree and found that 14 strains are homozygous for the deletion. Some of these strains are DBA/1J, DBA/2J, FVB/NJ, LG/J, MRL/MpJ, NOD/LtJ, and SJL/J. However, the deletion was not found in any of the wild-derived inbred strains analyzed. Haplotype analysis suggested that the deletion originates from a unique mutation event that occurred more than 100 years ago, preceding the development of the first inbred strain (DBA), from a Mus musculus domesticus source. As GPR84 ostensibly plays a role in the biology of myeloid cells, it could be relevant 1) to consider the existence of this Gpr84 nonsense mutation in several mouse strains when choosing a mouse model to study immune processes and 2) to consider reevaluating data obtained using such strains.
Key words: spontaneous mutations, G protein–coupled receptors, inbred mice, mouse models
Due to several sequencing projects involving mouse inbred strains, mutations with no obvious phenotype (referred as “quiet”) are getting progressively discovered (Stevens et al. 2007). These are spontaneous mutations that become fixed in certain inbred colonies (strains or substrains), a phenomenon known as genetic drift. Here, we report the characterization of a nonsense mutation in the Gpr84 (G protein–coupled receptor 84) gene (Yousefi et al. 2001) that is present in 14 classical inbred strains. GPR84, a receptor for medium-chain free fatty acids (FFAs), is highly expressed in monocytes/macrophages and granulocytes and has been suggested to play a role in inflammatory processes, particularly linking fatty acid metabolism to immunological regulation (Wang et al. 2006). Given that some of the strains carrying this putative loss-of-function mutation are widely used as mouse models in various immunological disorders, including autoimmune diabetes (NOD/LtJ), experimental autoimmune encephalomyelitis (EAE; SJL/J), and collagen-induced arthritis (DBA/1J), data obtained from theses strains should be reevaluated in this new context.
Materials and Methods
Mutation Analysis
Mouse genomic DNA from inbred strains was obtained from the Jackson Laboratory DNA Resource service (Bar Harbor, ME) and from the RASF-S Genetic Services at MD Anderson (Smithville, TX). One pair of primers (Gpr84-FOR: gcaagttctcataccatctccc; Gpr84-REV: AGCCCAAGCACAAAGTAGATG) was used to amplify a 619-bp fragment of Gpr84 exon 2. PCR reactions contained 50ng of genomic DNA, 5 pmol of each primer, 1.5 units of AmpliTaq DNA polymerase, 1.5 mmol/L MgCl2, 0.2 mmol/L of each deoxyribonucleotide, and 2 μL of 10× PCR buffer (Applied Biosystems, Foster City, CA). Samples were subjected to 1 cycle of denaturation (95 °C, 60 s), followed by 35 cycles of denaturation (95 °C, 35 s), annealing (58 °C, 45 s), and extension (68 °C, 45 s). The PCR products were purified with QIAquick PCR Purification columns (Qiagen, Valencia, CA) and sequenced using the ABI-PRISMTM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin Elmer, Waltham, MA). Sequencing was carried out using an ABI 3130XL DNA sequencer (Perkin Elmer).
Results
A Frameshift Deletion in the Gpr84 Gene Is Shared by Several Classical Inbred Strains
As part of a positional cloning effort aimed to identify the gene responsible for a spontaneous mutation on distal chromosome 15, we sequenced all exons (1 and 2) of Gpr84 using genomic DNA from one homozygous mutant mouse. A 2-bp frameshift deletion was found in exon 2 at position 103 308 576bp (NCBI Build 37; Figure 1), resulting in a premature stop codon and a predicted truncated protein (124 aa in the mutant form vs. 396 aa in the wild type). This truncated protein, if expressed, would encode for a product lacking from the 4th to the 7th transmembrane domains. Since Gpr84 −/− mice were already described as having no obvious phenotype other than in vitro differences in T cell response (Venkataraman and Kuo 2005), we then hypothesized that this nonsense mutation could be a phenotypically “quiet” mutation (Stevens et al. 2007) that happened to be present in the genetic background of our mutant mice (i.e., our positional cloning project). Knowing that this spontaneous mutation arose in outbred albino mice, we started sequencing a few inbred strains originally developed from Swiss outbred mice. To this end, we sequenced Gpr84 exon 2 from FVB/NJ, NOD/LtJ, SENCARB/Pt, SJL/J, and SWR/J and found out that FVB/NJ, NOD/LtJ, and SJL/J mice were homozygous for the 2-bp deletion, supporting the quiet mutation hypothesis and the idea that Gpr84 is a loss-of-function tolerant gene. In order to expand our analysis, representative strains from the 7 groups described in the family tree developed by Petkov et al. (2004), as well as a few strains not present in this tree, were selected for further sequencing. Out of a total of 58 strains analyzed, we found that 14 were homozygous for the deletion. These strains are BDP/J, DBA/1J, DBA/2J, I/LnJ, P/J, and SM/J (group 6); FVB/NJ, NOD/LtJ, NOR/LtJ, and SJL/J (group 2); LG/J, MRL/MpJ, and PL/J (group 1); and the newly developed SKHIN/Sprd inbred hairless strain, derived from SKH1 outbred mice (Perez et al. 2012; Table 1 and Figure 2). We also found the deletion in the homozygous state in several individuals from the SKH1 (Crl:SKH1-hr) outbred hairless stock, and segregating in ICR and CD-1 outbred stocks. Interestingly, the deletion was not observed in wild-derived inbred strains like LEWES/EiJ, PERA/EiJ, TIRANO/EiJ, WSB/EiJ, and ZALENDE/EiJ (Mus musculus domesticus); MAI/Pas and PWK/PhJ (Mus musculus musculus); CAST/EiJ (Mus musculus castaneus); MSM/Ms (Mus musculus molossinus); and SEG/Pas (Mus spretus), suggesting that the Gpr84 mutation was present in the ancestors of the classical inbred strains. Unfortunately, the lack of specificity of the currently available commercial antibodies did not allow us to determine if the mutation alters protein expression by western blot analyses.
Figure 1.
A homozygous frameshift deletion in exon 2 of Gpr84. The mouse Gpr84 gene is located in chromosome 15: 103 308 236–103 310 438bp (ideogram, cytogenetic band F3) and contains 2 exons (Mouse Genome Build 37). The chromatograms are showing wild-type (WT) and mutant partial sequences from exon 2 obtained from C57BL/6J and DBA/2J, respectively. The 2-bp deletion is located at 103 308 576bp and generates a premature stop codon and a predicted truncated protein lacking transmembrane domains 4–7. Abbreviations: C = cytosine; T = thymine.
Table 1.
Inbred strains with Gpr84 exon 2 sequenced for this study
| 129P1/ReJ | AKR/J | C57BL/10J | FVB/NJ | NOD/LtJ | SEG/Pas | TALLYHO/J |
| 129P2/OlaHsd | ALS/LtJ | C57BLKS/J | I/LnJ | NON/LtJ | SENCARA/PtJ | TIRANO/EiJ |
| 129P3/J | BALB/cJ | C57BR/cdJ | KK/HIJ | NOR/LtJ | SENCARB/PtJ | WSB/EiJ |
| 129S1/SvImJ | BALB/cByJ | C57L/J | LEWES/EiJ | NZB/BINJ | SENCARC/PtJ | ZALENDE/EiJ |
| 129S6/SvEvBrdJ | BDP/J | C58/J | LG/J | NZO/HILtJ | SJL/J | |
| 129S7/SvEvTac | BUB/BnJ | CAST/EiJ | MAI/Pas | P/J | SKHIN/Sprd a | |
| 129T2/SvEmsJ | C3H/HeJ | CBA/J | MRL/MpJ | PERA/EiJ | SM/J | |
| 129X1/SvJ | C3H/HeNCrl | DBA/1J | MSM/Ms | PL/J | SSIN/Sprd | |
| A/J | C57BL/6J | DBA/2J | NMRIb | PWK/PhJ | SWR/J |
Fifty-eight strains were analyzed and 14 (highlighted) found to be homozygous for the 2-bp deletion in Gpr84.
For strains ending with the “J” lab code, DNA was acquired from The Jackson Laboratory Mouse DNA Resource (http://www.jax.org/dnares/index.html); for strains ending with the “Tac” Lab Code, DNA was acquired from Taconic (http://www.taconic.com); for strains ending with the “Hsd” Lab Code, DNA was acquired from Harlan (http://www.harlan.com/); for strains ending with the “Crl” Lab Code, DNA was acquired from Charles River (http://www.criver.com/); for strains ending with the “Pas” Lab Code, DNA was a gift from Dr. Jean Jaubert at Unité de Génétique Fonctionnelle de la Souris, Institut Pasteur, Paris, France; and for strains ending with the “Sprd” Lab Code, DNA was acquired from the Genetic Services at MD Anderson Cancer Center, Smithville, Texas.
MSM/Ms DNA was a gift from Dr. Tomoji Mashimo at Institute of Laboratory Animals, Kyoto University, Japan and NMRI inbred DNA was a gift from Dr. Ernst-Martin Füchtbauer at the Department of Molecular Biology and Genetics, Aarhus, Denmark. All DNAs acquired or prepared between 2009 and 2012.
aInbred by F.B. from outbred SKH1 mice (Perez et al. 2012).
bInbred by Jörg Schmidt from outbred NMRI mice.
Figure 2.
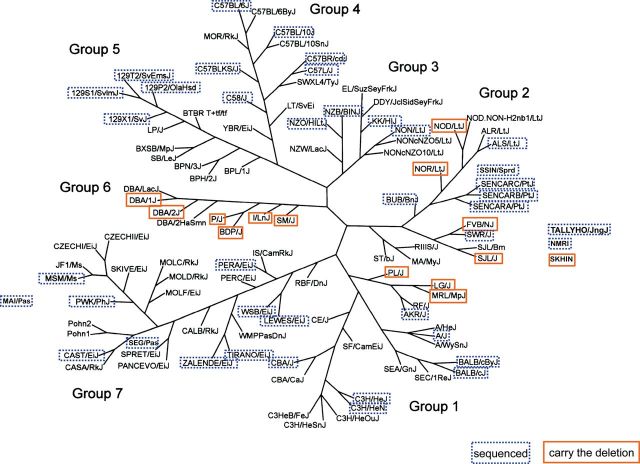
Distribution of the Gpr84 deletion among classical inbred strains. The strains carrying the 2-bp deletion are clustered in groups 1 (Bagg albino derivatives), 2 (Swiss mice), and 6 (Little’s DBA and related strains) of the mouse family tree proposed by Petkov et al. (2004). Out of the 58 strains analyzed, 14 were homozygous for the deletion, including BDP/J, DBA/1J, DBA/2J, I/LnJ, P/J, and SM/J (group 6); FVB/NJ, NOD/LtJ, NOR/LtJ, and SJL/J (group 2); and LG/J, MRL/MpJ, and PL/J (group 1). Strains not included in the original tree were added close to related strains but not connected with lines. Since DBA/1 and DBA/2 are the oldest of all inbred strains, it is expected that they passed the mutation onto the related strains of group 6. The presence of the mutation outside the DBA group (i.e., albino strains in groups 1 and 2) could be explained by unknown contributions from DBA mice (or other members of group 6) to the ancestors of the Swiss mice or, alternatively, by DBA strains and Swiss mice inheriting the mutation independently from the same ancestral mouse. The clustering analysis used by Petkov and collaborators for constructing the parsimony tree of mouse strains was performed by the neighbor-joining method (Petkov et al. 2004).
SNP Haplotype Analyses Suggest That the Gpr84 Deletion Is Identical by Descent
To trace the origin of the mutation, we carried out a Single Nucleotide Polymorphism (SNP) haplotype analyses of the distal region of chromosome 15 where Gpr84 is located using the Mouse Phenome Database (MPD) SNP variation resource and the Wellcome Trust Sanger Institute (Mouse Genomes Project) SNP and Indel Query. Comparisons were first made between 10 strains carrying the Gpr84 deletion from which SNP genotype data were available at MPD (Broad2 strain list; Daly and Wade 2012). No polymorphic SNPs were retrieved between DBA/2J and DBA/1J, FVB/NJ, I/LnJ, LG/J, MRL/MpJ (~75% LG genome), NOD/LtJ, NOR/LtJ, SJL/J, and SM/J in a ~300-kb region flanking Gpr84 (Figure 3) suggesting a common haplotype. Further analysis showed that this common haplotype block is ~1.7Mb in size, from position 101.8 to 103.5Mb. Conversely, other classical inbred strains not carrying the deletion do not share this SNP block, even those closely situated in the same group (e.g., FVB/NJ and SWR/J; Figure 3). These data suggest that the Gpr84 deletion is identical by descent (a unique mutation event) and is, at least, 100 years old, preceding the development of DBA in 1909. Further analysis of the SNP variation among the available strains in the Broad2 set allowed us to determine that the 10 strains carrying the deletion also share an almost identical ~300-kb block with some noncarrier M. m. domesticus–derived strains like PERA/EiJ and PERC/EiJ (founders trapped in Peru) and ZALERNE/EiJ (founders trapped in Switzerland). A smaller haplotype block (~32kb) flanking the Gpr84 gene was also shared among the strains carrying the deletion and noncarrier strains A/J, C3H/HeJ, CBA/J, and WSB/EiJ (M. m. domesticus; Figure 3). These data suggest that the Gpr84 mutation appeared on a chromosome 15 derived from a M. m. domesticus source. This is not surprising considering that classical inbred strains are largely derived from the M. m. domesticus subspecies (Frazer et al. 2007; Yang et al. 2007).
Figure 3.
Distribution of SNPs flanking Gpr84 on distal chromosome 15. We queried the MPD for SNPs flanking Gpr84 and compared strains carrying the deletion with selected strains carrying the wild-type allele using the Broad2 strain list. The results of the query showed that the 10 strains carrying the deletion (in the Broad2 set) share a conserved SNP haplotype block spanning ~1.7Mb (101.8–103.5Mb). This figure shows only the SNPs in the distal part of chromosome 15 flanking Gpr84 (~300kb). Analyzing the SNP variation among the available strains, we could determine that the strains carrying the deletion also share an almost identical block with some Mus musculus domesticus–derived strains, particularly ZALERNE/EiJ (solid line box), and to a lesser extent with PERA/EiJ and PERC/EiJ. Notice that closely related strains like FVB/N (deletion allele) and SWR/J (wild-type allele), both directly derived from Swiss mice, do not share this haplotype block. A smaller common haplotype (~32kb) flanking Gpr84 is also shared with A/J, C3H/HeJ, and CBA/J classical strains, and WSB/EiJ (M. m. domesticus, dotted line box). The haplotype data suggest that the Gpr84 mutation is identical by descent and occurred on a chromosome 15 derived from a M. m. domesticus source. Data retrieved from http://phenome.jax.org/db/q?rtn=snp/ret1 (February 2013).
Taking into account that DBA/1 and DBA/2 substrains (also considered different strains), the oldest of all inbred strains, are homozygous for the deletion, it is conceivable that one of these substrains passed the mutation onto the other strains of group 6, all nonalbino strains with known DBA ancestors (Figure 2). However, it is more difficult to explain the presence of the mutation outside the DBA group (i.e., albino strains in groups 1 and 2), particularly because these strains have outbred Swiss ancestors, not known to be derived from any of the classical inbred strains (Chia et al. 2005). Even so, we cannot completely rule out contributions from early DBA mice (undocumented crosses) to the dealer mice that originated the stock of André de Coulon in Switzerland, direct ancestors of the Swiss mice (Chia et al. 2005). In fact, a dendogram of relatedness among 12 classical inbred strains based on 8.2 million SNPs shows that FVB/NJ, NOD/LtJ, and DBA/2J cluster together (Frazer et al. 2007). Alternatively, DBA strains and Swiss mice could have inherited the mutation independently from the same ancestral mouse, as classical laboratory strains are known to be derived from a few fancy mice with limited haplotype diversity (Yang et al. 2011).
Discussion
GPR84 is a transmembrane receptor that can bind to medium-chain FFAs in vitro (Wang et al. 2006) but whose endogenous ligand is still unknown. Expression of GPR84 is mainly seen in myeloid cells (e.g., monocytes, macrophages, microglia, and neutrophils) and is upregulated by immune stimuli (Yousefi et al. 2001; Wittenberger et al. 2001; Venkataraman and Kuo 2005; Bédard et al. 2007; Bouchard et al. 2007; Lattin et al. 2008; Ichimura et al. 2009; Oh and Lagakos 2011). Very recently, it was shown that Gpr84 mRNA expression is upregulated in adipose tissues from C57BL/6J males fed with a high-fat diet. The authors propose that Gpr84 mRNA expression is enhanced in adipocytes upon stimulation of TNF-α, released from macrophages infiltrating the adipose tissue (Nagasaki et al. 2012). In vitro studies using Gpr84-deficient T cells suggested that GPR84 plays a role in regulating early interleukin-4 gene expression in activated T cells (Venkataraman and Kuo 2005). Additionally, it was proposed that medium-chain FFAs could mediate Th1/Th2 balance upon direct interaction with GPR84 receptors, a potential link between metabolic disorders and autoimmune diseases (Oh and Lagakos 2011).
Some of the strains carrying the deletion in Gpr84 are widely used as mouse models. For example, DBA/2 mice are used in multiple research areas, including immunology (Cardona et al. 2003), neurobiology (Reichstein et al. 2007), skin cancer (Angel and DiGiovanni 1999), and glaucoma (Frankel 2009). On the other hand, the closely related DBA/1 strain is a classical model of collagen-induced arthritis (Jung et al. 2009). FVB/N is a popular inbred strain used for transgenic experiments by pronuclear microinjection and also as a model for skin cancer, due to their high susceptibility to chemically induced papillomas and squamous cell carcinomas (Hennings et al. 1993). SJL/J mice are used as a model for EAE and are known to have elevated levels of circulating T cells (Lindsey 1996). While NOD/Lt is a classical model of autoimmune diabetes (Driver et al. 2011), it is also the only strain in the Collaborative Cross (Threadgill and Churchill 2012) carrying the Gpr84 deletion. LG/J and MRL/MpJ are also models for autoimmune disease, although with late onset compared with mutant MRL/MpJ-Fas lpr/Fas lpr mice (Shirai and Klinman 1994; Clark et al. 2008). Interestingly, MRL/MpJ and LG/J display accelerated wound healing relative to other strains (Heber-Katz et al. 2004; Blankenhorn et al. 2009). At the same time, LG/J and SM/J mice are often compared for quantitative trait locus (QTL) analysis for body weight (Kenney-Hunt et al. 2006), obesity-, and diabetes-related traits (Cheverud et al. 2004).
In addition, a number of QTLs have been described in distal chromosome 15. For example, Pgia9 (proteoglycan-induced arthritis 9) was identified using a model for rheumatoid arthritis involving resistant DBA/2 and susceptible BALB/c strains (Glant et al. 2008). The collagen-induced arthritis susceptibility locus Cia37 was identified using crosses involving C57BL/10 and DBA/1J (Ahlqvist et al. 2007). The super-healing QTL Heal4 was identified using the MRL/MpJ-Fas lpr and C57BL/6 strains (Blankenhorn et al. 2003, 2009). Although not associated with any deleterious phenotype, we cannot rule out the involvement of the Gpr84 deletion in the QTLs described on distal chromosome 15 and neither potential epistatic interactions with other genes.
Conclusion
In conclusion, our finding of a putative loss-of-function mutation in the Gpr84 gene in several inbred strains should be taken into consideration when reviewing data obtained using these strains or when choosing to work with them. Future investigations should be conducted to determine if the mutation has any functional consequence in carrier strains versus those with the wild-type allele. In this regard, it would be beneficial to create a database inventorying these type of phenotypically quiet mutations in inbred strains, since it is known that modifier genes can have no obvious phenotype on their own but still change the phenotypic outcome of an independent locus (Hamilton and Yu 2012). What is more, these mutations can be linked with genes of interest (targeted loci or QTLs) as (undesirably) “passenger mutations” (Specht and Schoepfer 2001; (Kenneth et al. 2012; Mattapallil et al. 2012).
Funding
Cancer Center Support Grant (CCSG; NCI P30 CA16672).
Acknowledgments
We acknowledge the Molecular Biology Facility Core in Smithville, Texas for the DNA sequencing. This study made use of the Genetic Services supported by the NCI Cancer Center Support Grant to MD Anderson Cancer Center. We are grateful to Ernst-Martin Füchtbauer for the inbred NMRI DNA. Special thanks to Rick Wood for his support, to Joe Angel for critical reading of the manuscript, and to Petko Petkov for his useful comments on the results.
References
- Ahlqvist E, Bockermann R, Holmdahl R. 2007. Fragmentation of two quantitative trait loci controlling collagen-induced arthritis reveals a new set of interacting subloci. J Immunol. 178:3084–3090 [DOI] [PubMed] [Google Scholar]
- Angel JM, DiGiovanni J. 1999. Genetics of skin tumor promotion. Prog Exp Tumor Res. 35:143–157 [DOI] [PubMed] [Google Scholar]
- Bédard A, Tremblay P, Chernomoretz A, Vallières L. 2007. Identification of genes preferentially expressed by microglia and upregulated during cuprizone-induced inflammation. Glia. 55:777–789 [DOI] [PubMed] [Google Scholar]
- Blankenhorn EP, Bryan G, Kossenkov AV, Clark LD, Zhang XM, Chang C, Horng W, Pletscher LS, Cheverud JM, Showe LC, et al. 2009. Genetic loci that regulate healing and regeneration in LG/J and SM/J mice. Mamm Genome. 20:720–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn EP, Troutman S, Clark LD, Zhang XM, Chen P, Heber-Katz E. 2003. Sexually dimorphic genes regulate healing and regeneration in MRL mice. Mamm Genome. 14:250–260 [DOI] [PubMed] [Google Scholar]
- Bouchard C, Pagé J, Bédard A, Tremblay P, Vallières L. 2007. G protein-coupled receptor 84, a microglia-associated protein expressed in neuroinflammatory conditions. Glia. 55:790–800 [DOI] [PubMed] [Google Scholar]
- Cardona PJ, Gordillo S, Díaz J, Tapia G, Amat I, Pallarés A, Vilaplana C, Ariza A, Ausina V. 2003. Widespread bronchogenic dissemination makes DBA/2 mice more susceptible than C57BL/6 mice to experimental aerosol infection with Mycobacterium tuberculosis. Infect Immun. 71:5845–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Ehrich TH, Hrbek T, Kenney JP, Pletscher LS, Semenkovich CF. 2004. Quantitative trait loci for obesity- and diabetes-related traits and their dietary responses to high-fat feeding in LGXSM recombinant inbred mouse strains. Diabetes. 53:3328–3336 [DOI] [PubMed] [Google Scholar]
- Chia R, Achilli F, Festing MF, Fisher EM. 2005. The origins and uses of mouse outbred stocks. Nat Genet. 37:1181–1186 [DOI] [PubMed] [Google Scholar]
- Clark AG, Mackin KM, Foster MH. 2008. Tracking differential gene expression in MRL/MpJ versus C57BL/6 anergic B cells: Molecular markers of autoimmunity. Biomark Insights. 3:335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M, Wade CM. 2012. SNP data in support of a haplotype map, 132,000+ locations for 89 inbred strains of mice [Bar Harbor (ME)]: MPD:Broad2. Mouse Phenome Database web site, The Jackson Laboratory; [Google Scholar]
- Driver JP, Serreze DV, Chen YG. 2011. Mouse models for the study of autoimmune type 1 diabetes: a NOD to similarities and differences to human disease. Semin Immunopathol. 33:67–87 [DOI] [PubMed] [Google Scholar]
- Frankel WN. 2009. Genetics of complex neurological disease: challenges and opportunities for modeling epilepsy in mice and rats. Trends Genet. 25:361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 448:1050–1053 [DOI] [PubMed] [Google Scholar]
- Glant TT, Szántó S, Vegvari A, Szabo Z, Kis-Toth K, Mikecz K, Adarichev VA. 2008. Two loci on chromosome 15 control experimentally induced arthritis through the differential regulation of IL-6 and lymphocyte proliferation. J Immunol. 181:1307–1314 [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Yu BD. 2012. Modifier genes and the plasticity of genetic networks in mice. PLoS Genet. 8:e1002644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber-Katz E, Chen P, Clark L, Zhang XM, Troutman S, Blankenhorn EP. 2004. Regeneration in MRL mice: further genetic loci controlling the ear hole closure trait using MRL and M.m. Castaneus mice. Wound Repair Regen. 12:384–392 [DOI] [PubMed] [Google Scholar]
- Hennings H, Glick AB, Lowry DT, Krsmanovic LS, Sly LM, Yuspa SH. 1993. FVB/N mice: an inbred strain sensitive to the chemical induction of squamous cell carcinomas in the skin. Carcinogenesis. 14:2353–2358 [DOI] [PubMed] [Google Scholar]
- Ichimura A, Hirasawa A, Hara T, Tsujimoto G. 2009. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat. 89:82–88 [DOI] [PubMed] [Google Scholar]
- Jung S, Shin HS, Hong C, Lee H, Park YK, Shin JH, Hong S, Lee GR, Park SH. 2009. Natural killer T cells promote collagen-induced arthritis in DBA/1 mice. Biochem Biophys Res Commun. 390:399–403 [DOI] [PubMed] [Google Scholar]
- Kenneth NS, Younger JM, Hughes ED, Marcotte D, Barker PA, Saunders TL, Duckett CS. 2012. An inactivating caspase 11 passenger mutation originating from the 129 murine strain in mice targeted for c-IAP1. Biochem J. 443:355–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney-Hunt JP, Vaughn TT, Pletscher LS, Peripato A, Routman E, Cothran K, Durand D, Norgard E, Perel C, Cheverud JM. 2006. Quantitative trait loci for body size components in mice. Mamm Genome. 17(6):526–537 [DOI] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, et al. 2008. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 4:5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey JW. 1996. Characteristics of initial and reinduced experimental autoimmune encephalomyelitis. Immunogenetics. 44:292–297 [DOI] [PubMed] [Google Scholar]
- Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR. 2012. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 53:2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Kondo T, Fuchigami M, Hashimoto H, Sugimura Y, Ozaki N, Arima H, Ota A, Oiso Y, Hamada Y. 2012. Inflammatory changes in adipose tissue enhance expression of GPR84, a medium-chain fatty acid receptor: TNFα enhances GPR84 expression in adipocytes. FEBS Lett. 586:368–372 [DOI] [PubMed] [Google Scholar]
- Oh DY, Lagakos WS. 2011. The role of G-protein-coupled receptors in mediating the effect of fatty acids on inflammation and insulin sensitivity. Curr Opin Clin Nutr Metab Care. 14:322–327 [DOI] [PubMed] [Google Scholar]
- Perez C, Parker-Thornburg J, Mikulec C, Kusewitt DF, Fischer SM, Digiovanni J, Conti CJ, Benavides F. 2012. SKHIN/Sprd, a new genetically defined inbred hairless mouse strain for UV-induced skin carcinogenesis studies. Exp Dermatol. 21:217–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, et al. 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14:1806–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichstein D, Ren L, Filippopoulos T, Mittag T, Danias J. 2007. Apoptotic retinal ganglion cell death in the DBA/2 mouse model of glaucoma. Exp Eye Res. 84:13–21 [DOI] [PubMed] [Google Scholar]
- Shirai A, Klinman DM. 1994. The genetic basis of autoimmune disease in MRL-lpr/lpr mice. Int Rev Immunol. 11:179–192 [DOI] [PubMed] [Google Scholar]
- Specht CG, Schoepfer R. 2001. Deletion of the alpha-synuclein locus in a subpopulation of C57BL/6J inbred mice. BMC Neurosci. 2:11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JC, Banks GT, Festing MF, Fisher EM. 2007. Quiet mutations in inbred strains of mice. Trends Mol Med. 13:512–519 [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. 2012. Ten years of the Collaborative Cross. Genetics. 190:291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman C, Kuo F. 2005. The G-protein coupled receptor, GPR84 regulates IL-4 production by T lymphocytes in response to CD3 cross-linking. Immunol Lett. 101:144–153 [DOI] [PubMed] [Google Scholar]
- Wang J, Wu X, Simonavicius N, Tian H, Ling L. 2006. Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J Biol Chem. 281:34457–34464 [DOI] [PubMed] [Google Scholar]
- Wittenberger T, Schaller HC, Hellebrand S. 2001. An expressed sequence tag (EST) data mining strategy succeeding in the discovery of new G-protein coupled receptors. J Mol Biol. 307:799–813 [DOI] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. 2007. On the subspecific origin of the laboratory mouse. Nat Genet. 39:1100–1107 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, et al. 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 43:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Cooper PR, Potter SL, Mueck B, Jarai G. 2001. Cloning and expression analysis of a novel G-protein-coupled receptor selectively expressed on granulocytes. J Leukoc Biol. 69:1045–1052 [PubMed] [Google Scholar]