Abstract
Periodic ovulation in rats, mice and hamsters is the result of a surge in LH that depends on circadian gating signals emerging from the master circadian clock within the suprachiasmatic nucleus (SCN) and rising ovarian oestrogen levels. These two signals converge into the anteroventral periventricular nucleus (AVPV) and lead to the release of kisspeptin, which is responsible for surges of GnRH and, in turn, of LH release. How the AVPV integrates circadian and reproductive signals remains unclear. Here we show that the female rat AVPV itself shows circadian oscillations in the expression of the clock genes PER1 and BMAL1, which lie at the core circadian clockwork of mammals. In ovariectomized (OVX) females treated with estradiol (E2) these oscillations are in synchrony with the AVPV rhythmic expression of the KISS1 gene and the gene that codes for the arginine-vasopressin (AVP) receptor AVPr1a. Whereas clock gene oscillations are independent of oestrogen levels, circadian expression of Kiss1 and Avpr1a (also referred to as V1a) mRNA are respectively blunted and absent in ovariectomized animals without E2 replacement. Because AVP is believed to be a critical SCN transmitter to gate the LH surge, our data suggest that a there is a circadian oscillator located in the AVPV, and that such a putative oscillator could time, in an oestrogen dependent manner, the sensitivity to circadian signals emerging from the SCN and the release of kisspeptin.
Keywords: kisspeptin, vasopressin, circadian oscillators, ovulation
Introduction
In mammals, periodic ovulation results from the conjunction of at least two oscillations: that of ovarian hormone concentration, principally estradiol (E2), and that of the circadian system, with its master pacemaker located in the suprachiasmatic nucleus (SCN) of the hypothalamus, and extra-SCN oscillators in the brain and peripheral organs. Ovulation is triggered when high E2 enables the release of kisspeptin (coded by the KISS1 gene) from the anteroventral periventricular nucleus (AVPV). Kisspeptin stimulates gonadotropin-releasing hormone (GnRH) release from the medial preoptic area (MPO), which in turn drives pituitary luteinizing hormone (LH) release into systemic blood, the key endocrine signal leading to the rupture of the ovarian follicle (for reviews, see (1–3)). Both a circadian signal from the SCN and rising levels of ovarian oestrogen are necessary for the LH surge to take place; however, the pathways and transmitters by which the SCN communicates time to reproductive targets and how these targets decode circadian and oestrogen signals remains unclear (3).
The SCN projects to the MPO; these projections are not homogenous but instead are topographically organized. In the rat, the AVPV receives input mainly from the dorsomedial (dm) subdivision of the SCN—also known as the shell (4). Indeed, kisspeptin-expressing cells in the AVPV are predominantly innervated by SCN arginine-vasopressin (AVP) fibres, which originate primarily in dmSCN neurons, but not by SCN fibres containing vasoactive intestinal polypeptide (VIP), which originate primarily in ventrolateral (vl) SCN neurons (5, 6). Other MPO targets of the SCN, such as GnRH cells themselves, receive input from the vlSCN, and specifically from VIP-containing neurons (7–9). Although neuroanatomical lesions cannot assess the independent function of the vl- and dmSCN, these two subregions can be desynchronized from each other in the rat, by exposure to an 11:11 light-dark (LD) cycle (LD22), which causes the vl- and dmSCN to stably oscillate with different circadian periods (10). This paradigm allows distinguishing circadian outputs that differentially rely on the activity of either subregion (11–14).
Work in our laboratory showed that a circadian rhythm of Kiss1 mRNA expression exists in the AVPV of ovariectomized (OVX), E2-primed (OVX+E2) mice and rats, and that in the rat, the phase of this rhythm is coordinated by projections from the dorsomedial (dm) SCN (12, 15). A major output of the dmSCN is AVP (16), and previous work has shown that infusion of AVP to the hypothalamus of OVX+E2 rats can induce an LH surge in SCN-lesioned rats (17); it can also enhance LH release in SCN-intact animals, but this enhancement depends on the time of day of AVP administration (18). Taken together, these results suggest that AVP from the dmSCN represents an SCN output signal needed for ovulation, but also that the response to this circadian gating signal is modulated in a time-of-day dependent manner downstream from the SCN. Therefore, we hypothesized that accurate circadian timing of ovulation may result from the synchronous release of the circadian triggering signal by the master clock and a circadian gating of the response to this signal by the AVPV.
Here we test this hypothesis by first showing a rhythm in clock gene expression within the AVPV of OVX+E2 rats. Through the desynchronization of the vl- and dmSCN, we show that this oscillation is synchronized with clock gene oscillations within AVP-rich dorsomedial SCN neurons, and is associated with a circadian rhythm in the expression of the AVP-receptor Avpr1a mRNA and Kiss1 mRNA within the AVPV. In the absence of E2, the Avpr1a mRNA is undetectable whereas Kiss1 expression remains rhythmic but highly muted.
These results suggest that there is a functioning circadian clock in the AVPV, which times receptivity of the AVPV to AVP from the SCN, and that phase coincidence between the SCN and AVPV circadian oscillators is required for normal ovulation timing. They also suggest that the coincidence between the circadian and reproductive signals is encoded by the modulation of receptor expression within the AVPV. That is, time of day is encoded by an SCN output, but receptivity to that signal is impaired without both appropriate E2-levels and matching of correct time of day as judged internally in the AVPV.
Materials and Methods
Animals
Eight-week old female Wistar rats were purchased from Charles River and used for all the experiments. All experiments were performed according to the NIH Guide for Care and Use of Laboratory Animals and were approved by the University of Washington Institutional Animal Care and Use Committee.
Activity Cycles and Forced Desynchrony
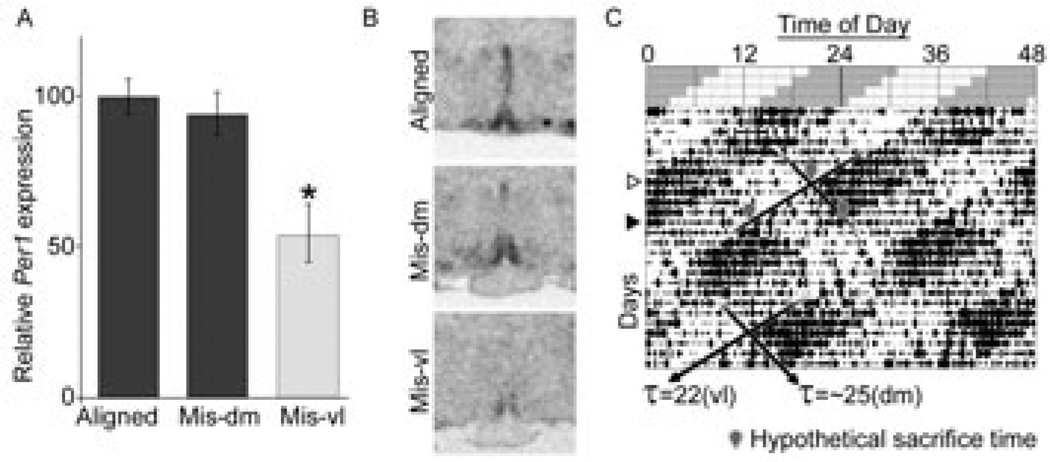
Animal activity monitoring and forced desynchrony was carried out as reported previously (10, 12). Briefly, rats under LD22, show two locomotor activity rhythms within the same individual. One locomotor activity rhythm has a period of 22 h and is tied to the circadian oscillation of clock gene expression of the vlSCN; the second locomotor activity rhythm has a period of ~25 h and is associated with clock gene expression within the dmSCN (shown previously in males and females (10, 12); gene expression patterns in the SCN are not recapitulated here). This forced desynchrony protocol provides a method of identifying the relative influence of SCN subregions on a given extra-SCN oscillation without the potential confounds of surgery or genetic manipulations. Animals were singly housed either under a 12:12 LD cycle (LD24 animals) or desynchronized under LD22. In both cases, animals were housed with 50–150 lux light during the light phase and red light not brighter than 1 lux during the dark phase. LD22 animals were housed under the 11:11 LD cycle for 1–2 months. Locomotor activity was recorded using 2 perpendicular infrared beams crossed through the centre of the cage approximately 2 cm above the bedding, with beam breaks recorded digitally with ClockLab (Actimetrics, Wilmette, IL). Analysis of activity was carried out using the software “El Temps” (Dr. Antoni Díez-Noguera, University of Barcelona) and the graphs imported into Adobe Photoshop to prepare final Figures. Only LD22 animals showing 2 statistically significant rhythms using Chi-squared-periodogram analysis were used for any analysis. The two rhythms in LD22 rats move in and out of phase with each other and define days in which the two rhythms are “aligned” and days in which they are “misaligned” (Fig. 3). Desynchronized animals were grouped into either aligned or misaligned groups for tissue collection, but this did not change their housing or handling, only the time of collection relative to their activity cycles.
Figure 3.
Effect of forced desynchrony of vl- and dmSCN oscillators on AVPV Per1 expression in OVX+E2 rats. (A) Per1 expression is significantly diminished at the onset of the vlSCN-associated locomotor activity bout in misaligned days (Mis-vl), but not at the onset of dmSCN-associated locomotor activity bout (Mis-dm), as compared to expression measured at the onset of locomotor activity on days of alignment (Aligned), when the peak expression is expected. One-way ANOVA shows significant effect of phase (P = 0.007). *Statistically different from Aligned and Mis-dm (P < 0.05, Tukey post-hoc comparisons). (B) Representative radiomicrographs of labelled AVPVs from each phase. (C) Representative actogram of the locomotor-activity output under forced desynchrony. Schematic top section shows light (pale): dark (grey) cycle; below is real activity of one desynchronized rat under this schedule (black vertical ticks represent 10-minute bins of locomotor activity). Note that one locomotor rhythm begins in the dark phase of the LD cycle (period = 22 h) while one does not and shows a period of ~25 h (period differences highlighted with black diagonal lines). White triangle to the left highlights a day of alignment, where both locomotor activity rhythms begin at the same time; black triangle highlights day of maximum misalignment, when one locomotor bout begins as the other ends. Grey outlined balloons point to hypothetical times of sacrifice for animals under this schedule to construct the comparisons in A and B, with the top balloon showing alignment, lower left showing Mis-vl, and lower right showing Mis-dm.
Ovariectomy, E2 treatment
All ovariectomies were carried out under isoflurane anaesthesia, using buprenorphine as an analgesic. Peritoneal incisions were closed with biodegradable suture while skin was closed with wound clips. Animals were allowed to heal from ovariectomy for two weeks before further experimental manipulation. Two days before sacrifice, E2-treated animals were implanted with silastic capsules (1-cm long, 1.57-mm internal diameter, 3.18-mm outer diameter) filled with 20% E2/80% cholesterol by mass, and capped with silastic glue. This capsule size and concentration has been shown to yield 1.5–2 times the physiological concentration of E2 during proestrus in rats (17). Capsules were soaked overnight in 70% ethanol before being subcutaneously implanted into OVX rats under isoflurane anaesthesia. Controls were not given an implant.
Tissue collection
All animals were sacrificed by decapitation. Brains were collected immediately and frozen in −30°C methyl butane; brains were then stored at −80°C until processed. All brains were cut into 16-µm sections in a cryostat, mounted on slides and frozen until histological processing.
In situ hybridization for Kiss1, Per1, Bmal1, and Avpr1a mRNA
In situ hybridization (ISH) was carried out as described previously (19). Briefly, 35S-labeled riboprobe was transcribed from a mouse Kiss1 template (15), a rat Per1 template (10), a Bmal1 template from mouse (96% identity with rat) (10), or a Avpr1a template generated de novo. The Avpr1a template was generated by PCR-amplification from liver genomic DNA using the following primer pair based on the rat AVPr1a sequence (GenBank U3945.1): 5’-CGGTCGCCTTCTTCCAAGTATTAC-3’ (forward) and 5’-AAATGCTCTTCACGCTGCTGAC-3’ (reverse). The gene has a single exon in the rat, so no intronic information is encoded in the PCR amplicon. The resulting product was inserted into pCRII Cloning Vector (Invitrogen) and cloned in Top10 Chemically Competent E. coli cells (Invitrogen). The resulting cloned plasmid was amplified and isolated by MaxiPrep (Qiagen) and sent to GeneWiz (www.genewiz.com) for sequencing using the T7 and SP6 primers. A BLAST search of the result showed 99% identity with the rat AVPr1a gene identified above. Sense and antisense probes were both constructed in parallel, using the same radiation source and run in the same ISH trial to assess specificity, with antisense, but not sense, probes showing labelling in the AVPV and SCN (data not shown).
For ISH, after pre-hybridization washes, slides were incubated overnight at 52°C with 4*107 cpm/ml; after post-hybridization washes and dehydration in an alcohol series, they were air-dried and exposed to autoradiographic films for 4 days. All autoradiographic films were scanned and digital images were scored using ImageJ by two independent experimenters masked to the experimental condition. Kiss1 labelling was scored by assigning each slice an area value based on the whole AVPV radiolabeled area above a preset darkness threshold and multiplying that area by the average optical density within that area. Each animal’s score was the mean of the three strongest radiolabeled images. These slices were then used as reference to localize the area of interest for Per1, Bmal1, and Avpr1a radiolabeled sections; all labelled sections for these probes were alternate sections within 64 µm (four 16-um sections) of the scored Kiss1–labelled slices from the same animals. All scores are reported as a percentage of the average score of the highest-scoring condition within the groups being compared. One-way ANOVA was used to assess effect of time, and two-way ANOVA was used for experiments in which multiple factors existed (treatment and time). Tukey tests were then used for post-hoc analysis.
Because many of the micrographs show fairly subtle absolute changes in intensity and size of labelling, and because the reproductions here are small, we have used Adobe Photoshop to uniformly enhance the contrast of the photomicrographs shown in figures here. This is done only to make more clear the changes over time represented by the token micrographs in the figures; the enhancement happened after scoring, did not affect scoring, and does not distort the original images beyond enhancing contrast uniformly across the entire group of images.
Results
The AVPV shows circadian expression of clock genes, AVPr1a and KISS1
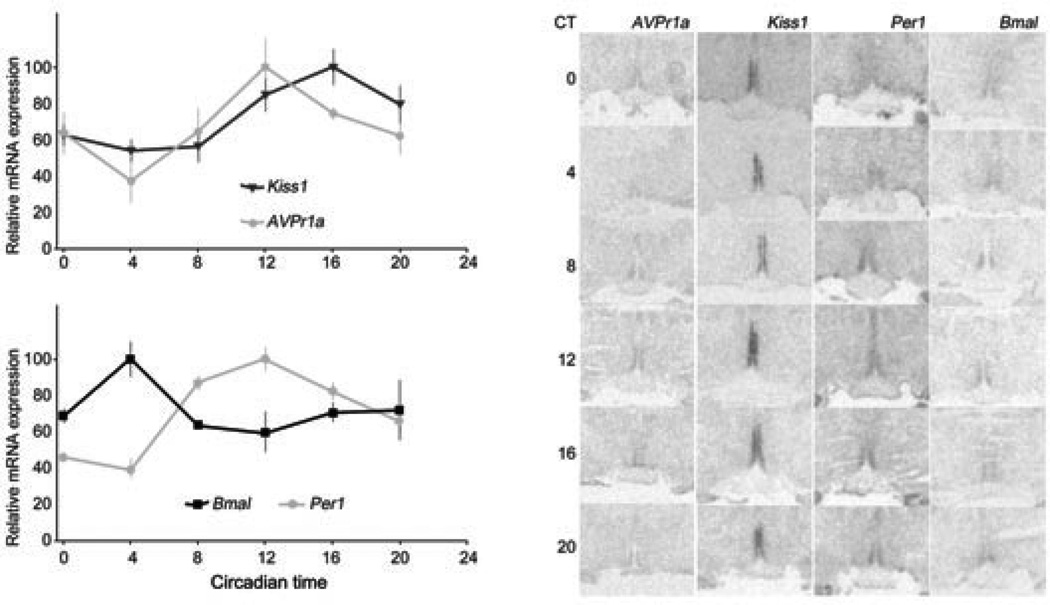
We assessed the expression of AVPr1a and KISS1 within the AVPV of OVX+E2 rats sacrificed every 4 hours over a 24-h period (n = 4–5 per time point). Sacrifices started 24 h after release from the 12:12 LD cycle into constant darkness (DD), which occurred 24 h after E2 implantation. Circadian time (CT) is based on the extrapolated time of lights-off, which represents CT12. A one-way ANOVA found a significant effect of time for Avpr1a (F = 2.96, P = 0.033), with a peak at CT12 (the time of locomotor activity onset under DD). Kiss1 mRNA also showed an effect of time (F = 4.34, P = 0.006), with a peak one time-point later than Avpr1a, at CT16, similar to previous findings (12) (Fig. 1A).
Figure 1.
Relative mRNA expression measured every 4 h for 24 h from 4–5 animals released into DD from a 12:12 LD cycle 24 h prior to the first sacrifices. All 24-h profiles show a significant rhythm (one-way ANOVA, see text for details). Avpr1a peaks one time point before Kiss1 does (A), at the same time as Per1 does (B). Per1 oscillates in antiphase with Bmal1 (B). Asterisks on each curve denote points that are significantly different with each other after post-hoc comparisons. Note that the Bmal1-Per1 oscillations are out of phase with the same gene oscillations expected in the SCN. Representative AVPV radiomicrographs for each mRNA and time are shown (C).
We also measured mRNA expression levels for the core clock genes PER1 and BMAL1 in the same animals. We found that both had a significant rhythm in mRNA expression (Per1: F = 9.09, P = 0.0002; Bmal1: F = 3.11, P = 0.032), with their peaks in antiphase with each other (Fig. 1B). Per1 shows a peak at CT12 and a trough at CT4, while Bmal1 shows the inverse. Because these rhythms were taken after 24 h without oscillating environmental cues they are truly circadian. Representative radiomicrographs for each mRNA labelled at each time are shown in Fig. 1C.
Effect of E2 treatment on peak and trough gene expression in the AVPV
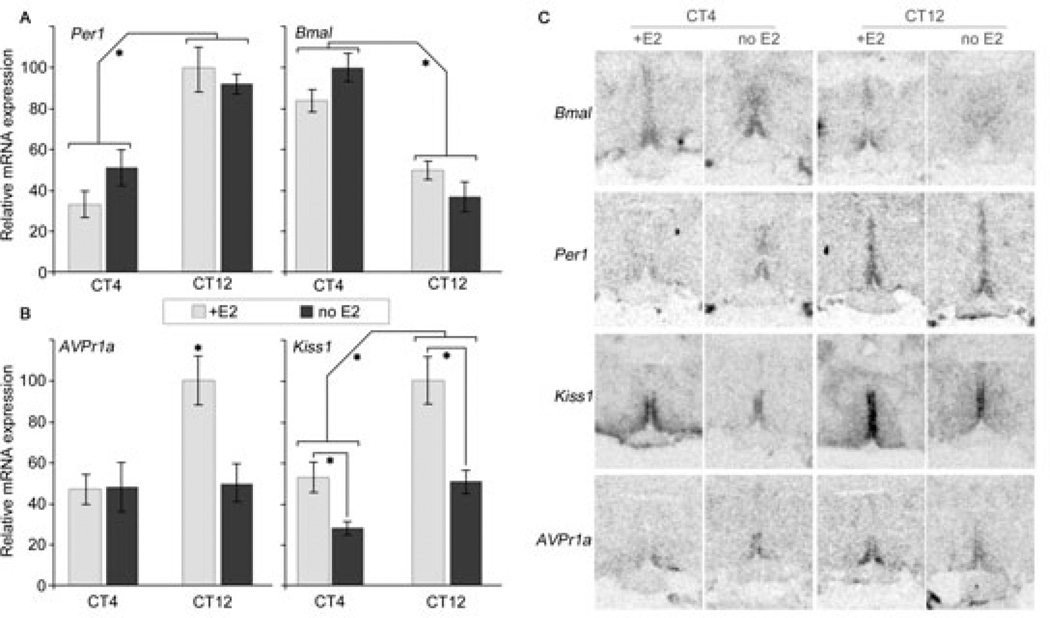
To determine whether the above circadian rhythms are dependent on E2, we sacrificed a second group of rats at the predicted time of peak and trough for each rhythm found above (CT4 and CT12; Kiss1 expression peak is at CT16 but Kiss1 levels are near peak value at CT12, which was previously the reported peak (12), so we use CT12 here as a proxy for peak time), OVX+E2 and OVX animals were sacrificed at each time point (n = 4–5 per time point per E2 condition), and gene expression assessed by ISH as before. A two-way ANOVA for time and E2 treatment on Avpr1a expression revealed a significant effect of time (P = 0.026), E2 (P = 0.041), and a significant interaction (P = 0.038) (Fig. 2A, left). By contrast, assessment of Kiss1 expression revealed a significant effect of time and E2 (P = 0.0003, 0.0002 respectively), but did not find a significant interaction (P = 0.137) (Fig. 2A, right). Post-hoc analysis of Avpr1a reveals that only CT12 with E2 is significantly different from the other three points, which are not statistically different from each other (P<0.05). Post-hoc analysis of Kiss1 reveals that CT12 levels differ from CT4 levels for each E2 treatment (P<0.05). The changes in Avpr1a and Kiss1 mRNA levels after removal of E2 could potentially reflect phase shifts of an otherwise unchanged rhythm waveform; this is highly unlikely though, as in both cases we detected either trough levels (Avpr1a) or levels that were at or below trough levels (Kiss1).
Figure 2.
Effect of E2 treatment on expected peak (CT12) and trough (CT4) AVPV mRNA expression. Avpr1a shows a significant peak at CT12 but only in the presence of E2 (A, left). Kiss1 shows a significant peak at CT12 with or without E2, but the peak is significantly smaller without E2 (A, right). Per1 and Bmal1 (B) show peak and trough at opposite times, with significant effects of time but not of E2 treatment. Representative radiomicrographs for each mRNA and treatment are shown (C). * Brackets identify significant main effects of the two-way ANOVA or statistically different bars; see text for details on statistics.
Assessment of Per1 and Bmal1 both revealed a significant effect of time (P <0.0001, P = 0.0007 respectively) but no effect of E2 treatment, and no interaction (P = 0.506, 0.325 for E2, and 0.1115, 0.350 for interaction, for Per1 and Bmal1 respectively) (Fig. 2B). Representative radiomicrographs for each mRNA labelled at each time and condition are shown in Fig. 2C.
AVPV remains synchronized to the dmSCN under forced desynchrony
Our previous work has shown that the Kiss1 rhythm in the AVPV is synchronized to the activity of the dmSCN (12) and not the vlSCN. To test if the clockwork uncovered here is also synchronized to the dmSCN, we assessed Per1 expression in the AVPV of desynchronized rats. Because Per1 showed a peak in the AVPV at CT12 (the onset of locomotor activity) we sacrificed one group of desynchronized female rats at the onset of locomotor activity in days of alignment, which mimic SCN clock gene expression LD24 animals (10). For rats on days of misalignment, which have two locomotor onsets—one corresponding to rhythms in each SCN subregion—we sacrificed one group of animals before the dmSCN-associated locomotor activity onset and another group before the vlSCN-associated locomotor activity onset (see Fig. 3C for a diagram). We find no detectable difference in Per1 peak expression intensity in brains taken at the onset of dmSCN-associated locomotor activity on days of alignment and misalignment. In contrast, on days of misalignment, Per1 levels at the time of the vlSCN-associated locomotor activity onset (that is, 11–12 h out of phase with the dmSCN-associated locomotor activity onset) were significantly lower than at the time of the dmSCN-associated locomotor activity onset [n = 4 per time point, one-way ANOVA for phase found a significant effect (F = 8.98, P = 0.007)]. Post-hoc Tukey comparisons found a difference between the AVPV Per1 expression before the vlSCN-associated activity onset on misaligned days and the expression before the dmSCN-associated activity onset on aligned or misaligned days (P < 0.05) (Fig. 3A and representative radiomicrographs on Fig. 3B). By contrast, there was no detectable post-hoc difference between the expression before the activity onset in aligned days and before the dmSCN-associated locomotor activity in misaligned days (P > 0.05). Thus the locomotor activity onset of the dmSCN appears to predict the peak of Per1 expression in the AVPV, whereas the vlSCN does not. This finding parallels our previous findings on the expression rhythm of Kiss1 in the AVPV and further supports the hypothesis that the AVPV contains an extra-SCN circadian oscillator whose phase appears locked to the dmSCN but not vlSCN,
Discussion
The results above suggest that the AVPV may contain an extra-SCN oscillator (or oscillators) entrained to the dmSCN, and that clock outputs, but not the core clock genes involved in the circadian transcriptional-translational feedback loop, are sensitive to E2 concentrations. The autonomy of this putative AVPV circadian oscillator remains to be determined; experiments looking at oscillations ex vivo and the persistence of oscillations in SCN-lesioned animals would confirm the AVPV’s role as an independent oscillator. We hypothesize that this AVPV circadian oscillator could enable the AVPV to act as a coincidence detector of E2 cycles and circadian time to guide the neural activation of ovulation, with both high E2 and the correct circadian phase necessary to stimulate the pre-ovulatory release of kisspeptin. The fact that either SCN lesions or ovariectomy abolishes the pre-ovulatory LH surge (1, 3), combined with the apparent role of the AVPV-kisspeptin population as the positive driver of the GnRH surge, suggests that detection of the circadian signal and rising levels of oestrogen must coincide at the appropriate time to gate the GnRH and subsequent LH surges; our data suggest that this detector may well reside in the AVPV, and point to a pathway and mechanism for future exploration. Further, they suggest that precise timing of the LH surge may rely on the ability of extra-SCN oscillators to ensure coincidence between a central circadian signal and its response, through phase-dependent receptivity to the AVP signal from the SCN.
Systems in which an extra-SCN oscillator is linked to a known rhythmic functional output are rare. One such example is a circadian oscillator located within the olfactory bulb (OB) known to regulate olfactory responsiveness in a circadian fashion (20). What is more, the SCN-AVPV-MPO circuit of ovulatory timing-control is linked to easily measured physiological and behavioural outputs. The circadian olfactory response modulated by the OB persists in the absence of the SCN master circadian pacemaker (20). In contrast, SCN lesions abolish the LH surge and ovulation (17, 21), suggesting these reproductive processes rely on a more centralized regulation. Our findings not only shed light on the neural regulation of ovulation but they also offer a new framework on which to study the regulation and function of extra-SCN circadian oscillators.
The AVPV has been shown to have circadian expression patterns in Kiss1. The clock gene oscillations we show suggest that these rhythms may be in part driven by a population of canonical circadian oscillators within the female AVPV, with the typical antiphase oscillation of Per1 and Bmal1. The phase of these oscillators is apparently set by the dmSCN but delayed—with a peak at CT12—relative to the phase of the SCN pacemaker—with a peak at CT8 (22)—; a similar phase mismatch has been found between the SCN and other extra-SCN oscillators (23). Previous work has shown that the AVPV expressed the clock-controlled gene Dbp (24), but to our knowledge oscillations in expression of the core clock genes PER1 and BMAL1, constituting the core of the feedback loop driving circadian oscillations, had not been shown.
We recapitulate rhythms already found in AVPV Kiss1 expression, but show that while they are dependent on E2 as shown by our group previously (12, 15), they remain present but muted in low E2 animals. This suggests that the peak in Kiss1 expression may result from an E2 amplification of a pre-existing clock-regulated expression of Kiss1. The idea that the AVPV is integrating E2 and circadian information to control Kiss1 expression is supported by the fact that ERα seems to operate through the circadian transcription factor DBP to drive Kiss1 mRNA expression (24). We have previously reported a loss of circadian Kiss1 expression in mice without E2 (15). This result may reflect a more robust oscillation in Kiss1 expression in the rat, which after E2 treatment also shows a higher amplitude oscillation than the mouse (12, 15). The presence of circadian expression of Kiss1 in the absence of E2 suggests that, at least in the rat, kisspeptin may have daily functions as yet unappreciated. Of note, both our previous and current studies measured Kiss1 RNA levels but not kisspeptin levels, and the possibility exist that E2 may exert species-specific posttranscriptional and posttranslational effects on KISS1 gene products.
We also demonstrate that Avpr1a mRNA expression seems to be an E2-dependent clock output of the AVPV; that is, however the clock and E2 levels interact to impact AVPr1a expression they both play a role shaping it. This suggests AVPV receptivity to AVP is at some level clock-controlled. Given previous work showing a time-dependent effect of hypothalamic-infused AVP on the LH surge (18), our findings are consistent with a model where the AVPV’s circadian control of AVPr1a is disinhibited by high E2, providing a potential mechanism for the integration of E2 and circadian information in the circuit..
Recently, work in the hamster suggested that the AVPV responded to AVP both during the morning and evening, and that circadian responsiveness was more keenly modulated, downstream, at the level of GnRH neurons (5). If such findings hold in the rat they would suggest yet another layer of coincidence detection. In addition, it would suggest time-dependent post-transcriptional modifications of AVPr1a expression in the AVPV on top of the time-dependent and E2-dependent changes in mRNA levels shown here. In either case, these data strengthen the argument that circadian timing of the HPG relies on a circuit with multiple redundant checks on coincident signalling.
We do not assess whether all of the measured rhythms are occurring in the same cells; however, previous work indicates that kisspeptin cells in the AVPV receive AVP projections from the SCN (5, 6). The most parsimonious model would place the molecular clockwork in the same cells that express Avpr1a and Kiss1 in a circadian fashion, but future work could confirm whether this is the case and whether AVPr1a and/or KISS1 indeed represent clock-controlled genes. Regardless of whether the molecular clockwork is within the same cells, it would be sufficient for the AVPV to act as the coincidence-detector region we suggest.
We cannot rule out the possibility that the SCN is only sending a pro-ovulatory signal when some upstream centre relays high E2 information to the SCN (25), nor that the apparent gating—or entraining—signal(s) from the SCN to the AVPV is not AVP, but some other transmitter(s). If AVP were the entraining signal, it would presumably be driving the rhythmicity of its own detection. Such auto-tuning systems are common in biology. Another alternative is that the AVPV reception of circadian information from the SCN serves internal cohesion of oscillating cells in the AVPV, to better gate the peak of kisspeptin release when E2 levels rise. More investigation into the exact mechanism of entrainment and coincidence detection is certainly warranted, evincing once again this system’s power as a model to untangle such presumably broad issues as SCN-to-extra-SCN oscillator entrainment, redundancy, and signalling. For instance, this system could be used to investigate the impact of abrupt phase shifts of the LD cycle on the desynchronization or disruption of downstream physiological and behavioural outputs that are typically synchronized, such as ovulation and sexual receptivity.
Acknowledgements
This work was supported by NIH: R01MH075016, R03HD061853 to HOD; NSF IOS-1145796 NSF to HOD, DDIG 0909716 to HOD and BLS, Gates Millennium Scholarship and McNair Fellowship to JJG
References
- 1.de la Iglesia HO, Schwartz WJ. Timely ovulation: Circadian regulation of the female hypothalamo-pituitary-gonadal axis. Endocrinology. 2006;147(3):1148–1153. doi: 10.1210/en.2005-1311. [DOI] [PubMed] [Google Scholar]
- 2.Pinilla L, Aguilar E, Dieguez C, Millar RP, Tena-Sempere M. Kisspeptins and reproduction: physiological roles and regulatory mechanisms. Physiol Rev. 2012;92(3):1235–1316. doi: 10.1152/physrev.00037.2010. [DOI] [PubMed] [Google Scholar]
- 3.Christian CA, Moenter SM. The neurobiology of preovulatory and estradiol-induced gonadotropin-releasing hormone surges. Endocr Rev. 2010;31(4):544–577. doi: 10.1210/er.2009-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2001;433(3):312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- 5.Williams WP, 3rd, Jarjisian SG, Mikkelsen JD, Kriegsfeld LJ. Circadian control of kisspeptin and a gated GnRH response mediate the preovulatory luteinizing hormone surge. Endocrinology. 2010;152(2):595–606. doi: 10.1210/en.2010-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vida B, Deli L, Hrabovszky E, Kalamatianos T, Caraty A, Coen CW, Liposits Z, Kallo I. Evidence for suprachiasmatic vasopressin neurones innervating kisspeptin neurones in the rostral periventricular area of the mouse brain: regulation by oestrogen. J Neuroendocrinol. 2010;22(9):1032–1039. doi: 10.1111/j.1365-2826.2010.02045.x. [DOI] [PubMed] [Google Scholar]
- 7.de la Iglesia HO, Blaustein JD, Bittman EL. The suprachiasmatic area in the female hamster projects to neurons containing estrogen receptors and GnRH. Neuroreport. 1995;6(13):1715–1722. doi: 10.1097/00001756-199509000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Van der Beek EM, Horvath TL, Wiegant VM, van den Hurk R, Buijs RM. Evidence for a direct neuronal pathway from the suprachiasmatic nucleus to the gonadotropin-releasing hormone system: combined tracing and light and electron microscopic immunocytochemical studies. J Comp Neurol. 1997;384(4):569–579. doi: 10.1002/(sici)1096-9861(19970811)384:4<569::aid-cne6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 9.van der Beek EM, Wiegant VM, van der Donk HA, van den Hurk R, Buijs RM. Lesions of the suprachiasmatic nucleus indicate the presence of a direct vasoactive intestinal polypeptide-containing projection to gonadotrophin-releasing hormone neurons in the female rat. J Neuroendocrinol. 1993;5(2):137–144. doi: 10.1111/j.1365-2826.1993.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 10.de la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14(9):796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 11.Wotus C, Lilley TR, Neal A, Suleiman N, Schmuck S, Smarr BL, de la Iglesia HO. Forced desynchrony reveals independent contributions of suprachiasmatic oscillators to the daily plasma corticosterone rhythm in rats. PLoS ONE. 2013 doi: 10.1371/journal.pone.0068793. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smarr BL, Morris E, de la Iglesia HO. The dorsomedial suprachiasmatic nucleus times circadian expression of Kiss1 and the luteinizing hormone surge. Endocrinology. 2012;153(6):2839–2850. doi: 10.1210/en.2011-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz MD, Wotus C, Liu T, Friesen WO, Borjigin J, Oda GA, de la Iglesia HO. Dissociation of circadian and light inhibition of melatonin release through forced desynchronization in the rat. Proc Natl Acad Sci U S A. 2009;106(41):17540–17545. doi: 10.1073/pnas.0906382106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee ML, Swanson BE, de la Iglesia HO. Circadian timing of REM sleep is coupled to an oscillator within the dorsomedial suprachiasmatic nucleus. Curr Biol. 2009;19(10):848–852. doi: 10.1016/j.cub.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA, Kauffman AS. Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin-releasing hormone/luteinizing hormone surge. Endocrinology. 2009;150(8):3664–3671. doi: 10.1210/en.2009-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore RY, Speh JC, Leak RK. Suprachiasmatic nucleus organization. Cell Tissue Res. 2002;309(1):89–98. doi: 10.1007/s00441-002-0575-2. [DOI] [PubMed] [Google Scholar]
- 17.Palm IF, Van Der Beek EM, Wiegant VM, Buijs RM, Kalsbeek A. Vasopressin induces a luteinizing hormone surge in ovariectomized, estradiol-treated rats with lesions of the suprachiasmatic nucleus. Neuroscience. 1999;93(2):659–666. doi: 10.1016/s0306-4522(99)00106-2. [DOI] [PubMed] [Google Scholar]
- 18.Palm IF, van der Beek EM, Wiegant VM, Buijs RM, Kalsbeek A. The stimulatory effect of vasopressin on the luteinizing hormone surge in ovariectomized, estradiol-treated rats is timedependent. Brain Res. 2001;901(1–2):109–116. doi: 10.1016/s0006-8993(01)02309-5. [DOI] [PubMed] [Google Scholar]
- 19.de la Iglesia HO. In situ hybridization of suprachiasmatic nucleus slices. In: Rosato E, editor. Methods Mol Biol. Totowa, NJ: Humana Press; 2007. pp. 513–531. [DOI] [PubMed] [Google Scholar]
- 20.Granados-Fuentes D, Tseng A, Herzog ED. A circadian clock in the olfactory bulb controls olfactory responsivity. J Neurosci. 2006;26(47):12219–12225. doi: 10.1523/JNEUROSCI.3445-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coen CW, MacKinnon PC. Lesions of the suprachiasmatic nuclei and the serotonin-dependent phasic release of luteinizing hormone in the rat: effects on drinking rhythmicity and on the consequences of preoptic area stimulation. J Endocrinol. 1980;84(2):231–236. doi: 10.1677/joe.0.0840231. [DOI] [PubMed] [Google Scholar]
- 22.Yan L, Takekida S, Shigeyoshi Y, Okamura H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience. 1999;94(1):141–150. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]
- 23.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22(1):350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Kaga S, Tsubomizu J, Fujisaki J, Mochiduki A, Sakai T, Tsukamura H, Maeda K, Inoue K, Adachi AA. Circadian transcriptional factor DBP regulates expression of Kiss1 in the anteroventral periventricular nucleus. Mol Cell Endocrinol. 2011;339(1–2):90–97. doi: 10.1016/j.mce.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 25.de la Iglesia HO, Blaustein JD, Bittman EL. Oestrogen receptor-alpha-immunoreactive neurones project to the suprachiasmatic nucleus of the female Syrian hamster. J Neuroendocrinol. 1999;11(7):481–490. doi: 10.1046/j.1365-2826.1999.00341.x. [DOI] [PubMed] [Google Scholar]