Abstract
Group II introns ai1 and ai2 of the Saccharomyces cerevisiae mitochondrial COXI gene encode proteins having a dual function (maturase and reverse transcriptase) and are mobile genetic elements. By construction of adequate donor genomes, we demonstrate that each of them is self-sufficient and practises homing in the absence of homing-type endonucleases encoded by either group I introns or the ENS2 gene. Each of the S. cerevisiae group II self-mobile introns was tested for its ability to invade mitochondrial DNA (mtDNA) from two related Saccharomyces species. Surprisingly, only ai2 was observed to integrate into both genomes. The non-mobility of ai1 was clearly correlated with some polymorphic changes occurring in sequences flanking its insertion sites in the recipient mtDNAs. Importantly, studies of the behaviour of these introns in interspecific crosses demonstrate that flanking marker co-conversion accompanying group II intron homing is unidirectional and efficient only in the 3' to 5' direction towards the upstream exon. Thus, the polar co-conversion and dependence of the splicing proficiency of the intron reported previously by us are hallmarks of group II intron homing, which significantly distinguish it from the strictly DNA-based group I intron homing and strictly RNA-based group II intron transposition.
Full text
PDF

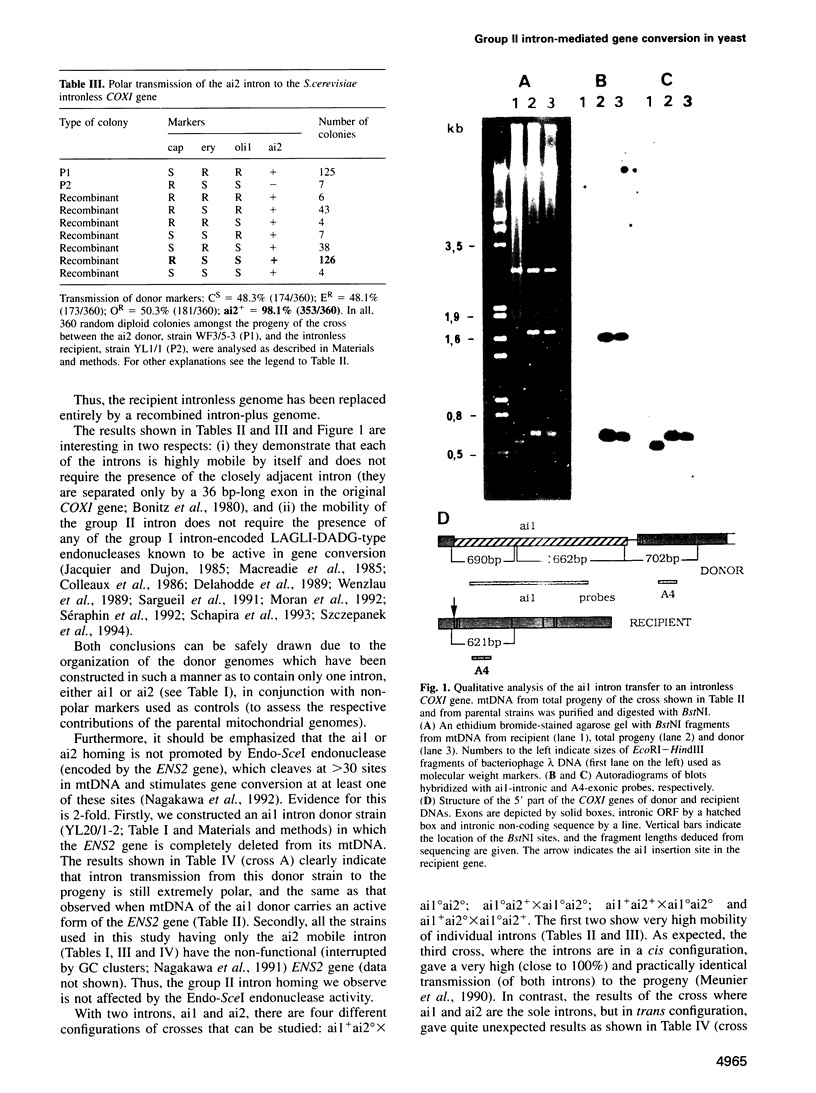
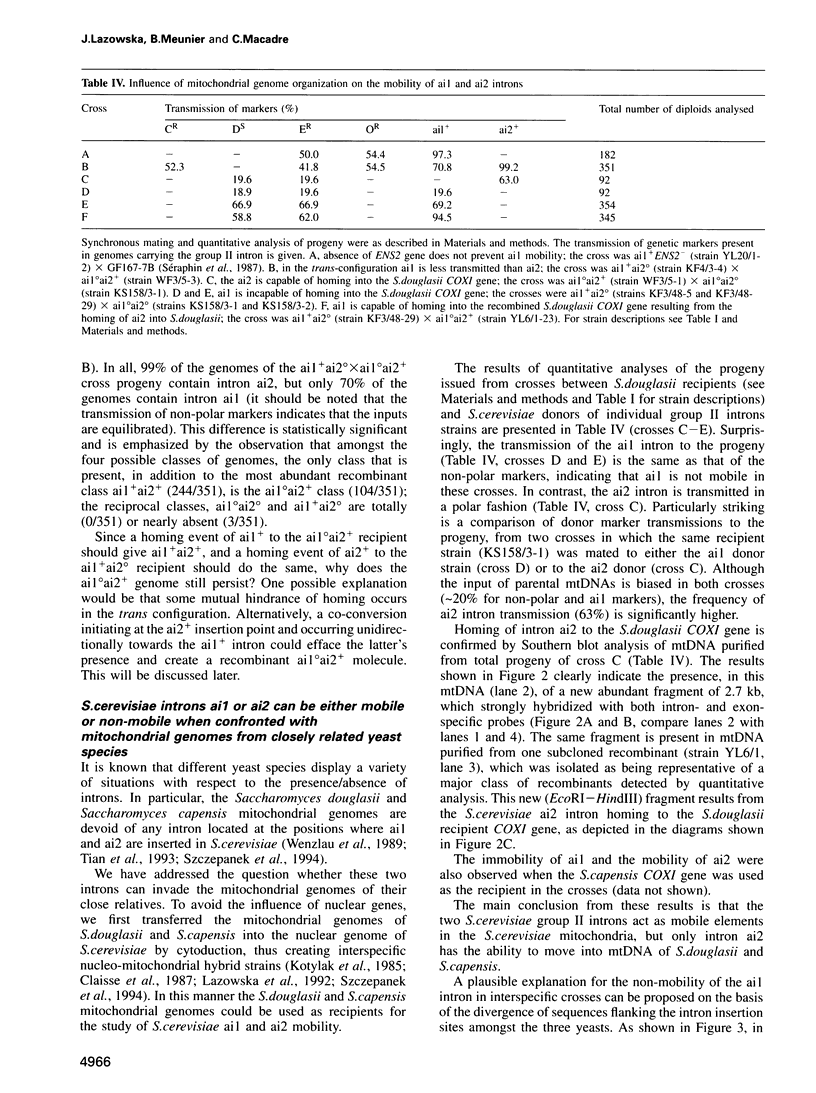





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belfort M. An expanding universe of introns. Science. 1993 Nov 12;262(5136):1009–1010. doi: 10.1126/science.7694364. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J Biol Chem. 1980 Dec 25;255(24):11927–11941. [PubMed] [Google Scholar]
- Carignani G., Netter P., Bergantino E., Robineau S. Expression of the mitochondrial split gene coding for cytochrome oxidase subunit I in S. cerevisiae: RNA splicing pathway. Curr Genet. 1986;11(1):55–63. doi: 10.1007/BF00389426. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Colleaux L., d'Auriol L., Betermier M., Cottarel G., Jacquier A., Galibert F., Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986 Feb 28;44(4):521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- Delahodde A., Goguel V., Becam A. M., Creusot F., Perea J., Banroques J., Jacq C. Site-specific DNA endonuclease and RNA maturase activities of two homologous intron-encoded proteins from yeast mitochondria. Cell. 1989 Feb 10;56(3):431–441. doi: 10.1016/0092-8674(89)90246-8. [DOI] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations--a review. Gene. 1989 Oct 15;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Dujon B., Slonimski P. P., Weill L. Mitochondrial genetics IX: A model for recombination and segregation of mitochondrial genomes in saccharomyces cerevisiae. Genetics. 1974 Sep;78(1):415–437. doi: 10.1093/genetics/78.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassbender S., Brühl K. H., Ciriacy M., Kück U. Reverse transcriptase activity of an intron encoded polypeptide. EMBO J. 1994 May 1;13(9):2075–2083. doi: 10.1002/j.1460-2075.1994.tb06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferat J. L., Le Gouar M., Michel F. Multiple group II self-splicing introns in mobile DNA from Escherichia coli. C R Acad Sci III. 1994 Feb;317(2):141–148. [PubMed] [Google Scholar]
- Jacquier A., Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985 Jun;41(2):383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- Jacquier A. Self-splicing group II and nuclear pre-mRNA introns: how similar are they? Trends Biochem Sci. 1990 Sep;15(9):351–354. doi: 10.1016/0968-0004(90)90075-m. [DOI] [PubMed] [Google Scholar]
- Kennell J. C., Moran J. V., Perlman P. S., Butow R. A., Lambowitz A. M. Reverse transcriptase activity associated with maturase-encoding group II introns in yeast mitochondria. Cell. 1993 Apr 9;73(1):133–146. doi: 10.1016/0092-8674(93)90166-n. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- Lazowska J., Szczepanek T., Macadre C., Dokova M. Two homologous mitochondrial introns from closely related Saccharomyces species differ by only a few amino acid replacements in their Open Reading Frames: one is mobile, the other is not. C R Acad Sci III. 1992;315(2):37–41. [PubMed] [Google Scholar]
- Levra-Juillet E., Boulet A., Séraphin B., Simon M., Faye G. Mitochondrial introns aI1 and/or aI2 are needed for the in vivo deletion of intervening sequences. Mol Gen Genet. 1989 May;217(1):168–171. doi: 10.1007/BF00330957. [DOI] [PubMed] [Google Scholar]
- Macreadie I. G., Scott R. M., Zinn A. R., Butow R. A. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985 Jun;41(2):395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns--a review. Gene. 1989 Oct 15;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Moran J. V., Wernette C. M., Mecklenburg K. L., Butow R. A., Perlman P. S. Intron 5 alpha of the COXI gene of yeast mitochondrial DNA is a mobile group I intron. Nucleic Acids Res. 1992 Aug 11;20(15):4069–4076. doi: 10.1093/nar/20.15.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M. W., Allmaier M., Eskes R., Schweyen R. J. Transposition of group II intron aI1 in yeast and invasion of mitochondrial genes at new locations. Nature. 1993 Nov 11;366(6451):174–176. doi: 10.1038/366174a0. [DOI] [PubMed] [Google Scholar]
- Nakagawa K., Morishima N., Shibata T. A maturase-like subunit of the sequence-specific endonuclease endo.SceI from yeast mitochondria. J Biol Chem. 1991 Jan 25;266(3):1977–1984. [PubMed] [Google Scholar]
- Nakagawa K., Morishima N., Shibata T. An endonuclease with multiple cutting sites, Endo.SceI, initiates genetic recombination at its cutting site in yeast mitochondria. EMBO J. 1992 Jul;11(7):2707–2715. doi: 10.1002/j.1460-2075.1992.tb05336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargueil B., Delahodde A., Hatat D., Tian G. L., Lazowska J., Jacq C. A new specific DNA endonuclease activity in yeast mitochondria. Mol Gen Genet. 1991 Feb;225(2):340–341. doi: 10.1007/BF00269867. [DOI] [PubMed] [Google Scholar]
- Schapira M., Desdouets C., Jacq C., Perea J. I-Sce III an intron-encoded DNA endonuclease from yeast mitochondria. Asymmetrical DNA binding properties and cleavage reaction. Nucleic Acids Res. 1993 Aug 11;21(16):3683–3689. doi: 10.1093/nar/21.16.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellem C. H., Lecellier G., Belcour L. Transposition of a group II intron. Nature. 1993 Nov 11;366(6451):176–178. doi: 10.1038/366176a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. On the origin of RNA splicing and introns. Cell. 1985 Sep;42(2):397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- Skelly P. J., Hardy C. M., Clark-Walker G. D. A mobile group II intron of a naturally occurring rearranged mitochondrial genome in Kluyveromyces lactis. Curr Genet. 1991 Jul;20(1-2):115–120. doi: 10.1007/BF00312773. [DOI] [PubMed] [Google Scholar]
- Szczepanek T., Macadre C., Meunier B., Lazowska J. Two homologous introns from related Saccharomyces species differ in their mobility. Gene. 1994 Feb 11;139(1):1–7. doi: 10.1016/0378-1119(94)90516-9. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Boulet A., Simon M., Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G. L., Macadre C., Kruszewska A., Szczesniak B., Ragnini A., Grisanti P., Rinaldi T., Palleschi C., Frontali L., Slonimski P. P. Incipient mitochondrial evolution in yeasts. I. The physical map and gene order of Saccharomyces douglasii mitochondrial DNA discloses a translocation of a segment of 15,000 base-pairs and the presence of new introns in comparison with Saccharomyces cerevisiae. J Mol Biol. 1991 Apr 20;218(4):735–746. doi: 10.1016/0022-2836(91)90262-5. [DOI] [PubMed] [Google Scholar]
- Tian G. L., Michel F., Macadre C., Lazowska J. Sequence of the mitochondrial gene encoding subunit I of cytochrome oxidase in Saccharomyces douglasii. Gene. 1993 Feb 28;124(2):153–163. doi: 10.1016/0378-1119(93)90389-k. [DOI] [PubMed] [Google Scholar]
- Wenzlau J. M., Saldanha R. J., Butow R. A., Perlman P. S. A latent intron-encoded maturase is also an endonuclease needed for intron mobility. Cell. 1989 Feb 10;56(3):421–430. doi: 10.1016/0092-8674(89)90245-6. [DOI] [PubMed] [Google Scholar]
- Xiong Y. E., Eickbush T. H. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988 Oct 21;55(2):235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Rago J. P., Netter P., Slonimski P. P. Pseudo-wild type revertants from inactive apocytochrome b mutants as a tool for the analysis of the structure/function relationships of the mitochondrial ubiquinol-cytochrome c reductase of Saccharomyces cerevisiae. J Biol Chem. 1990 Feb 25;265(6):3332–3339. [PubMed] [Google Scholar]