SUMMARY
Mammalian pyruvate dehydrogenase multi-enzyme complex (PDC) is a key metabolic assembly comprising a 60- meric pentagonal dodecahedral E2 core attached to which are 30 E1 heterotetramers and 6 E3 homodimers at maximal occupancy. Stable E3 integration is mediated by an accessory E3 binding protein (E3BP) located on each of the 12 E2 icosahedral faces. Here, we present evidence for a novel subunit organisation in which dihydrolipoamide dehydrogenase (E3) and E3BP form subcomplexes with a 1:2 stoichiometry implying the existence of a network of E3 ‘cross-bridges’ linking pairs of E3BPs across the surface of the E2 core assembly. We have also determined a low resolution structure for a truncated E3BP/E3 subcomplex using small angle xray scattering showing one of the E3BP lipoyl domains docked into the E3 active site. This new level of architectural complexity in mammalian PDC contrasts with the recently published crystal structure of human E3 complexed with its cognate subunit binding domain and provides important new insights into subunit organisation, its catalytic mechanism and regulation by the intrinsic PDC kinase.
INTRODUCTION
Many enzymes associate permanently or transiently into large, multifunctional complexes. The evolution of these multienzyme assemblies allows for substrate channeling and active site coupling: sequential catalytic reactions proceed through the rapid transfer of intermediates between individual components and without diffusion into the bulk medium. The mitochondrial 2-oxoacid dehydrogenase complexes are such a family of high molecular weight (4–10 MDa) multi-enzyme assemblies that serve as models for the study of protein-protein interactions and molecular recognition phenomena. They catalyze the oxidative decarboxylation of a vital group of 2-oxoacid intermediates in carbohydrate and amino acid metabolism. Principal members include the pyruvate dehydrogenase (PDC), 2-oxoglutarate dehydrogenase (OGDC) and branched-chain 2-oxoacid dehydrogenase complexes (BCOADC). PDC is responsible for overall glucose homeostasis in mammals since it controls the key committed step in carbohydrate utilization, namely the conversion of pyruvate to acetyl-CoA and NADH, linking glycolysis to the citric acid cycle. In recent years, genetic and physiological defects in PDC have been implicated in a wide range of diseases including various types of metabolic acidosis and mitochondrial myopathies (1); as major autoantigens in the autoimmune disease, primary biliary cirrhosis (2–4); in neurodegenerative disorders e.g. Alzheimer’s disease (5;6) and as primary targets for modification by environmental toxins and oxidative damage (7).
PDC consists of multiple copies of three different enzymes: pyruvate decarboxylase (E1), dihydrolipoamide acetyltransferase (E2) and dihydrolipoamide dehydrogenase (E3). A fourth protein, E3 binding protein (E3BP, protein X) is present in many eukaryotic complexes, but has no known enzymatic function. PDC is regulated by a tightly-bound kinase (PDK) and a looselybound phosphatase (PDP). These act in concert to exercise acute control of its activity state (phosphorylated E1 is inactive (8)), thereby precisely regulating the flux of 2-carbon units entering the citric acid cycle. PDK is a major drug target since activation of PDC by limiting its phosphorylation has considerable potential for alleviating symptoms associated with impaired carbohydrate metabolism in severe diabetes.
The icosahedral core of mammalian PDC comprises 60 E2 enzymes and 12 copies of an E2-related protein, E3BP (9). Basic trimeric units of E2 form the 20 vertices of the icosahedron with E3BP bound in each of the 12 faces (10). Both E2 and E3BP have a similar, modular domain structure: human (h) E2 and E3BP consist of two (E2) or one (E3BP) N-terminal lipoyl domain(s) (LD) of ≈ 80 amino acids. Each lipoyl domain carries a lipoic acid moiety covalently linked to a lysine residue situated at the tip of a type I β-turn. The lipoyl domain is followed by a subunit binding domain (SBD) of ≈ 35 residues and a C-terminal domain of ≈ 250 residues that is essential for core formation. In E2 the C-terminal domain is also the catalytic domain, while the active site is absent from E3BP rendering it incapable of catalyzing the acetyltransferase reaction. All domains are interconnected by Ala and Pro rich linker regions of ≈ 30 amino acids in length, that impart the flexibility necessary for the lipoyl domains to visit all three active sites during catalysis.
In eukaryotes the E2:E3BP core of PDC provides the structural and mechanistic framework for the tight but non-covalent association of the heterotetrameric E1 and homodimeric E3 enzymes. The stability and overall subunit organization of the complex are governed by specific and tight proteinprotein interactions between E2-SBD and E1, and E3BP-SBD and E3, respectively. However, PDCs from Gram positive bacteria also have icosahedral cores but lack E3BP. Consequently, E1 and E3 have to compete for overlapping binding sites on E2; interaction of E2 with either E1 or E3 prevents complex formation with the other (11). Interestingly, PDC-deficient patients who are totally lacking the E3BP subunit possess partial complex activity (10–20% of controls) (8), apparently because the SBD of E2 has retained a limited ability to mediate low affinity E3 binding.
In B. stearothermophilus PDC, E1 and E3 both bind to the SBD of E2 with 1:1 ratios (11–15). B. stearothermophilus E3/E2-SBD and E1/E2-SBD crystal structures show unequivocally that association of a second E2-SBD to E1 or E3 is impossible. In the case of E1 the binding site for E2-SBD is located across the two-fold axis (16) whereas the binding site on E3 is close to the two-fold axis of symmetry (17). Occupation of both binding sites on E3 would result in steric clashes in one of the loop regions (17). A similar arrangement in the crystal structure of human (h) E3 bound to E3BP-SBD has been reported very recently (18;19).
Intriguingly, however, the reported ratios for each of the constituent enzymes of mammalian PDC suggest the possibility of formation of 2:1 stoichiometric subcomplexes in this case. Thus, at maximal occupancy, 30 E1 molecules can associate with the 60-meric E2 core while 12 E3BP molecules associate with 6 E3 dimers (9).
Overexpressed hE3BP forms large aggregates and is unsuited to biophysical studies. Instead, we investigated the interaction of E3 with two different E3BP constructs consisting of the LD and/or SBD(s) (XDD and XSBD, respectively). A range of biochemical and biophysical approaches including native polyacrylamide gel electrophoresis (PAGE), analytical ultracentrifugation (AUC) and isothermal titration calorimetry (ITC) were used to show cross-bridge formation of hE3 with its cognate SBD in solution. A low-resolution structure for this subcomplex was obtained by small angle x-ray scattering (SAXS).
EXPERIMENTAL PROCEDURES
Plasmid construction
The construction of vectors used for the overexpression of N-terminally His-tagged human E3 and truncated versions of E3BP will be described elsewhere. Briefly, fulllength hE3 and residues 59–216 of mature hE3BP were cloned into pET14b (Novagen). Residues 166–230 of hE3BP were cloned into pET30a (Novagen). The E3BP constructs comprise either both the lipoyl and subunit binding domains or the subunit binding domain (SBD) on its own, and are referred to as the X di-domain (XDD) or X subunit binding domain (XSBD), respectively, throughout.
Recombinant proteins
E3, XSBD and XDD were overexpressed in Escherichia coli BL21 DE3 pLysS (Stratagene). Cells were grown in LB to an OD600 of 0.6–0.8 and induced with 1 mM IPTG for 3–5 h at 30°C. At induction, lipoic acid to a final concentration of 50 μg/ml was added to XDD cultures in order to maximize lipoylation. Cells were harvested by centrifugation at 15300 g for 10 minutes and lysed in 20 mM Hepes buffer, pH 8.0 supplemented with Complete® EDTA-free protease inhibitors (Roche) using the Bugbuster® kit (Novagen) according to manufacturer’s instructions. Insoluble cell debris was pelleted by centrifugation at 15300 g for 15 minutes.
All proteins were purified using immobilized Zn2+-chelate chromatography on a BioCAD 700E workstation (Applied Biosystems). A 20MC column was preloaded with 0.1 M ZnCl2 and equilibrated using binding buffer (20 mM Hepes, 100 mM NaCl, 10 mM imidazole, pH 8.0). Bound protein was washed with binding buffer and eluted in a linear 10–500 mM gradient of imidazole in 100 mM NaCl, 20 mM Hepes, pH 7.0. E3 was further purified by gel filtration on a Sephacryl S-300 column (Amersham) in 2 mM EDTA, 50 mM Tris-HCl, pH 7.5. DNA contamination of XSBD and XDD preparations was removed using HQ anion exchange chromatography. XSBD and XDD were buffer exchanged into 20 mM Tris-HCl, pH 7.5, loaded onto the column and following an extensive wash step eluted in a linear 0–1 M NaCl gradient. The final step of XSBD and XDD purification was gel filtration on Sephacryl S-100 (XSBD) and S-300 (XDD) columns (Amersham), respectively, in 2 mM EDTA, 50 mM Tris-HCl, pH 7.5.
XDD/E3 complex was reconstituted from the purified proteins at a stoichiometric ratio of 3:1 (XDD:E3) using a final size exclusion step on Sephacryl-S300 in 2 mM EDTA, 50 mM Tris-HCl buffer, pH 7.5 to remove unbound XDD. The protein concentration of XSBD and XDD was determined using their calculated extinction coefficients at 280 nm of 9800 M−1 and 11380 M−1, respectively. E3 was quantitated using FAD absorbance at 450 nm (ɛ = 22600 M−1).
Native polyacrylamide gel electrophoresis
Mixtures of XSBD:E3 at ratios from 4:1 to 1:3, and XDD:E3 from 10:1 to 1:3 were prepared, keeping the amount of E3 used constant at 160 pmol. The samples were incubated at 25°C for 15 minutes and 5–10 μg of protein were loaded onto Tris-glycine gels (5% stacking, 8% resolving gel) and subjected to native PAGE. Gels were stained with Coomassie Brilliant Blue R-250.
Analytical ultracentrifugation
Sedimentation velocity (SV) experiments were conducted at 4°C in a Beckman Coulter Optima XL-I analytical ultracentrifuge (Palo Alto, USA) using an An-60 Ti rotor. A rotor speed of 45000 rpm was selected and interference optics scans were collected every minute until sedimentation was complete. The concentration of E3 in the XDD:E3 mixtures was fixed at 4.9 μM. The XDD concentrations were adjusted to achieve XDD:E3 ratios of 4:1 to 1:3. Samples (380 μl) were loaded into 12 mm charcoal-filled epon double sector centerpieces. SV analysis of the purified XDD/E3 complex was conducted as described above, covering a concentration range of 6–44 μM. The data were analyzed using the program SEDFIT (20) which allows the subtraction of radial and time-independent noise, and direct modeling of the sedimentation boundary as a continuous distribution of discrete, non interacting species (c(s) analysis). Sedimentation coefficients were further evaluated using finite element analysis in SEDFIT which finds the sedimentation coefficients that best fit the Lamm equation. Buffer density and viscosity as well as protein partial specific volumes were calculated using the program SEDNTERP (21).
Sedimentation equilibrium (SE) analysis of XDD:E3 stoichiometric mixtures was conducted using the same samples analyzed by SV at rotor speeds of 8500, 12000 and 16000 rpm and the baseline determined by overspeeding to 49000 rpm. 20 replicate data sets were recorded using absorbance optics with a step size of 0.001 cm. Samples (30 μl) were loaded into 3 mm double sector charcoal-filled epon centerpieces. Attainment of equilibrium was ascertained with WinMATCH (www.biotech.uconn.edu/auf/) until no net movement of protein was observed in scans recorded three hours apart. SE data were analyzed using global analysis in the Beckman XL-A/XL-I software implemented in MicroCal ORIGIN. SE analysis of purified XDD/E3 complex was carried out as described above using rotor speeds of 10500, 15000 and 18000 rpm. The data were analyzed as before, but included fitting of the second virial coefficient, B. A starting estimate for B of XDD/E3 was calculated using the program COVOL (22) based on molecule dimensions and surface charge. The values of Mw,app and B at infinite dilution were determined by plotting 1/Mw,app against concentration (in g/ml), where is the inverse of the y intercept and Bo can be calculated from the slope.
Microcalorimetry
Isothermal titration calorimetry (ITC) measurements were carried out in a VP-ITC microcalorimeter (MicroCal Inc., MA, USA) at 25°C as described by Jung et al. (13). All proteins were dialyzed overnight in 25 mM Tris-HCl, pH 7.0. E3 at a concentration of 40.7 μM was injected in 10 μl aliquots into the reaction cell containing 6.2 μM XDD. Data were analyzed using non-linear regression in the MicroCal ORIGIN software package, assuming a simple binding model.
Small angle x-ray scattering
SAXS experiments were performed on beamline X33 of the EMBL outstation Hamburg at storage ring DORIS III of the Deutsches Elektronen Synchrotron (DESY). The scattering curves of purified XDD/E3 complex at concentrations of 1.5, 6.3, 9.6 and 22.6 mg/ml were recorded at 10°C using a sample-to-detector distance of 2.6 m, covering a range of momentum transfer of 0.01 < s < 0.42 Å−1 (s = 4π sinθ/λ), where 2 θ is the scattering angle and λ= 1.5 Å, the wavelength of the x-rays. The data were normalized to the incident beam and corrected for the detector response. The scattering of the buffer was subtracted and the difference curves scaled for concentration using the program PRIMUS (23). The final scattering curve was obtained by merging the low angle region of the low concentration curve with the high angle region of the high concentration curve in order to eliminate possible interparticle interaction effects in the low angle region of the high concentration data. The maximum dimension, Dmax and particle distance distribution function p(r) were obtained by indirect Fourier transformation using the program GNOM (24;25). The radius of gyration, Rg was obtained by employing both the Guinier approximation and GNOM. Ab initio shape reconstruction was done using the programs DAMMIN (26) and GASBOR (27;28) without imposing symmetry restraints. DAMMIN restores the structure as a collection of densely packed beads in a dummy atom model inside a search volume (a sphere with diameter Dmax). The program starts from a random configuration and uses a simulated annealing algorithm to explore the landscape of acceptable low resolution structures. GASBOR represents particles as collections of dummy residues (DR) using one DR per amino acid residue. The DRs are randomly distributed within the search volume and simulated annealing is used to obtain a chain-compatible spatial distribution. Ten GASBOR models were superimposed, averaged and the resulting pdb file used as search space for a final DAMMIN reconstruction.
Homology, hydrodynamic and rigid body modeling
A homology model of the human XDD/E3 complex was built using SWISSMODEL (29;30) based on the structures determined by protein crystallography or NMR for E3/E2-SBD from B. stearothermophilus (PDB 1EBD) (17), E3 from Saccharomyces cerevisiae (PDB 1JEH) (31), and human E2- LD (PDB 1FYC) (32). Models of XDD/E3 with lipoyl domains in different positions were generated (see results) and their sedimentation coefficients calculated using the program HYDROPRO (33;34). HYDROPRO calculates the hydrodynamic parameters of rigid particles from their atomic structures. For rigid body modeling with the program Rayuela (35), the complex was defined by five “domains”: E3, XSBD1, XSBD2, XLD1 and XLD2. Rayuela allows translation and rotation of each individual domain, calculates a SAXS curve as well as sedimentation coefficient for each structure and compares it to the experimental data. Using a simulated annealing algorithm, it determines the lowest energy conformation for each domain. The best model obtained with Rayuela, as judged by its fit to the experimental data, was used for superimposition onto the ab initio model using the program SITUS (36;37).
RESULTS
Native polyacrylamide gel electrophoresis
Initial binding studies were carried out by mixing either XSBD or XDD with E3 at different molar ratios and subjecting them to native PAGE. The addition of XSBD or XDD to E3 results in a noticeable bandshift which increases as more XSBD or XDD is added. At a stoichiometry of 2:1 all of the E3 present is shifted into the lower mobility band corresponding to the XSBD/E3 (Fig. 1A) or XDD/E3 (Fig. 1B) complex. At ratios lower than 2:1, excess E3 is seen (Fig. 1), while at stoichiometries higher than 2:1, excess XDD remains (Fig. 1B). At the pH used to conduct native PAGE (pH 8.8), XSBD moves towards the cathode and thereby out of the gel due to its high pI of 9.9. No intermediates in complex formation were observed.
Fig. 1. Native PAGE of different XSBD:E3 and XDD:E3 stoichiometries.
E3 was incubated with stoichiometric amounts of XSBD (A) or XDD (B). As the amount of XSBD/XDD increases a shift from the position corresponding to E3 to that for the complex (←) is observed, indicating formation of a tight complex. At stoichiometries above 2:1 free XDD is seen in addition to complex, whereas free E3 is observed at stoichiometries below 2:1. Free XSBD migrates towards the cathode and out of the gel due to its high pI (9.9).
Analytical ultracentrifugation
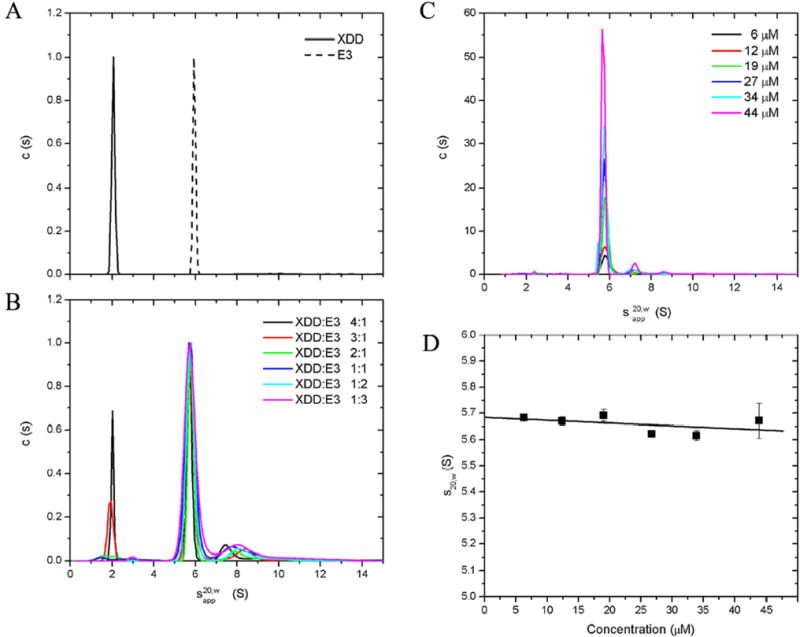
Sedimentation velocity (SV) experiments were conducted for uncomplexed E3 and XDD as well as for different XDD:E3 stoichiometries and the sedimentation profiles fitted using c(s) analysis in SEDFIT (20). The c(s) profiles of free XDD and E3 show single peaks with apparent sedimentation coefficients of 2.0 S and 5.9 S, respectively (Fig. 2A). Fig. 2B clearly shows the complete disappearance of the XDD peak at a stoichiometry of 2:1. The E3 and XDD/E3 peaks overlap completely, although the peak position is slightly reduced with an apparent sedimentation coefficient of 5.7 S. A globular protein of higher molecular weight is expected to sediment faster (and therefore exhibit a higher sedimentation coefficient) than a protein of lower molecular weight. Therefore, it can be inferred that the complex has an extended shape which offsets the gain in molecular weight with respect to free E3, and causes it to sediment at the same speed as E3. Similarly, the elongated shape and thus the large hydrodynamic radius of XDD/E3 can also be inferred from gel filtration chromatography (Supplemental material). Its apparent molecular weight, Mwapp, of 300 kDa on a standardized column is considerably higher than predicted from the amino acid sequence (144 kDa).
Fig. 2. Sedimentation velocity of XDD:E3 stoichiometric mixtures and purified XDD/E3 complex.
c(s) analysis of SV data for uncomplexed E3 and XDD (A) as well as different XDD:E3 stoichiometric mixtures (B). c(s) data analysis for a concentration range of purified XDD/E3 complex (C) and determination of the concentration independent sedimentation coefficient, for XDD/E3 (D).
The minor broad peak around 7.5–8 S observed at all ratios corresponds to aggregate species and is greatly reduced if the complex is purified to uniformity by gel filtration prior to AUC analysis (Fig. 2C). c(s) analysis of SV profiles from the purified complex also show that a single species is present in solution bar a small amount of aggregate that is present at high concentrations only, a result substantiated by native PAGE (results not shown). No free protein or intermediates are observed at any concentration, in accordance with the expected tight interaction between XDD and E3. Finite element analysis of SV data with a single-species model yielded the true sedimentation coefficients for XDD/E3 at all experimental concentrations. These were extrapolated to infinite dilution to give a sedimentation coefficient for XDD/E3 independent of concentration of = 5.68 ± 0.05 S (Fig. 2D).
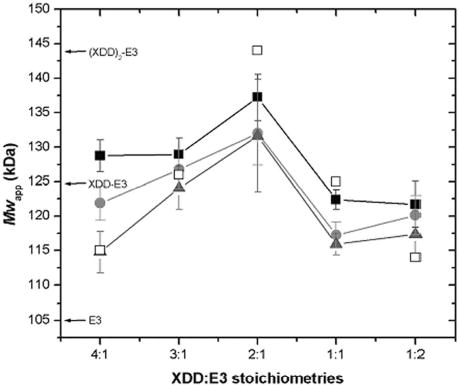
The apparent average whole cell molecular weight, Mw,app, was determined for each XDD:E3 stoichiometric mixture using sedimentation equilibrium (SE). The Mw,app peaks, as expected, at a stoichiometry of 2:1 (Fig. 3) when all protein present is associated into complex, owing to the tight interaction between XDD and E3. At all other molar ratios, free XDD or E3 is observed, thus depressing Mw,app. The value of Mw,app at a molar ratio of 2:1 (XDD:E3) of 137 kDa is lower than expected when compared to the calculated molecular weight of 144 kDa. This is attributable to the non-ideal behavior of the complex. SE data for XDD/E3 complex purified by gel filtration were also analyzed: agreement with the expected molecular weight was substantially improved when non-ideality arising from molecular shape and surface charge was accounted for by inclusion of the second virial coefficient (Bo) in the data analysis. The values obtained experimentally for Bo (7.2 x 10−4, 1.7 x 10−4 and 2.9 x 10−4 ml mol g−2 for rotor speeds 10500, 15000 and 18000 rpm, respectively) correspond very well to B calculated with COVOL (22) of 1.7 x 10−4 ml mol g−2.
Fig. 3. Sedimentation equilibrium of XDD:E3 stoichiometries.
Whole cell weight average molecular weights, Mw,app were determined for XDD:E3 mixtures by SE at 8500 (▪), 12000 (•) and 16000 rpm (▴). Mw,app calculated on the basis of a very strong interaction between XDD and E3 are also shown (□). The molecular weights of E3, XDD-E3 and (XDD)2-E3 calculated from the amino acid sequence are indicated.
When non-ideality is accounted for the molecular weight estimates for XDD/E3 are 151 kDa, 142 kDa and 141 kDa for rotor speeds 10500, 15000 and 18000 rpm, respectively (results not shown). These values compare very well to the calculated Mw of 144 kDa for a 2:1 complex of XDD:E3. When non-ideality was included in the data analysis for the 2:1 stoichiometric XDD:E3 mixture, Mw,app was increased to 147 kDa, 142 kDa and 141 kDa for rotor speeds 8500, 12000 and 16000 rpm, respectively, in excellent agreement with the results for the purified complex and the calculated molecular weight. For all other stoichiometric mixtures of XDD:E3, Mw,app decreased with increasing rotor speed which is indicative of the presence or formation of high molecular weight aggregate, as shown also by the SV results in Fig. 2B. Only for the 1:1 mixture was Mw,app consistently lower than the calculated molecular weight. This was due to the choice of fitting limits: the aggregate was steeply distributed and thus effectively excluded from the subsequent analysis, resulting in an underestimate of Mw,app. Because stoichiometric mixtures other than 2:1 contained more than one species (plus aggregate), the exponential describing the heavier component was disproportionately affected by the inevitable loss of data from the cell base, resulting in a corresponding reduction in Mw,app.
Isothermal titration calorimetry
The heats of interaction of XDD with E3 were determined using ITC. Complex formation is exothermic (i.e. negative peaks in the ITC output) (Fig. 4A). Differential thermal binding curves were obtained by integration of the data and analyzed using standard non-linear regression in order to obtain estimates of the binding stoichiometry, equilibrium association constant and enthalpy of binding (Fig. 4B). The data confirm the tight binding of XDD to E3 (Ka = 2.8 x 107 M−1) with a stoichiometry of 2:1. Binding is characterized by a large, favorable enthalpy change (ΔH = −12.1 kcal/mol) and a small negative entropy change (TΔS = −1.7 kcal/mol). This experiment was confirmed by similar ITC analysis of full-length E3BP:E3 interaction (data not shown) which shows that full-length E3BP binds E3 with similar affinity and identical stoichiometry.
Fig. 4. Isothermal titration calorimetry of XDD and E3.
(A) Raw data obtained from a series of 10 μl injections of E3 into XDD at 25°C. (B) Binding isotherms created by plotting the areas under the peaks in (A) against the molar ratio of E3 injected. The best fit shown was obtained by least-squares fitting using a simple binding model and gives a stoichiometry of 0.5 (E3:XDD) which is equivalent to 2:1 (XDD:E3).
Homology and hydrodynamic modeling
Homology modeling was used in combination with AUC and SAXS (see below) to gain more insight into the XDD/E3 structure. A model of the human XDD/E3 complex was built from homology models (generated with SWISSMODEL (29;30)) for E3, E3BP lipoyl and subunit binding domains. The crystal structure of hE3 was solved very recently (38) (PDB ID 1ZMC). Superimposition of these coordinates with those of the homology model gave a very low rms deviation of 0.6 Å: the two structures are almost identical, differing primarily in the NAD domain. The structure of the B. stearothermophilus E2- SBD/E3 complex (17) was used to position the human E3 and XSBD homology models with respect to each other. The second XSBD molecule was positioned exploiting the E3 two-fold symmetry axis. Several models with different positioning of the two lipoyl domains LD1 and LD2 were generated, while taking into account available biochemical and structural data. In an extended conformation, both LD1 and LD2 were located approximately 65 Å from the SBDs, in accordance with previous EM data (10;39;40) and overall PDC dimensions. The resulting model XDD/E3extn has a maximum dimension, Dmax of 227 Å. The lack of structural information for part of the linker region and the paucity of related sequences meant that it was not it was not possible to model this section of the linker using either homology or secondary structure modeling. A second complex model with both lipoyl domains docked in the E3 active sites and a Dmax of 134 Å (XDD/E3dock) was also generated. In the process Lys97 was positioned 14 Å from E3 active site residues Cys45 and Cys50, accommodating the lipoate cofactor linked to Lys97 (32), in agreement with data from mutation studies on E3 mechanism (41;42). Although other E3 residues involved in the binding of the lipoyl domain are not known, access to the E3 active site is only possible from one side of the molecule. The FAD and NAD+ cofactors otherwise obstruct the interaction between the lipoamide and the cysteine pair (31). An intermediate model was generated with one lipoyl domain present in the extended conformation and the second docked in the active site (XDD/E3int, Dmax = 156 Å). Sedimentation coefficients of 5.3 S, 5.9 S and 6.5 S were calculated for XDD/E3extn, XDD/E3int and XDD/E3dock, respectively, using the program HYDROPRO (33;34). Accordingly, the XDD/E3int model with one docked and one extended lipoyl domain gave the best agreement with the experimentally determined sedimentation coefficient of 5.68 S and also agrees best with the experimentally determined Dmax of 158 Å (see below).
Small angle x-ray scattering and rigid body modeling
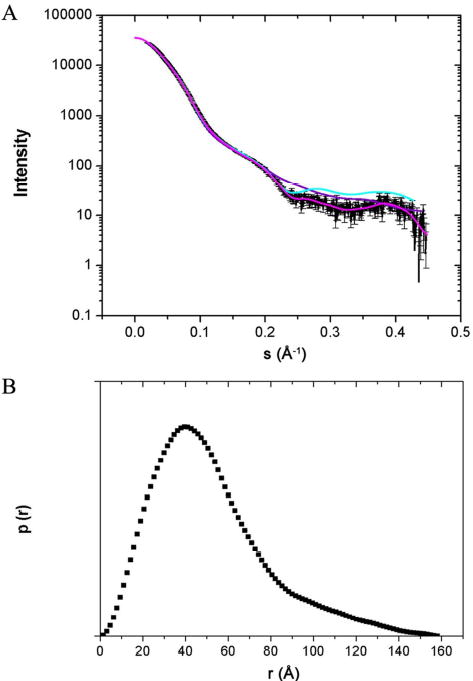
Small angle x-ray scattering (SAXS) curves for purified XDD/E3 complex (Fig. 5A) were acquired at three different protein concentrations to account for the effects of interparticle interference. The radius of gyration Rg determined using the Guinier approximation was 38 ± 1 Å. The particle distribution function, p(r) (Fig. 5B) gave Dmax of 158 ± 5 Å and Rg = 41 ± 1 Å which agrees well with the value obtained using the Guinier approximation. Ab initio shape restoration produced an elongated, asymmetric structure (Fig. 6). Rigid body modeling was used in combination with ab initio modeling to analyze the structure of the XDD/E3 complex: using translations and rotations, the lowest energy conformation of each “domain” (E3, XSBD1, XSBD2, XLD1, XLD2) within the complex was determined using the program Rayuela (35) as judged by fits to the SAXS data. This model was superimposed upon the ab initio model (Fig. 6). While most of the complex model fits well into the SAXS envelope, it was not possible to fit residues 265–297 which form part of the E3 NAD domain. The differences in structure between human E3, its yeast counterpart and our homology model are not significant enough to alter the fit of the high resolution structure within the SAXS envelope. However, E3 is thought to adopt a more extended conformation in solution when compared to the crystal structure (MS, OB, JGL, unpublished material).
Fig. 5. Small angle x-ray scattering of XDD/E3.
(A) Experimental scattering curve for XDD/E3 (–), the shape scattering curve (–) and curves calculated for the models obtained from ab initio (–) and rigid body modeling (–). (B) The distance distribution p(r) was calculated using GNOM. Error bars are too small to be visible.
Fig. 6. Superimposition of XDD/E3 models from ab initio and rigid body modeling.
The ab initio model is shown as a blue wire mesh. Homology models of the five “domains” used for rigid body modeling are: E3 (orange), XSBD1 (lime), XSBD2 (green), XLD1 (magenta), and XLD2 (yellow). Orthogonal views are shown.
DISCUSSION
We have shown for the first time using a number of independent approaches that two E3BP-derived molecules interact with a single E3 homodimer, thus enabling the formation of 2:1 stoichiometric “cross-bridges” in human PDC. These findings are in direct contrast to the crystal structures obtained for the B. stearothermophilus E3/E2SBD (17) and hE3/E3BP-SBD complexes (18;19) where only one SBD is associated with E3 due to steric hindrance by a loop in E3BP which prevents binding of a second SBD. Comparison of the crystal structures determined for unbound and bound hE3 and yeast E3 (16;31;38) yields only very small differences in coordinates. The highly homologous (54% identical) hE3 and yeast E3 may crystallize similarly. As our results were determined in solution we propose that either crystal packing and/or crystallization buffer compositions affect the structures determined for eukaryotic E3 and E3/E3BP-SBD complex. Unfortunately, E3 is too large for structure determination via NMR spectroscopy. However, unpublished SAXS data from our laboratories suggest that the solution conformation of free hE3 is more elongated (Dmax = 130 Å) and flattened than that in the crystal structure. The solution conformations of other proteins, including yeast pyruvate decarboxylase (43) can differ significantly from their crystal structures (44–49). The 1:1 stoichiometry of bacterial PDC E2-SBD/E1 and E2-SBD/E3 has been confirmed via surface plasmon resonance (14) and ITC (13;15). Very recently, Brautigam and colleagues also published ITC data claiming a 1:1 stoichiometry in conjunction with the latest crystal structure (19). However, visual inspection of their data suggests a stoichiometry of approximately 1.3. Without further details of the authors’ experimental protocols used for ITC we cannot comment on this discrepancy. However, we used four independent methods in order to verify the 2:1 stoichiometry of binding between XDD and E3. This includes determination of the complex molecular weight by sedimentation equilibrium (see results) that is independent of protein concentration.
Since E3BP is thought to be located in the 12 faces of the icosahedral E2/E3BP core (10;40), its 2:1 stoichiometric relationship with the dimeric E3 enzyme suggests the potential for the formation of “cross-bridges” across the core surface (Fig. 7). A substitution model of E2 and E3BP within the human core has been proposed more recently by Hiromasa et al. (50): the authors suggested that E3BP replaces some of the E2 molecules, resulting in a core formed from 48 E2 molecules and 6 E3BP dimers. However, our model of cross-bridge formation is compatible with both models for the organization of the core assembly, although in the case of a substitution model for the core assembly the E3BP/E3 cross-bridges would obviously also be orientated along its edges. Studies to distinguish between the two different core models are currently underway in our laboratories.
Fig. 7. A model for “cross-bridge” formation in eukaryotic PDC.
E3BP/E3 cross-bridges are shown in magenta/cyan, E2/E1 cross-bridges in green/blue. PDK and PDP/PRP are shown in purple. The E2/E3BP core is based on a cryo-EM structure (53).
Since 60 E2s can bind up to 30 E1 heterotetramers at maximum occupancy we speculate that similar E2/E1 cross-bridges form at the edges of the icosahedral core. Indeed, recent AUC and ITC analyses support this hypothesis (data not shown).
Current concepts of PDC subunit architecture envisage the E2:E3BP core forming the structural and mechanistic framework for the assembly to which the E1 and E3 enzymes bind tightly but non-covalently. The peripherally bound E1 and E3 enzymes are precisely positioned and oriented around its surface to facilitate optimal interaction of their active sites with the lipoyl ‘swinging arms’ of E2/E3BP during the catalytic cycle. In this model, E1 and E3 interactions with the ‘core’ assembly are passive in nature in the sense that they are not predicted to have a direct influence on the operation of the E2/E3BP core. However, the existence of a previously unrecognized level of ultrastructure resulting from a network of E3 (and E1) cross-bridges on the core surface is predicted to introduce a considerable degree of restraint into the movement of the N-terminal ‘swinging arms’ of E3BP (and E2), limiting in particular the degree of freedom enjoyed by E3BP and E2 lipoyl domains. Since lipoyl domains are, in effect, substrates for all three active sites within the PDC, this more restrictive environment highlights some interesting questions concerning PDC mechanism and efficiency. In particular, it suggests that E1 may interact exclusively or preferentially with E2 lipoyl domains while E3 may interact similarly with E3BP lipoyl domains, so it is unlikely that individual E2 or E3BP lipoyl domains are able to visit all three types of active site. This new structural feature is also compatible with the idea that specific subsets of lipoyl domains visit the various active sites of the constituent enzymes and defined routes of migration may be involved in mediating the transfer of acetyl groups and reducing equivalents between E2 and E3BP lipoyl domains during the catalytic cycle.
Another important aspect of PDC function that is influenced by its subunit organization is regulation of its activity state by an intrinsic PDC kinase (PDK) and loosely associated phosphatase (PDP). It is reported that only 1–3 molecules of PDC kinase (PDK) are bound per complex (51). Both regulatory enzymes associate with the inner E2 lipoyl domain (ILD) whereby the lipoyl cofactor provides an important part of the recognition site. PDP and PDK both act by de-/phosphorylation of E1 on any of three specific E1α serine residues. Phosphorylation of E1 renders the complex inactive. Since only one molecule of PDK is present per complex, the enzyme has to migrate around the surface of the complex in order to act on the entire population of bound E1 molecules. A hand-over-hand mechanism has been proposed where the dimeric PDK molecule migrates from one inner lipoyl domain to its nearest neighbor (52). Regular spacing of the E2 lipoyl domains linked by a system of E1 cross-bridges could greatly facilitate this type of movement and provide convenient access to all E1 molecules.
The low resolution x-ray scattering structure determined for the human XDD/E3 subcomplex conclusively verifies its 2:1 stoichiometry and high stability. Unexpectedly, the subcomplex proved to be highly asymmetric. These data are compatible with a model in which one lipoyl domain is peripherally extended away from the E3 dimer whereas the second lipoyl domain is docked into one of the E3 active sites that are located at the monomermonomer interface.
During XDD purification, the lipoyl group was maintained in its dithiol form by isolation in the presence of DTT. Reduced lipoate is the true substrate for the E3 enzyme and its reoxidation by E3 was prevented by the absence of NAD+. The redox state of the lipoamide cofactor was confirmed by modification with PEG 5000 maleimide, a sulphydryl group reagent, that leads to a dramatic increase in the apparent Mw of XDD, provided it is present in its reduced form (data not shown). Current studies focus on (a) the influence of the redox state of the lipoamide prosthetic group on the overall conformation of the XDD/E3 subcomplex, (b) refining our biophysical and structural analysis of the E2DD/E1 subcomplex and (c) assessing the implications of this previously unrecognized architectural feature on PDC function.
Acknowledgments
We would like to thank Heather Lindsay and Margaret Nutley for technical assistance, Marcelo Nollmann for help with Rayuela, and Dimitri Svergun and Manfred Rossle at the EMBL Hamburg for help with the data collection. We acknowledge support for beam time from EMBL. M. S. is the recipient of a Wellcome Trust Prize PhD studentship. This work was supported by the Wellcome Trust and the BBSRC.
References
- 1.Aral B, Benelli C, Ait-Ghezala G, Amessou M, Fouque F, Maunoury C, Creau N, Kamoun P, Marsac C. AmJHumGenet. 1997;61:1318–1326. doi: 10.1086/301653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung PSC, Van de Water J, Coppel RL, Nakanuma Y, Munoz S, Gershwin ME. JAutoimmun. 1996;9:119–128. doi: 10.1006/jaut.1996.0015. [DOI] [PubMed] [Google Scholar]
- 3.Jones DEJ. ClinSci. 1996;91:551–558. [Google Scholar]
- 4.Lissens W, De Meirleir L, Seneca S, Liebaers I, Brown GK, Brown RM, Ito M, Naito E, Kuroda Y, Kerr DS, Wexler ID, Patel MS, Robinson BH, Seyda A. HumMutat. 2000;15:209–219. doi: 10.1002/(SICI)1098-1004(200003)15:3<209::AID-HUMU1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 5.Gibson GE, Zhang H, Sheu KFR, Bogdanovich N, Lindsay JG, Lannfelt L, Vestling M, Cowburn RF. AnnNeurol. 1998;44:676–681. doi: 10.1002/ana.410440414. [DOI] [PubMed] [Google Scholar]
- 6.Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. AnnNeurol. 2005;57:695–703. doi: 10.1002/ana.20474. [DOI] [PubMed] [Google Scholar]
- 7.Bruschi SA, Lindsay JG, Crabb JW. PNAS. 1998;95:13413–13418. doi: 10.1073/pnas.95.23.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling MF, McEachern G, Seyda A, MacKay N, Scherer SW, Bratinova S, Beatty B, Giovannucci-Uzielli ML, Robinson BH. HumMolGenet. 1998;7:501–505. doi: 10.1093/hmg/7.3.501. [DOI] [PubMed] [Google Scholar]
- 9.Sanderson SJ, Miller C, Lindsay JG. EurJBiochem. 1996;236:68–77. doi: 10.1111/j.1432-1033.1996.00068.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhou ZH, McCarthy DB, O'Connor CM, Reed LJ, Stoops JK. ProcNatAcadSciUSA. 2001;98:14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessard IA, Perham RN. BiochemJ. 1995;306:727–733. doi: 10.1042/bj3060727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hipps DS, Packman LC, Allen MD, Fuller C, Sakaguchi K, Appella E, Perham RN. BiochemJ. 1994;297:137–143. doi: 10.1042/bj2970137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung HI, Bowden SJ, Cooper A, Perham RN. ProtSci. 2002;11:1091– 1100. doi: 10.1110/ps.4970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lessard IAD, Fuller C, Perham RN. Biochemistry. 1996;35:16863–16870. doi: 10.1021/bi961683r. [DOI] [PubMed] [Google Scholar]
- 15.Jung HI, Cooper A, Perham RN. EurJBiochem. 2003;270:4488–4496. doi: 10.1046/j.1432-1033.2003.03842.x. [DOI] [PubMed] [Google Scholar]
- 16.Frank RAW, Pratap J, Pei XY, Perham RN, Luisi BF. Structure. 2005;13:1119–1130. doi: 10.1016/j.str.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Mande SS, Sarfaty S, Allen MD, Perham RN, Hol WG. Structure. 1996;4:277–286. doi: 10.1016/s0969-2126(96)00032-9. [DOI] [PubMed] [Google Scholar]
- 18.Ciszak EM, Makal A, Hong YS, Vettaikkorumakankauv AK, Korochkina LG, Patel MS. JBiolChem. 2006;281:648–655. doi: 10.1074/jbc.M507850200. [DOI] [PubMed] [Google Scholar]
- 19.Brautigam CA, Wynn RM, Chuang JL, Machius M, Tomchick DR, Chuang DT. Structure. 2006;14:611–621. doi: 10.1016/j.str.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuck P. BiophysJ. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society for Chemistry; London: 1992. [Google Scholar]
- 22.Harding SE, Horton JC, Jones S, Thornton JM, Winzor DJ. BiophysJ. 1999;76:2432–2438. doi: 10.1016/S0006-3495(99)77398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. JApplCryst. 2003;36:1277–1282. [Google Scholar]
- 24.Semenyuk AV, Svergun DI. JApplCryst. 1991;24:537–540. [Google Scholar]
- 25.Svergun DI. JApplCryst. 1992;25:495–503. [Google Scholar]
- 26.Svergun DI. BiophysJ. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svergun DI, Petoukhov MV, Koch MHJ. BiophysJ. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petoukhov MV, Svergun DI. JApplCryst. 2003;36:540–544. [Google Scholar]
- 29.Guex N, Peitsch M. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 30.Schwede T, Kopp J, Guex N, Peitsch M. NuclAcids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyoda T, Suzuki K, Sekiguchi T, Reed LJ, Takenaka A. JBiochem. 1998;123:668–674. doi: 10.1093/oxfordjournals.jbchem.a021989. [DOI] [PubMed] [Google Scholar]
- 32.Howard MJ, Fuller C, Broadhurst RW, Perham RN, Tang JG, Quinn J, Diamond AG, Yeaman SJ. Gastroenterology. 1998;115:139–146. doi: 10.1016/s0016-5085(98)70375-0. [DOI] [PubMed] [Google Scholar]
- 33.Garcia de la Torre J, Huertas ML, Carrasco B. BiophysJ. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia de la Torre J. BiophysChem. 2001;93:159–170. doi: 10.1016/s0301-4622(01)00218-6. [DOI] [PubMed] [Google Scholar]
- 35.Nollmann M, Stark WM, Byron O. JApplCryst. 2005;38:874–887. [Google Scholar]
- 36.Wriggers W, Birmanns S. JStructBiol. 2001;133:193–202. doi: 10.1006/jsbi.2000.4350. [DOI] [PubMed] [Google Scholar]
- 37.Wriggers W, Chacon P. Structure. 2001;9:779–788. doi: 10.1016/s0969-2126(01)00648-7. [DOI] [PubMed] [Google Scholar]
- 38.Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT. JMolBiol. 2005;350:543–552. doi: 10.1016/j.jmb.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Zhou ZH, Liao WC, Cheng RH, Lawson JE, McCarthy DB, Reed LJ, Stoops JK. JBiolChem. 2001;276:21704–21713. doi: 10.1074/jbc.M101765200. [DOI] [PubMed] [Google Scholar]
- 40.Milne JLS, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu XW, Brooks BR, Perham RN, Henderson R, Subramaniam S. EMBO J. 2002;21:5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H, Patel M. JBiolChem. 1992;267:5128–5132. [PubMed] [Google Scholar]
- 42.Leung PSC, Iwayama T, Coppel RL, Gershwin ME. Hepatology. 1990;12:1321–1328. doi: 10.1002/hep.1840120612. [DOI] [PubMed] [Google Scholar]
- 43.Svergun DI, Petoukhov MV, Koch MH, Konig S. JBiolChem. 2000;275:297–302. doi: 10.1074/jbc.275.1.297. [DOI] [PubMed] [Google Scholar]
- 44.Trewhella J, Carlson VA, Curtis EH, Heidorn DB. Biochemistry. 1988;27:1121–1125. doi: 10.1021/bi00404a007. [DOI] [PubMed] [Google Scholar]
- 45.Vigh R, Cser L, Kilar F, Simon I. ArchBiochemBiophys. 1989;275:181–184. doi: 10.1016/0003-9861(89)90362-7. [DOI] [PubMed] [Google Scholar]
- 46.Kozak M, Jurga S. Acta BiochimPol. 2002;49:509–513. [PubMed] [Google Scholar]
- 47.Nakasako M, Fujisawa T, Adachi S, Kudo T, Higuchi S. Biochemistry. 2001;40:3069–3079. doi: 10.1021/bi002482x. [DOI] [PubMed] [Google Scholar]
- 48.Svergun DI, Barberato C, Koch MH, Fetler L, Vachette P. Proteins StructFunctGenet. 1997;27:110–117. [PubMed] [Google Scholar]
- 49.Vestergaard B, Sanyal S, Roessle M, Mora L, Buckingham RH, Kastrup JS, Gajhede M, Svergun DI, Ehrenberg M. MolCell. 2005;20:929–938. doi: 10.1016/j.molcel.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 50.Hiromasa Y, Fujisawa T, Aso Y, Roche TE. JBiolChem. 2004;279:6921– 6933. doi: 10.1074/jbc.M308172200. [DOI] [PubMed] [Google Scholar]
- 51.Yang D, Song J, Wagenknecht T, Roche TE. JBiolChem. 1997;272:6361– 6369. doi: 10.1074/jbc.272.10.6361. [DOI] [PubMed] [Google Scholar]
- 52.Liu S, Baker JC, Roche TE. JBiolChem. 1995;270:793–800. doi: 10.1074/jbc.270.2.793. [DOI] [PubMed] [Google Scholar]
- 53.Stoops JK, Cheng RH, Yazdi MA, Maeng CY, Schroeter JP, Klueppelberg U, Kolodziej SJ, Baker TS, Reed LJ. JBiolChem. 1997;272:5757–5764. doi: 10.1074/jbc.272.9.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]