Abstract

The loss of a coding nucleobase from the structure of DNA is a common event that generates an abasic (Ap) site (1). Ap sites exist as an equilibrating mixture of a cyclic hemiacetal and a ring-opened aldehyde. Aldehydes are electrophilic functional groups that can form covalent adducts with nucleophilic sites in DNA. Thus, Ap sites present a potentially reactive aldehyde as part of the internal structure of DNA. Here we report evidence that the aldehyde group of Ap sites in duplex DNA can form a covalent adduct with the N6-amino group of adenine residues on the opposing strand. The resulting interstrand DNA–DNA cross-link occurs at 5′-ApT/5′-AA sequences in remarkably high yields (15–70%) under physiologically relevant conditions. This naturally occurring DNA-templated reaction has the potential to generate cross-links in the genetic material of living cells.
Introduction
Interstrand cross-links are among the most deleterious types of damage that can be generated in cellular DNA.1,2 These lesions are complete blocks to DNA transcription and replication because the covalent linkage prevents strand separation.3,4 Concurrent modification of both strands in the duplex also presents severe challenges to cellular DNA repair systems.5−7 Accordingly, even small numbers of interstrand DNA–DNA cross-links can have profound biological consequences.1,2,8−10
We recently reported a new structural type of interstrand cross-link derived from the reaction of a DNA abasic (Ap) site with a guanine residue on the opposing strand of the duplex (Scheme 1).11,12 This cross-link is intriguing because Ap sites (1, Scheme 2) are perhaps the most common type of endogenous damage suffered by cellular DNA.13,14 Ap sites are generated by spontaneous depurination, nitrosative stress, and enzymatic base excision repair processes.14−18 As a result, DNA from normal mammalian tissue carries steady-state levels of 50 000–200 000 Ap sites per cell.16,19 Ap sites are also generated by exposure of DNA to a wide variety of mutagens, toxins, and anticancer drugs.14,18 Cross-links derived from this abundant lesion could have significance in biology and medicine.
Scheme 1.
Scheme 2.
In our previous work, dG-Ap cross-links were observed at 5′-CAp/5′-AG sequences (duplex A, Figure 1) in equilibrium yields of 2–3%, under physiologically relevant conditions.11,12 The evidence supported a mechanism involving attack of the exocyclic N2-amino group of guanine on the Ap aldehyde (Scheme 1).11,12 Two other canonical nucleobases, adenine and cytosine, also contain exocyclic amino groups that have been shown to react with aldehydes.20−23 However, our previous studies provided no indication regarding whether adenine or cytosine can participate in cross-link formation with an Ap site in duplex DNA.11,12 In fact, no cross-linking was observed at the adenine residue directly opposing the Ap site in duplex A (Figure 1).11,12 Cross-links involving adenine residues have been observed in DNA duplexes containing oxidized abasic sites;24−27 however, it is important to recognize that the structures and reactivities of oxidized abasic sites are distinct from that of the native Ap site considered here.
Figure 1.

DNA duplexes used in this study. Ap-containing duplexes were generated from the corresponding dU-containing duplexes by treatment with UDG. Locations where cross-links form in substantial yield are indicated with a (red \).
In order to grasp the full potential of Ap sites to generate cross-links in duplex DNA, we felt it was necessary to examine whether canonical nucleobases other than guanine can forge cross-links with Ap sites in DNA. Here we provide evidence for a previously unknown type of interstrand DNA–DNA cross-link involving the reaction of adenine residues with Ap sites in double-helical DNA. The dA-Ap cross-links described here formed in remarkably high yields (15–70%) under physiologically relevant conditions and we present evidence consistent with a cross-linking mechanism involving attack of the exocyclic N6-amino group of adenine on the aldehyde group of the Ap site. The widespread occurrence of Ap sites in cellular DNA combined with the high yields of cross-links observed here raise the possibility that this naturally occurring, DNA-templated reaction28−30 can generate cross-links in the genetic material of living cells.
Results and Discussion
Generation and Stability of Interstrand Cross-Links in DNA Duplexes Containing 5′-ApT/5′-AA Sequences
When considering nucleobase sequences in duplex DNA that might support formation of dA-Ap cross-links, we first recognized that any nucleobase involved in Ap-derived cross-link formation likely must be located near the Ap site. This is because cross-linking by a distal nucleobase presumably would generate substantial and energetically unfavorable structural distortion in the duplex.31 With this in mind, we constructed models of Ap-containing DNA duplexes to determine sequence locations on the opposing strand that would position the N6-amino group of adenine in close proximity to the Ap site. In these models, we employed the simplifying assumption that Ap-containing duplexes retain B-DNA-like character.32 This approach successfully predicted cross-linking sequences in the case of dG-Ap cross-links.11,12 Measurments within these models indicated that the exocyclic N6-amino groups of both adenine residues opposing the Ap site in a 5′-ApT/5′-AA sequence were located near the Ap aldehyde residue (Figure 2). As noted in the Introduction, however, our previous studies showed that adenine residues directly opposing an Ap site (e.g., duplex A) did not engage in cross-link formation.11,12 Overall, this analysis led us to the prediction that an adenine residue offset one base to the 3′-side of the Ap site might have potential to engage the Ap-aldehyde in cross-link formation (Figure 2).
Figure 2.

Molecular model illustrating the proximity of the abasic site aldehyde and N6-amino group of 2′-deoxyadenosine in a 5′-ApT sequence in B-DNA. The image was constructed using Pymol and is based on pdb entry 3bse.
Ap sites were produced at defined locations in DNA duplexes by treatment of the corresponding 5′-32P-labeled, 2′-deoxyuridine-containing duplexes with uracil DNA glycosylase (UDG).33,34 Efficient formation of Ap sites in the DNA was confirmed by piperidine treatment to generate the cleavage product 4 (Scheme 2; Figure 3, lanes 3, 7, and 12).14,18,35
Figure 3.
Interstrand cross-link formation in duplexes B–D. Lanes 1–5, duplex B; lanes 6–9, duplex C; and lanes 10–14, duplex D. Lanes 1 and 10 contain the 32P-labeled, uracil-containing oligodeoxynucleotides. Lanes 2, 6, and 11 contain the 32P-labeled UDG-treated (abasic-site-containing) duplexes without incubation. Lanes 3, 7, and 12 contain the 32P-labeled, UDG-treated (abasic-site-containing) duplexes subjected to piperidine workup (1 M, 95 °C, 25 min) to cleave the Ap site. Lanes 4, 8, and 13 contain the cross-linking reactions involving incubation of the abasic-site-containing duplexes for 120 h in HEPES buffer (50 mM, pH 7) containing NaCl (100 mM) at 37 °C. Lanes 5, 9, and 14 contain the methoxyamine-capping reactions involving incubation of the abasic-site-containing duplexes in HEPES buffer (50 mM, pH 7) containing NaCl (100 mM) and CH3ONH2·HCl (2 mM) at 37 °C. The 32P-labeled oligodeoxynucleotides were resolved by electrophoresis on a 20% denaturing polyacrylamide gel, and the radioactivity in each band quantitatively measured by phosphorimager analysis. The numbers on the left side of the image represent our structural assignment of the bands and referring to the structure numbers in Schemes 2 and 3.
Incubation of duplex B containing a central 5′-GApT/5′-AAC sequence in HEPES buffer (50 mM, pH 7, 100 mM NaCl) at 37 °C, followed by denaturing polyacrylamide gel electrophoretic analysis, revealed a 15.1 ± 0.5% yield of a slow-migrating band in the region of the gel where the cross-linked duplex was expected to appear (Figure 3, lane 4).11,12 We also observed bands corresponding to the intact Ap-containing oligonucleotide 1 and small amounts of a product inferred to be the elimination product 3, based upon its gel mobility.14,18,36,37 Duplexes C and D, equipped with 3′-overhanging ends, generated slow-migrating bands whose gel mobility was retarded relative to the putative cross-link generated by duplex B (Figure 3, lanes 8 and 13). These results provided evidence that the slow-migrating bands were indeed cross-linked duplexes containing both full-length strands of the parent duplex and ruled out cross-link formation by the more strongly electrophilic 3′-4-hydroxy-2-pentenal-5-phosphate end group 3. Inclusion of the aldehyde-trapping agent methoxyamine in the reactions substantially inhibited formation of the slow-migrating bands (Figure 3, lanes 5, 9, and 14), consistent with involvement of the Ap-aldehyde residue in cross-link formation (2, Schemes 2 and 3).11,12,38,39 It is noteworthy that cross-link formation in duplexes B–D occurred in good yields (13–18%) at physiological pH and did not require conditions of reductive amination (e.g., pH 4–5 and NaCNBH3) often used to capture the imine products resulting from the reaction of aldehydes and amines.11,12,40−45 A time course experiment showed that easily detectable amounts of the cross-link were formed in duplex B within a few hours (Figure S1).
Scheme 3.
Cross-link formation was a robust reaction that was not highly sensitive to reaction conditions. For example, in duplex B, similar cross-link yields (16 ± 3%) were observed in HEPES buffer (50 mM, pH 7; 100 mM NaCl), cacodylate (50 mM, pH 7, 100 mM NaCl), NaH2PO4/citric acid (16.5/1.8 mM, pH 7.0, 100 mM NaCl), MOPS (50 mM, pH 7, 100 mM NaCl), and bis-Tris (50 mM, pH 7.4, 100 mM NaCl) and in the presence of the biological thiol glutathione (1 mM, in HEPES 50 mM, pH 7, 100 mM NaCl) (Figure S2).
The formation of low molecular weight imines and N-arylglycosides, analogous to those shown in Scheme 3, are reversible processes.46,47 This made it interesting to examine the stability of the cross-link generated in duplex B. We found that, once formed, the cross-link in duplex B was stable under a variety of conditions including pH 3 (sodium acetate, 25 mM, 24 °C, 30 min), pH 10 (potassium phthalate, 24 °C, 25 mM, 30 min) or mild piperidine workup (0.1 M, 60 °C, 15 min) (Figure S3). On the other hand, the cross-link was decomposed by heat (90 °C, 15 min) or treatment with the aldehyde-trapping reagent methoxyamine (200 mM, 30 min, 60 °C; Figure S3).
Evidence That Cross-linking Involves the Adenine Residue Offset One Base to the 3′-Side of the Ap site in the 5′-ApT/5′-AA Sequence
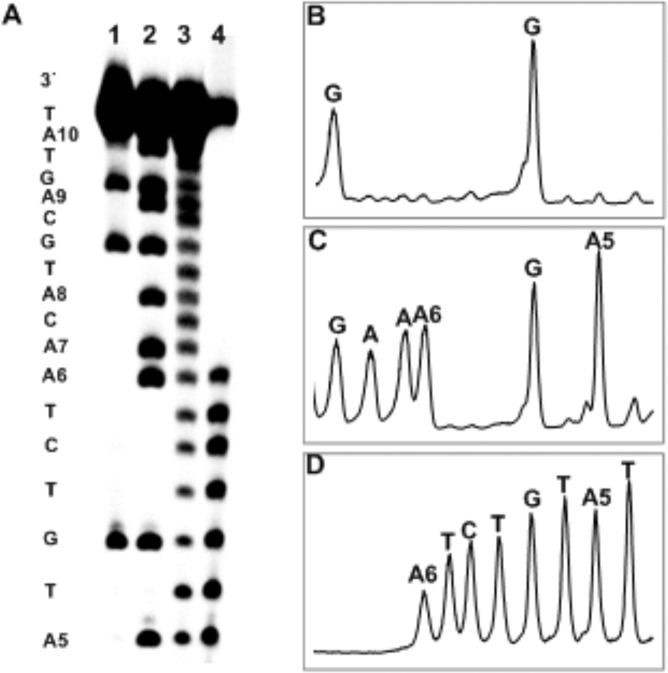
To identify the site at which the Ap-aldehyde was attached to the opposing strand, we carried out hydroxyl radical footprinting of a cross-linked duplex containing the 5′-ApT/5′-AA sequence.26,48 The longer duplex E (Figure 1) was used to move the DNA bands of interest away from a region of the gel where bands were obscured by a “salt flare”. In this duplex, the strand opposing the Ap-containing oligodeoxynucleotide was 5′-32P-labeled. The cross-link was generated as described above, isolated from the gel, and subjected to cleavage by a mixture of iron-EDTA-H2O2.26,48 In this type of experiment, the site of cross-linking appears as an interruption in the “ladder” of strand cleavage products generated by the iron-EDTA-H2O2 reagent, because cleavages beyond the cross-link yield large, slow-migrating DNA fragments that are connected to the opposing strand.26,48 In the present case, a clear disruption in the ladder of cleavage products was observed at the 5′-adenine residue in the 5′-GApT/5′-AAC sequence (residue A6 in duplex E, Figures 1 and 4). This provided evidence that cross-linking involves attachment of the Ap-aldehyde to the adenine residue that is offset one base to the 3′-side of the Ap site.
Figure 4.
Hydroxyl radical footprinting of duplex I to locate the site of cross-link attachment. Duplex I contains the same core sequence as that in duplex B. (A) Lane 1 is a Maxam–Gilbert G-specific cleavage (sequencing) reaction of the labeled oligodeoxynucleotide strand in duplex I. Lane 2 is an A+G specific cleavage (sequencing) reaction of the labeled oligodeoxynucleotide strand in duplex I. Lane 3 is the hydroxyl radical footprinting reaction of the single stranded nonuracil containing oligodeoxynucleotide. Lane 4 is the hydroxyl radical footprinting reaction of the slow-migrating, cross-link band generated by incubation of duplex I HEPES buffer (50 mM, pH 7) containing NaCl (100 mM) at 37 °C. The 32P-labeled oligodeoxynucleotides were resolved on a 20% denaturing sequencing gel, and the radioactivity in each band quantitatively measured by phosphorimager analysis. (B) Maxam–Gilbert G-specific cleavage (sequencing) reaction of the labeled oligodeoxynucleotide strand in duplex I. (C) A+G specific cleavage (sequencing) reaction of the labeled oligodeoxynucleotide strand in duplex I. (D) Hydroxyl radical footprinting reaction of the slow-migrating, cross-link band.
We next examined the effect of substituting different nucleobases for the key adenine residue in the 5′-GApT/5′-AAC sequence. Substitution of this adenine residue in duplex B with a guanine or thymine residue, respectively, gave duplexes F and G that did not produce significant yields of a slow-migrating cross-link band (Figure 5). Similarly, duplex H containing 2-aminopurine at this position did not efficiently generate the cross-link (Figure 5). The substitution of 2-aminopurine for the adenine residue amounts to the rather subtle relocation of a single exocyclic amino group from the major groove to the minor groove of the DNA duplex (see structures in Figure 1). Thus, the failure of duplex H to generate significant yields of cross-link allowed us to infer that the exocyclic N6-amino group of dA in the 5′-GApT/5′-AAC sequence was intimately involved in the cross-linking reactions described here. Substitution of cytosine for adenine at the cross-linking site also did not yield cross-links (Figure S4). Cytosine places an exocyclic amino group in the major groove of DNA similar to adenine, but the amino group of dC is located ∼0.7 Å closer to the sugar–phosphate backbone than that of dA (the amino group of dA is closer to the helical axis).
Figure 5.
Nucleobase replacement experiments reveal that the underlined adenine residue in the 5′-GApT/5′-AAC sequence of duplex B is critical for cross-link formation. Replacement of this adenine residue in duplex B with a guanine or thymine residue in duplexes E and F, respectively, abrogates cross-link formation (lanes 13 and 18). Replacement of the crucial adenine residue with 2-aminopurine in duplex G yields only small amounts of a slower-moving band (lane 8). Lanes 1–5, duplex B, lanes 6–9, duplex G, lanes 10–14, duplex E, and lanes 15–19, duplex F. Lanes 1, 10, and 15 contain 32P-labeled uracil-containing oligodeoxynucleotide duplexes. Lanes 2, 6, 11, and 16 contain the 32P-labeled UDG-treated (abasic-site-containing) duplexes without incubation. Lanes 3, 7, 12, and 17 contain the abasic-site-containing duplexes subjected to piperidine workup (1 M, 95 °C, 25 min) to cleave the Ap site. Lanes 4, 8, 13, and 18 contain the cross-linking reactions involving incubation of the abasic-site-containing duplexes in HEPES buffer (50 mM, pH 7) containing NaCl (100 mM) at 37 °C. Lanes 5, 9, 14, and 19 contain the methoxyamine-capping reactions involving incubation of the abasic-site-containing duplexes incubated in HEPES buffer (50 mM, pH 7) containing NaCl (100 mM), and CH3ONH2·HCl (2 mM) at 37 °C. The 32P-labeled oligodeoxynucleotides were resolved by electrophoresis on a 20% denaturing polyacrylamide gel and the radioactivity in each band quantitatively measured by phosphorimager analysis.
Mass Spectrometric Analysis of the Cross-linked Duplex B
The results described above were broadly consistent with a cross-linking mechanism involving reversible reaction of the exocyclic N6-amino group on dA with the Ap aldehyde to generate hemiaminal, imine, cyclic hydroxyalkylhemiaminal, or enamine linkages at 5′-GApT/5′-AAC sequences in duplex DNA (structures 6-9). MALDI-MS analysis of the cross-linked duplex B gave a strong [M-H]− signal at m/z 12718.66. This value is consistent with the imine 7, cyclic hydroxylalkylhemiaminal 8, or enamine 9 (Figure S5; calcd [M – H]− = 12719.4, 20 ppm dev). Literature precedents describing the properties of structurally related small molecules indicate that the cyclic hydroxylalkylhemiaminal structure 8 likely is favored.21,22,49,50
LC-MS/MS analysis provided additional insight regarding the chemical structure of the dA-Ap cross-link. The cross-linked duplex B (without the 5′-32P label) was excised from a 20% denaturing polyacrylamide gel, eluted from the gel slice, digested with a four-enzyme cocktail consisting of nuclease P1, alkaline phosphatase, and phosphodiesterases I and II, and subjected to LC-MS/MS analysis using previously reported experimental conditions.51−53 Selected-ion chromatograms revealed two peaks eluting at 24.1 and 26.1 min displaying the relevant m/z 368→252 transition anticipated for the neutral loss of 2-deoxyribose from the putative dA-AP cross-link (Figure 6). Further cleavage of the m/z 252 ion gave rise to a dominant fragment ion at m/z 136 in MS/MS/MS (Figure S6), which is identical to the MS/MS observed for the [M + H]+ ion of dA, suggesting that the initial m/z 368→252 transition arises via loss of the cross-linked 2-deoxyribose adduct from dA (Figure 6C). The presence of multiple peaks in the LC-MS/MS ion chromatogram was consistent with the expectation that the anticipated cross-link remnant resulting from the digestion of 8 will exist as an equilibrating mixture of pyranose and furanose isomers (Scheme S1).18,54
Figure 6.
LC-ESI-MS and MS/MS analysis of the mixture generated by digestion of the cross-linked oligodeoxynucleotide generated by incubation of duplex B in HEPES buffer (50 mM, pH 7) containing NaCl (100 mM) at 37 °C. (A) selected-ion chromatogram monitoring the neutral loss of 2-deoxyribose from the dA-AP cross-link. (B) MS/MS arising from the fragmentation of the ion at m/z 368. (C) Possible structures for the ions observed at m/z 368, 252, and 136 in these experiments.
High Yield dA-Ap Cross-Link Formation in the 5′-CApT/5′-AAG Sequence
We examined cross-link formation in duplex I that contained a central 5′CApT/5′-AAG sequence. This experiment was intriguing to us because, in this sequence context, the Ap site has the potential to generate cross-links with either the guanine or adenine residue on the opposing strand (residues colored red in duplex I shown in Figure 1). Incubation of duplex I in HEPES buffer (50 mM, pH 7, 100 mM NaCl) at 37 °C produced a remarkable 71.5 ± 3.7% yield of a slow-migrating band in the region of the denaturing gel where the cross-linked duplexes migrate (Figure 7). Similar to the experiments described above in the context of the 5′-GApT/5′-AAC sequence, the yield of the slow-moving band in the 5′CApT/5′-AAG sequence also was greatly diminished when the aldehyde-capping reagent methoxyamine was added to the reaction mixtures. Formation rate and stability of the cross-link in this sequence was similar to that described above for the 5′-GApT/5′-AAC sequence (Figures S7–S9). These results along with Nanospray TOF-MS data (Figure S11) confirmed that the slow-migrating band generated in the 5′CApT/5′-AAG sequence was an interstrand cross-linked duplex involving the Ap-aldehyde residue. Footprinting of a cross-linked duplex containing the 5′CApT/5′-AAG core sequence indicated that the cross-link attachment was to the underlined adenine residue in the 5′CApT/5′-AAG sequence (Figure S10). The footprinting results showed no detectable cross-linking to the dG residue in this sequence. Thus, the remarkable cross-link yield in this sequence appears to result from efficient formation of a dA-Ap cross-link rather than simultaneous formation of dG-Ap and dA-Ap cross-links.
Figure 7.

Interstrand cross-link formation in duplex I. Lane 1, 5′-32P-labeled uracil-containing precursor of oligodeoxynucleotide duplex I. Lane 2, 5′-32P-labeled abasic-site-containing oligodeoxynucleotide duplex I without incubation. Lane 3, 5′-32P-labeled abasic-site-containing oligodeoxynucleotide duplex I cleaved by treatment with piperidine (1 M, 95 °C, 25 min). Lane 4, Duplex I incubated in HEPES buffer (50 mM, pH 7) containing NaCl (100 mM) at 37 °C. Lane 5, Duplex I incubated with CH3ONH2 (2 mM) in HEPES buffer (50 mM, pH 7) containing NaCl (100 mM) at 37 °C. The 32P-labeled oligodeoxynucleotides were resolved on a polyacrylamide gel, and the radioactivity in each band quantitatively measured by phosphorimager analysis.
LC-MS/MS analysis of the products generated by nuclease P1 digestion12,51−53 of the cross-linked duplex I revealed the presence of a tetranucleotide with the Ap-aldehyde conjugated with the underlined adenine from the 5′-CApT/5′-AAG sequence (Figure S12), while the corresponding tetranucleotide with the Ap-aldehyde being attached to the guanine residue was not detectable (data not shown). Additionally, LC-MS/MS analysis of the products formed from the digestion with a four-enzyme cocktail12 showed that the dA-Ap remnant is present at a level that is ∼2 orders of magnitude higher than its dG-AP counterpart (Figures S13–14). These results mesh with the footprinting data described above and indicate that the cross-linking occurs predominantly between the Ap-aldehyde and the underlined adenine in the 5′-CApT/5′-AAG sequence.
Conclusions
The results described here identified a novel cross-link involving reaction of a DNA Ap site with an adenine residue on the opposing strand of the double helix. It is now clear that both dG and dA can engage in cross-linking with an Ap site and two distinct cross-linking sequence motifs, 5′-CAp/5′-AG and 5′-ApT/5′-AA, have been defined as locations for the formation of Ap-derived cross-links.11,12 The difference in the favored locations of dA and dG residues in these cross-links, with the cross-linking nucleobase displaced to the 3′- and 5′-side of the Ap site, respectively, is a natural consequence of the helical twist of duplex DNA coupled with the fact that the N6-amino group of adenine is located in the major groove while the N2-amino group of guanine resides in the minor groove (Figure S15).
Our results were consistent with a mechanism involving attack of the exocyclic N6-amino group of adenine on the aldehyde residue of the Ap site to generate a hemiaminal intermediate (6, Scheme 3). The mass spectrometric data is consistent with subsequent dehydration of 6 to give the imine (7), cyclic hydroxyalkylhemiaminal (8), or enamine (9) product. The detailed chemical structure of the dA-Ap cross-link remains to be rigorously established using 2D-NMR, but chemical precedents involving structurally related small molecules suggest that the cyclic hydroxyalkylhemiaminal (8) likely will be the predominant form.21,22,49,50 The substantial stability of the dA-Ap cross-link generated in duplex B is generally consistent with a cyclic hydroxyalkylhemiaminal 8, in which the reactive imine group of 7 has been masked by intramolecular attack of the hydroxyl group. Despite the stability of the cross-link to many of the conditions examined here, the reaction is reversible, and it will be important to determine whether Ap-derived cross-links have sufficient stability to block DNA processing enzymes such as helicases and polymerases.
It may be important to emphasize that the dA-Ap cross-links described here formed in good yields (13–70%) under physiologically relevant conditions (pH 7, 100 mM NaCl) and did not require conditions of reductive amination (e.g., pH 4–5 and NaCNBH3) often used to stabilize and increase the yield of the imine products resulting from the reaction of aldehydes and amines.11,12,40−45 In the sequences examined to date, the yields of dA-Ap cross-links were much higher than those observed for the dG-Ap cross-links, when both reactions are conducted under the same, physiologically relevant conditions (pH 7, in the absence of NaCNBH3).11,12 The sequence 5′-CApT/5′-AAG (duplex I) that, in principle, can form both dG-Ap and dA-Ap cross-links, in fact, generates a remarkably high yield (∼70%) of the dA-Ap cross-link. It seems likely that, in some duplexes such as I, multiple cross-linked species exist in dynamic equilibrium. The yield of each cross-linked species at equilibrium will be determined by the inherent abilities of each nucleobase to generate imine linkages with the Ap aldehyde and by the ability of the DNA duplex to position the amino and aldehyde groups in a manner that favors imine formation. Thus, formation of these Ap-derived cross-links is conceptually related to template-directed synthesis with dynamic combinatorial imine libraries, in which equilibrium “sorting processes” involving reversible imine formation among a mixture of various aldehydes and amines ultimately generate higher yields of the imine products that form the most stable ensembles with a templating molecule present in the reaction.55,56 Our results established that the equilibrium yields of the dA-Ap cross-link can be quite high in some sequence contexts, and future work will systematically explore the effects of sequence variation on cross-link yields.
The widespread occurrence of Ap sites14−19 along with the facile nature of the cross-linking reactions described here make it interesting to consider the possibility that Ap-derived cross-links can form in genomic DNA. In cells, processes such as repair13,57,58 and strand cleavage (as shown in Scheme 2)59,60 will compete with the formation of Ap-derived cross-links. As a result, the majority of Ap sites in genomic DNA almost certainly do not go forward to form interstrand DNA–DNA cross-links. Nonetheless, because interstrand cross-links are especially noxious lesions,1,2,8−10 even low yields of endogenous Ap-derived cross-links could prove significant in several diverse areas. First, endogenous DNA damage is thought to be important in the etiology of both human aging and sporadic cancers.61 Because cross-links present severe challenges to replication, transcription, and repair, endogenously formed cross-links could be especially significant. Unrepaired cross-links that block transcription and replication may promote cell death and senescence that contribute to the aging of tissues.3,4,62−64 At the same time, cellular attempts to repair cross-links are error prone and introduce cancer-causing mutations into the genome.6,65−67 In addition, the realization that Ap sites can generate cross-links in duplex DNA may help explain why pathways such as nucleotide excision repair and recombination that are typically associated with the repair of more complex lesions are required for eukaryotic cells to survive the introduction of Ap sites into their genome.13,57 Finally, Ap-derived cross-links could be relevant to difficulties inherent in the amplification and sequencing of ancient DNA.68 Ancient DNA samples contain a plethora of lesions including oxidative base damage, Ap sites, and strand breaks,68 but it has been suggested that a high density of interstrand cross-links ultimately limit the ability to amplify and sequence the DNA in these samples.69 The structural nature of the cross-links in ancient DNA has not been elucidated, but Ap-derived cross-links are good candidates for these blocking lesions that hinder sample processing.70
Overall, these findings establish Ap-derived interstrand cross-links as a family of DNA lesions that can form in diverse sequence contexts. Studies are currently underway to define the structure, occurrence, biochemical properties, and biological consequences of these lesions.
Acknowledgments
We are grateful to the National Institutes of Health for support of this work (ES021007 to K.G and Y.W.). We thank Professor Marc Greenberg (Johns Hopkins University) for helpful discussions during the course of this work. MALDI mass spectrometry was conducted at the Charles W. Gehrke Proteomics Center at the University of Missouri, and we thank Beverly DaGue for assistance with these experiments.
Supporting Information Available
Experimental procedures and data on the formation and properties of cross-linked duplexes B and I. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
⊥ Cairo University, Faculty of Pharmacy, Pharmacognosy Department, Kasr El-Aini, Cairo, Egypt 11562.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Schärer O. D. ChemBioChem 2005, 6, 27–32. [DOI] [PubMed] [Google Scholar]
- Noll D. M.; Mason T. M.; Miller P. S. Chem. Rev. 2006, 106, 277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth V. D.; Carlsson R.; Johansson F.; Erixon K.; Jenssen D. DNA Repair 2012, 11, 976–985. [DOI] [PubMed] [Google Scholar]
- Derheimer F. A.; Hicks J. K.; Paulsen M. T.; Canman C. E.; Ljungman M. Mol. Pharmacol. 2009, 75, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy P. A.; Liu J.; Majumdar A.; Liu S.-t.; Seidman M. M. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 23–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.; Li L. Environ. Mol. Mutagen. 2010, 51, 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavin E. M.; Smeaton M. B.; Miller P. S. Environ. Mol. Mutagen. 2010, 51, 604–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana-Schwencke N.; Henriques J. A.; Chanet R.; Moustacchi E. Proc. Natl. Acad. Sci. U.S.A. 1982, 79, 1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman K. F.; Ward A. M.; Matkovic M. E.; Folias A. E.; Moses R. E. Mutat. Res. 2001, 487, 73–83. [DOI] [PubMed] [Google Scholar]
- Reddy M. C.; Vasquez K. M. Radiat. Res. 2005, 164, 345–356. [DOI] [PubMed] [Google Scholar]
- Dutta S.; Chowdhury G.; Gates K. S. J. Am. Chem. Soc. 2007, 129, 1852–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. M.; Price N. E.; Wang J.; Fekry M. I.; Dutta S.; Seiner D. R.; Wang Y.; Gates K. S. J. Am. Chem. Soc. 2013, 135, 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S.; Guillet M. DNA Repair 2004, 3, 1–12. [DOI] [PubMed] [Google Scholar]
- Gates K. S. Chem. Res. Toxicol. 2009, 22, 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.; Wang C.; Deen W. M.; Dedon P. C. Chem. Res. Toxicol. 2003, 16, 1044–1055. [DOI] [PubMed] [Google Scholar]
- De Bont R.; van Larebeke N. Mutagenesis 2004, 19, 169–185. [DOI] [PubMed] [Google Scholar]
- Guillet M.; Bioteux S. Mol. Cell. Biol. 2003, 23, 8386–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates K. S.; Nooner T.; Dutta S. Chem. Res. Toxicol. 2004, 17, 839–856. [DOI] [PubMed] [Google Scholar]
- Nakamura J.; Swenberg J. A. Cancer Res. 1999, 59, 2522–2526. [PubMed] [Google Scholar]
- McGhee J. D.; von Hippel P. H. Biochemistry 1975, 14, 1281–1296. [DOI] [PubMed] [Google Scholar]
- Hermida S. A. S.; Possari E. P. M.; Souza D. B.; de Arruda Campos I. P.; Gomes O. F.; Di Mascio P.; Medeiros M. H. G.; Loureiro A. P. M. Chem. Res. Toxicol. 2006, 19, 927–936. [DOI] [PubMed] [Google Scholar]
- Upadhyaya P.; Hecht S. S. Chem. Res. Toxicol. 2008, 21, 2164–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw Y. F. M.; Crane L. E.; Lange P.; Shapiro R. Biochemistry 1980, 19, 5525–5531. [DOI] [PubMed] [Google Scholar]
- Sczepanski J. T.; Heimstra C. N.; Greenberg M. M. Bioorg. Med. Chem. 2011, 19, 5788–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczepanski J. T.; Jacobs A. C.; Greenberg M. M. J. Am. Chem. Soc. 2008, 130, 9646–9647. [DOI] [PubMed] [Google Scholar]
- Sczepanski J. T.; Jacobs A. C.; Majumdar A.; Greenberg M. M. J. Am. Chem. Soc. 2009, 131, 11132–11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L.; Greenberg M. M. J. Am. Chem. Soc. 2009, 131, 15225–15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J. T.; Lynn D. G. J. Am. Chem. Soc. 1992, 114, 9197–9198. [Google Scholar]
- Silverman A. P.; Kool E. T. Chem. Rev. 2006, 106, 3775–3789. [DOI] [PubMed] [Google Scholar]
- Niu J.; Hili R.; Liu D. R. Nat. Chem. 2013, 5, 282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins P. B.; Millard J. T.; Woo J.; Weidner M. F.; Kirchner J. J.; Sigurdsson S. T.; Raucher S. Tetrahedron 1991, 47, 2475–2489. [Google Scholar]
- Chen J.; Dupradeau F.-Y.; Case D. A.; Turner C. J.; Stubbe J. Nucleic Acids Res. 2008, 36, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T.; Ljunquist S.; Siegert W.; Nyberg B.; Sperens B. J. Biol. Chem. 1977, 252, 3286–3294. [PubMed] [Google Scholar]
- Varshney U.; van de Sande J. H. Biochemistry 1991, 30, 4055–4061. [DOI] [PubMed] [Google Scholar]
- Maxam A. M.; Gilbert W. Methods Enzymol. 1980, 65, 499–560. [DOI] [PubMed] [Google Scholar]
- Bayley C. R.; Brammer K. W.; Jones A. S. J. Chem. Soc. 1961, 1903–1907. [Google Scholar]
- Sugiyama H.; Fujiwara T.; Ura A.; Tashiro T.; Yamamoto K.; Kawanishi S.; Saito I. Chem. Res. Toxicol. 1994, 7, 673–683. [DOI] [PubMed] [Google Scholar]
- Rosa S.; Fortini P.; Karran P.; Bignami M.; Dogliotti E. Nucleic Acids Res. 1991, 19, 5569–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talpaert-Borle M.; Liuzzi M. Biochim. Biophys. Acta 1983, 740, 410–16. [DOI] [PubMed] [Google Scholar]
- Kurtz A. J.; Dodson M. L.; Lloyd R. S. Biochemistry 2002, 41, 7054–7064. [DOI] [PubMed] [Google Scholar]
- Manoharan M.; Andrade L. K.; Cook P. D. Org. Lett. 1999, 1, 311–314. [DOI] [PubMed] [Google Scholar]
- Hochgürtel M.; Biesinger R.; Kroth H.; Piecha D.; Hofmann M. W.; Krause S.; Schaaf O.; Nicolau C.; Eliseev A. V. J. Med. Chem. 2003, 46, 356–358. [DOI] [PubMed] [Google Scholar]
- Li X.; Zhan Z.-Y. J.; Knipe R.; Lynn D. G. J. Am. Chem. Soc. 2002, 124, 746–747. [DOI] [PubMed] [Google Scholar]
- Angelov T.; Guainazzi A.; Schärer O. D. Org. Lett. 2009, 11, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozekov I. D.; Turesky R. J.; Alas G. R.; Harris C. M.; Harris T. M.; Rizzo C. J. Chem. Res. Toxicol. 2010, 23, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes E. H.; Jencks W. P. J. Am. Chem. Soc. 1963, 85, 2843–2848. [Google Scholar]
- Holton S.; Runquist O. J. Org. Chem. 1961, 26, 5193–5195. [Google Scholar]
- Luce R. A.; Hopkins P. B. Methods Enzymol. 2001, 340, 396–412. [DOI] [PubMed] [Google Scholar]
- Amann N.; Wagenknecht H.-A. Tetrahedron Lett. 2003, 44, 1685–1690. [Google Scholar]
- Capon B.; Connett B. E. J. Chem. Soc. 1965, 4492–4497. [DOI] [PubMed] [Google Scholar]
- Lai C.; Cao H.; Hearst J. E.; Corash L.; Luo H.; Wang Y. Anal. Chem. 2008, 80, 8790–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H.; Hearst J. E.; Corash L.; Wang Y. Anal. Chem. 2008, 80, 2932–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wang Y. Anal. Chem. 2003, 75, 6306–6313. [DOI] [PubMed] [Google Scholar]
- Tomasz M.; Lipman R.; Lee M. S.; Verdine G. L.; Nakanishi K. Biochemistry 1987, 26, 2010–2027. [DOI] [PubMed] [Google Scholar]
- Meyer C. D.; Joiner C. S.; Stoddart J. F. Chem. Soc. Rev. 2007, 36, 1705–1723. [DOI] [PubMed] [Google Scholar]
- Ramström O.; Lehn J.-M. Nat. Rev. Drug Discovery 2001, 1, 26–36. [DOI] [PubMed] [Google Scholar]
- Otterlei M.; Kavli B.; Standal R.; Skjelbred C.; Bharati S.; Krokan H. E. EMBO J. 2000, 19, 5542–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. K.; Vande Berg B. J.; Prasad R.; Molina J. T.; Beard W. A.; Tomkinson A. E.; Wilson S. H. J. Biol. Chem. 1998, 273, 21203–21209. [DOI] [PubMed] [Google Scholar]
- Lindahl T.; Andersson A. Biochemistry 1972, 11, 3618–3623. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Sczepanski J. T.; Greenberg M. M. J. Am. Chem. Soc. 2012, 134, 16734–16741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers J. H. J. N. Engl. J. Med. 2009, 361, 1475–1485. [DOI] [PubMed] [Google Scholar]
- Schumacher B.; Garinis G. A.; Hoeijmakers J. H. J. Trends Genet. 2007, 24, 77–85. [DOI] [PubMed] [Google Scholar]
- Bergstrahl D. T.; Sekelsky J. Trends Genet. 2007, 24, 70–76. [DOI] [PubMed] [Google Scholar]
- Garinis G. A.; van der Horst G. T. J.; Vijg J.; Hoeijmakers J. H. J. Nat. Cell Biol. 2008, 10, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. V.; Schärer O. D. Environ. Mol. Mutagen. 2010, 51, 552–566. [DOI] [PubMed] [Google Scholar]
- Zheng H.; Wang X.; Warren A. J.; Legerski R. J.; Nairn R. S.; Hamilton J. W.; Li L. Mol. Cell. Biol. 2003, 23, 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R. B.; Alberti M.; Hearst J. E.; Chua M. A.; Saffran W. A. J. Biol. Chem. 2001, 276, 31551–31560. [DOI] [PubMed] [Google Scholar]
- Mitchell D.; Willerslev E.; Hansen A. Mutat. Res. 2005, 571, 265–276. [DOI] [PubMed] [Google Scholar]
- Hansen A. J.; Mitchell D. L.; Wiuf C.; Paniker L.; Brand T. B.; Binladen J.; Gilichinsky D. A.; Rønn R.; Willerslev E. Genetics 2006, 173, 1175–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-links derived from oxidative DNA damage and lipid peroxidation products also must be considered, see:; a Stone M. P.; Cho Y. J.; Huang H.; Kim H. Y.; Kozekov I. D.; Kozekova A.; Wang H.; Minko I. G.; Lloyd R. S.; Harris T. M.; Rizzo C. J. Acc. Chem. Res. 2008, 41, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Huang H.; Solomon M. S.; Hopkins P. B. J. Am. Chem. Soc. 1992, 114, 9240–9241. [Google Scholar]; c Ding H.; Majumdar A.; Tolman J. R.; Greenberg M. M. J. Am. Chem. Soc. 2008, 130, 17981–17987. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Regulus P.; Duroux B.; Bayle P.-A.; Favier A.; Cadet J.; Ravanat J.-L. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 14032–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Chen H.-J. C.; Chen Y.-C. Chem. Res. Toxicol. 2009, 22, 1334–1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.