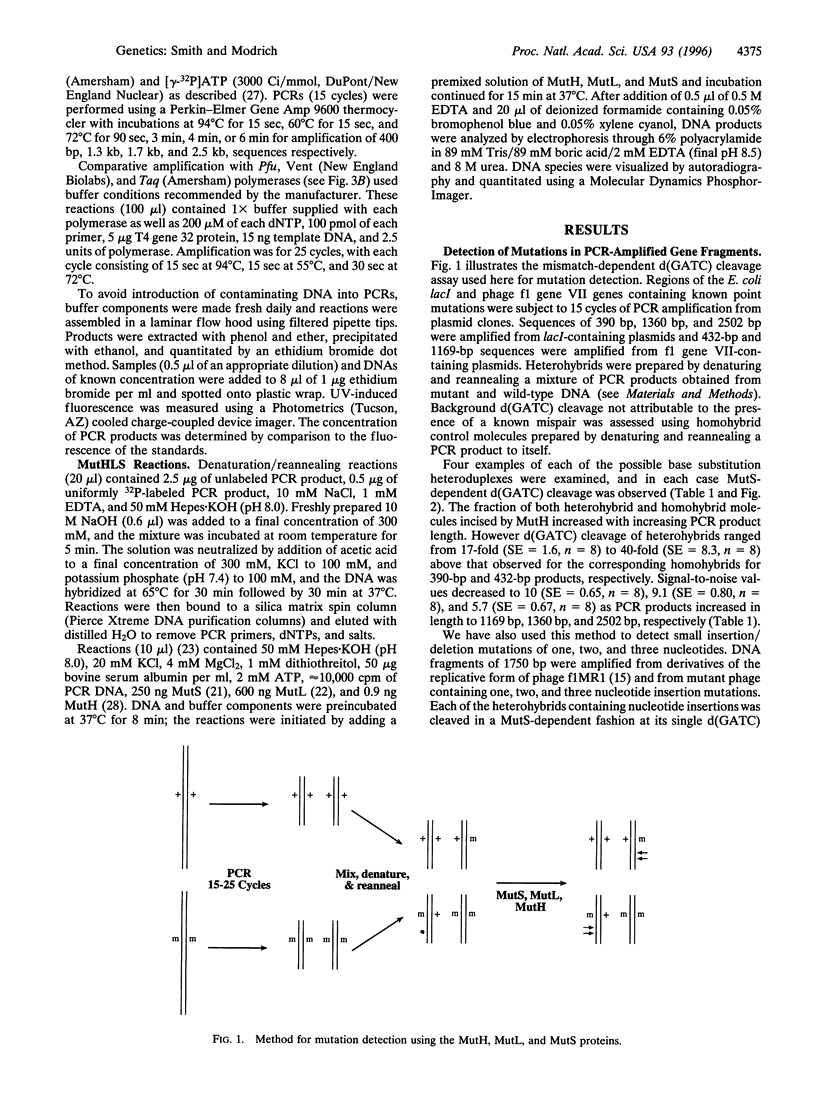
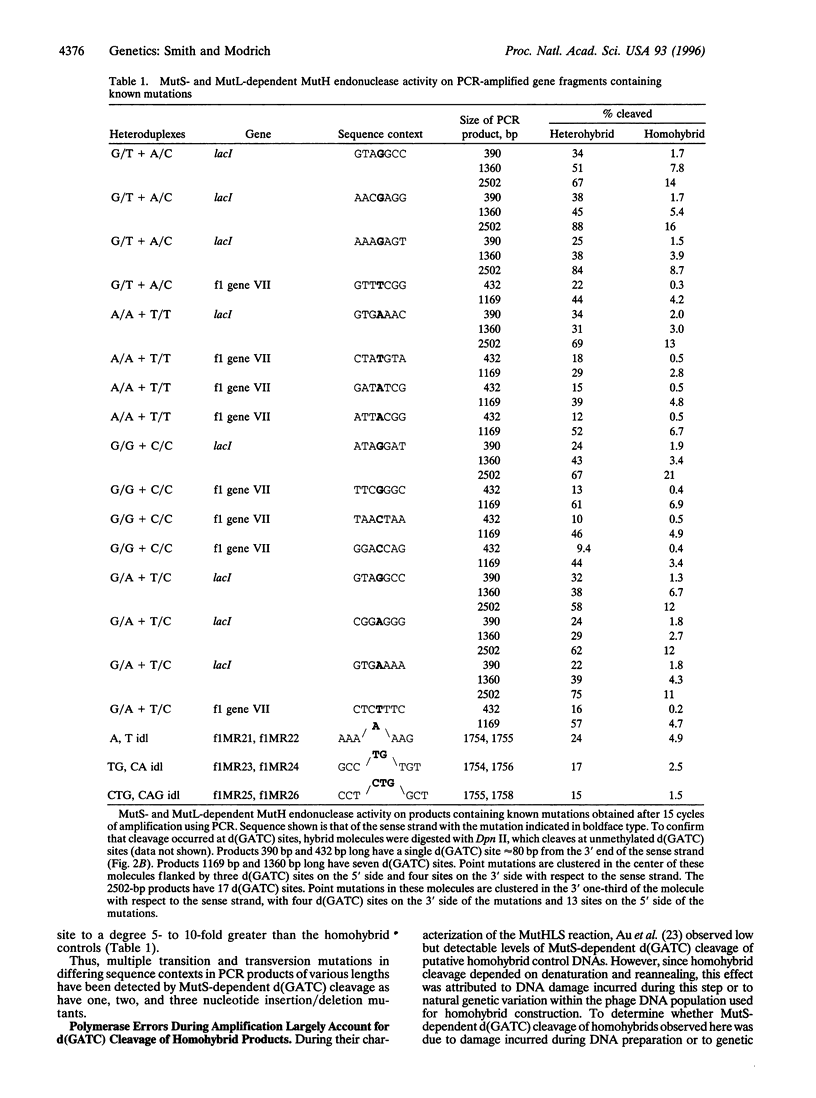
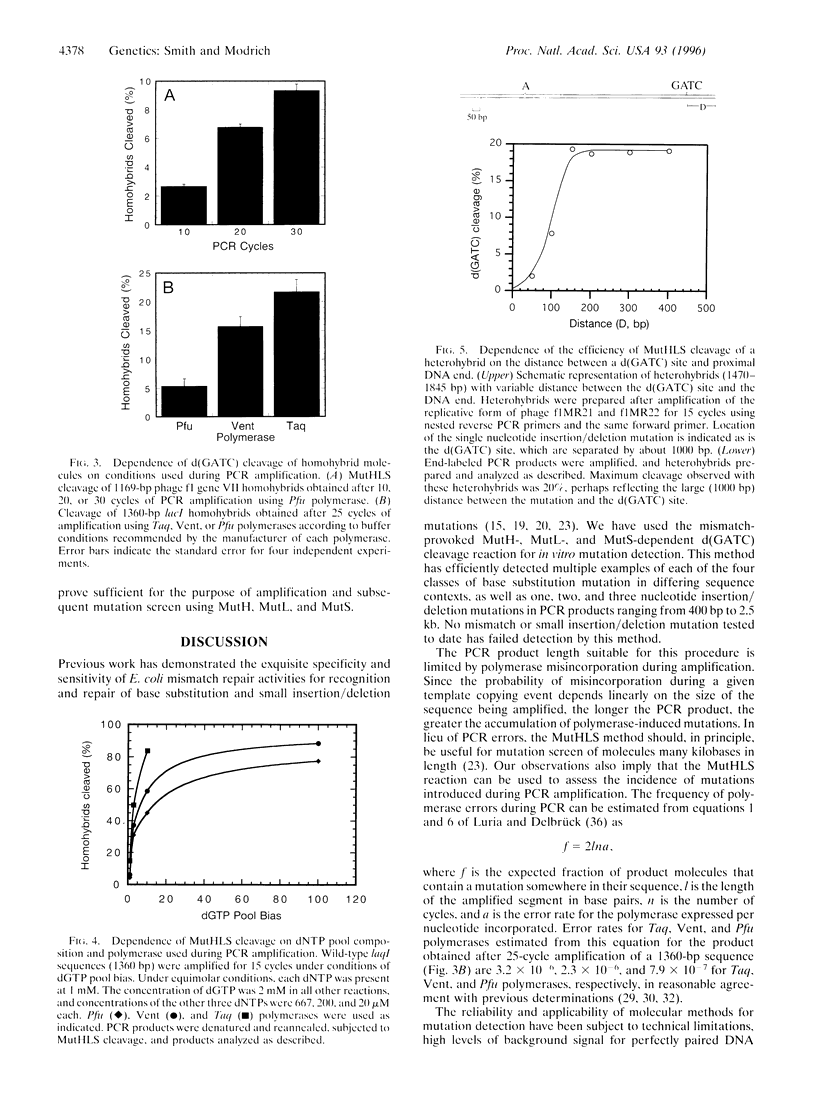
Abstract
Escherichia coli methyl-directed mismatch repair is initiated by MutS-, MutL-, and ATP-dependent activation of MutH endonuclease, which cleaves at d(GATC) sites in the vicinity of a mismatch. This reaction provides an efficient method for detection of mismatches in heteroduplexes produced by hybridization of genetically distinct sequences after PCR amplification. Multiple examples of transition and transversion mutations, as well as one, two, and three nucleotide insertion/deletion mutants, have been detected in PCR heteroduplexes ranging in size from 400 bp to 2.5 kb. Background cleavage of homoduplexes is largely due to polymerase errors that occur during amplification, and the MutHLS reaction provides an estimate of the incidence of mutant sequences that arise during PCR.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Au K. G., Clark S., Miller J. H., Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K. G., Welsh K., Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992 Jun 15;267(17):12142–12148. [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert K. A., Kunkel T. A. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990 Jul 11;18(13):3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L. A., Taylor G. R., Banks R., Baumberg S. MutS binding protects heteroduplex DNA from exonuclease digestion in vitro: a simple method for detecting mutations. Nucleic Acids Res. 1994 Jul 11;22(13):2710–2711. doi: 10.1093/nar/22.13.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R. Fidelity of replication of phage phi X174 DNA by DNA polymerase III holoenzyme: spontaneous mutation by misincorporation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4946–4950. doi: 10.1073/pnas.76.10.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R. A., Caskey C. T. Identification and localization of mutations at the Lesch-Nyhan locus by ribonuclease A cleavage. Science. 1987 Apr 17;236(4799):303–305. doi: 10.1126/science.3563511. [DOI] [PubMed] [Google Scholar]
- Grilley M., Welsh K. M., Su S. S., Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989 Jan 15;264(2):1000–1004. [PubMed] [Google Scholar]
- Hsu I. C., Yang Q., Kahng M. W., Xu J. F. Detection of DNA point mutations with DNA mismatch repair enzymes. Carcinogenesis. 1994 Aug;15(8):1657–1662. doi: 10.1093/carcin/15.8.1657. [DOI] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey-Hoyle M., Steege D. A. Mutational analysis of an inherently defective translation initiation site. J Mol Biol. 1992 Apr 20;224(4):1039–1054. doi: 10.1016/0022-2836(92)90468-y. [DOI] [PubMed] [Google Scholar]
- Jiricny J., Su S. S., Wood S. G., Modrich P. Mismatch-containing oligonucleotide duplexes bound by the E. coli mutS-encoded protein. Nucleic Acids Res. 1988 Aug 25;16(16):7843–7853. doi: 10.1093/nar/16.16.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A. On the fidelity of DNA replication. Effect of divalent metal ion activators and deoxyrionucleoside triphosphate pools on in vitro mutagenesis. J Biol Chem. 1979 Jul 10;254(13):5718–5725. [PubMed] [Google Scholar]
- Lahue R. S., Au K. G., Modrich P. DNA mismatch correction in a defined system. Science. 1989 Jul 14;245(4914):160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lishanski A., Ostrander E. A., Rine J. Mutation detection by mismatch binding protein, MutS, in amplified DNA: application to the cystic fibrosis gene. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2674–2678. doi: 10.1073/pnas.91.7.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Hsu I. C. Detection of single DNA base mutations with mismatch repair enzymes. Genomics. 1992 Oct;14(2):249–255. doi: 10.1016/s0888-7543(05)80213-7. [DOI] [PubMed] [Google Scholar]
- Lundberg K. S., Shoemaker D. D., Adams M. W., Short J. M., Sorge J. A., Mathur E. J. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991 Dec 1;108(1):1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashal R. D., Koontz J., Sklar J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat Genet. 1995 Feb;9(2):177–183. doi: 10.1038/ng0295-177. [DOI] [PubMed] [Google Scholar]
- Matteson R. J., Biswas S. J., Steege D. A. Distinctive patterns of translational reinitiation in the lac repressor mRNA: bridging of long distances by out-of-frame translation and RNA secondary structure, effects of primary sequence. Nucleic Acids Res. 1991 Jul 11;19(13):3499–3506. doi: 10.1093/nar/19.13.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P., Korpela J., Tenkanen T., Pitkänen K. Fidelity of DNA synthesis by the Thermococcus litoralis DNA polymerase--an extremely heat stable enzyme with proofreading activity. Nucleic Acids Res. 1991 Sep 25;19(18):4967–4973. doi: 10.1093/nar/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Novack D. F., Casna N. J., Fischer S. G., Ford J. P. Detection of single base-pair mismatches in DNA by chemical modification followed by electrophoresis in 15% polyacrylamide gel. Proc Natl Acad Sci U S A. 1986 Feb;83(3):586–590. doi: 10.1073/pnas.83.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B. O., Marinus M. G. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R., Li G. M., Longley M. J., Fang W. H., Papadopoulos N., Jen J., de la Chapelle A., Kinzler K. W., Vogelstein B., Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993 Dec 17;75(6):1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- Wagner R., Debbie P., Radman M. Mutation detection using immobilized mismatch binding protein (MutS). Nucleic Acids Res. 1995 Oct 11;23(19):3944–3948. doi: 10.1093/nar/23.19.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh K. M., Lu A. L., Clark S., Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987 Nov 15;262(32):15624–15629. [PubMed] [Google Scholar]
- Winter E., Yamamoto F., Almoguera C., Perucho M. A method to detect and characterize point mutations in transcribed genes: amplification and overexpression of the mutant c-Ki-ras allele in human tumor cells. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7575–7579. doi: 10.1073/pnas.82.22.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youil R., Kemper B. W., Cotton R. G. Screening for mutations by enzyme mismatch cleavage with T4 endonuclease VII. Proc Natl Acad Sci U S A. 1995 Jan 3;92(1):87–91. doi: 10.1073/pnas.92.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]