Abstract
Autophagy is a conserved homeostatic process by which cells degrade and recycle cytoplasmic contents and organelles. Recently autophagy has come to prominence as a factor in many disease states, including inflammatory bowel diseases. In this review we explore the recent discoveries in autophagy and how these relate to the special conditions experienced by the gut mucosa. We will pay particular attention to autophagy as an innate immune process and its role in the development and education of the adaptive immune system.
Keywords: Autophagy, Crohns disease, bacterial infection, systems biology, mucosal immunology
Introduction to autophagy
Homeostasis is the process by which organisms strive to maintain a constant internal environment, despite perturbations in their external circumstances. However, even multicellular organisms with their sophisticated multi-organ systems are unable to guarantee a constant supply of nutrients to all their cells. In addition, cells must also be able to dispose of and recycle damaged or unnecessary components. Therefore autophagy, or self-eating, is a highly conserved process throughout the eukaryotic domain. In higher eukaryotes, with a large number of terminally differentiated, non-dividing cells, autophagy takes on a new importance, since these cells must live without the ability to dilute their contents by division or growth. Thus long-lived, differentiated cells, such as neurons, must be able to turn over their damaged or toxic contents, or risk losing these cells entirely. Originally identified as a response to starvation in yeast, autophagy is now recognized as one of the key processes in cellular homeostasis and response to a variety of stresses, from starvation to infection.
Following identification of the lysosome in the 1950s, it was realized that there existed other specific membrane compartments which eventually fused with lysosomes and yet were distinct from them. These vesicles contained cytoplasmic contents within a single or double-walled membrane. It was quickly recognized that these vesicles formed in the cytoplasm, apparently at random and their cargo was also largely randomly assorted. From these morphological studies arose many questions about the role of autophagy and how it might differ from other vesicular handling and trafficking processes. Genetic studies of autophagy initially benefitted greatly from the tractability of the model organism, Saccharomyces cerevisiaeand ATG1 was cloned in 1997 (1). Within a span of 10 years the total grew to the 31 ATG (autophagy) genes known today (2). Homologues of the key yeast genes were found in mammals and equivalent mammalian models were rapidly established. The field exploded with the creation of reporter constructs fusing GFP to the mammalian Atg8 homologue, LC3, allowing facile fluorescent and real-time microscopy analysis (3).
Today, with a number of knockout mice models, a vast array of cloned and characterized ATG genes and use of cutting-edge microscopy and screening techniques, autophagy is one of the most exiting fields in cellular biology. Autophagy has been placed at the core of a number of disease states, including cancer, neurodegeneration, tuberculosis and Crohns disease. In this review we aim to give the reader both an overview of the field of autophagy, the key pathways and molecules involved, as well an in-depth look at how autophagy relates to inflammatory bowel disease. This final section will touch upon the current state-of-the-art in autophagy, host-microbiome interactions and innate immunity.
Autophagy – subtypes and terminology
Autophagy as a phenomenon can be subdivided into several classes, based upon both cargo selection, size of autophagosome and the use of specific components. In general the term autophagy is used to refer to macro-autophagy, the recycling of cytoplasmic contents via random encapsulation within an autophagosome. However, other types of autophagy have been recognized (Table 1), all sharing the core concept of delivering cargo to the lysosome for recycling. In this review we will restrict our discussion largely to macro-autophagy, since this appears to be the process with which IBD susceptibility is linked and represents the best studied area to date.
Table 1.
Autophagy subclasses
| Major category | Subtype name | Major purpose | Cargo selection | Notes |
|---|---|---|---|---|
| Macroautophagy | Constitutive / basal | Homeostatic | Non-selective | |
| Induced | Stress response – nutrient depletion, toxic protein accumulation or aggregation | Non-selective | It has been proposed that some induced autophagy may be selective, e.g. ribophagy | |
| Xenophagy | Response to infection | Selective – signals largely unknown | Mammalian cells appear to utilize additional specific proteins | |
| Mitophagy | Removal of damaged mitochondria | Selective – signals thought to relate to fusion/fission and membrane integrity | Signals and specific proteins largely uncharacterised | |
| Microautophagy | Chaperone-mediated autophagy | Homeostatic | Selective – cargoes bearing a pentapeptide motif are recognized by hsc70 | |
| Piecemeal autophagy of the nucleus | Homeostatic | Presumed to be selective, since only a subset of nuclear contents are degraded |
Autophagy – the major components and their regulation
There are 31 known autophagy-related genes in yeast and this list does not include all the other proteins involved in vacuolar traffic and fusion, such as Rabs and SNAREs. Thus it is clear that autophagy is a complex and highly regulated process. Despite such complexity the core autophagic machinery is well-understood and largely shared throughout the eukaryotic domain.
The primary regulatory axis for starvation-induced autophagy is the mTOR pathway, which integrates growth factor signals, cellular energy state and other stress signals to control cell growth. Inhibition of mTOR signaling, using rapamycin for example, potently and rapidly induces autophagy. In yeast this is mediated via Atg1 and it is thought that Ulk1 and Ulk2 play this role in mammalian cells (4). The presence of two Atg1 homologues in mammals reflects a general increase in system complexity and specialization over the yeast system, despite the conserved core. Mammalian mTOR forms two complexes, mTORC1 and 2, with mTORC1 being the nutrient sensitive complex responsible for regulation of ribosome and protein biogenesis and cell growth (5). Until recent publications by Hosokawa and Jung, little was known about how mTORC1 regulated autophagy, despite this complex being the target of rapamycin, one of the most commonly-used autophagic stimuli. Hosokawa et al (6) and Jung et al (7) elegantly demonstrate that the mammalian Atg13 homologue, together with ULK1/2 and FIP200 associates with mTORC1 in a nutrient-dependant manner. This association results in phosphorylation of ULK1/2 and Atg13, a process which occurred rapidly upon provision of nutrients. Thus mTORC1 acts to suppress autophagy by sequestering the ULK1/2-Atg13-FIP200 complex. In many ways this is similar to the regulation observed in yeast, although some aspects are altered – in particular it appears that mammalian Atg13 is a regulator, rather than being the signal mediator from TOR to Atg1. In addition Jung et al report that ULK1 and 2 have different affinities for the mTORC1 complex, and that at least three Atg13 splicing variants exist. Finally, another Atg13-binding protein, termed Atg101, has been shown to stabilize Atg13 and bind ULK1 (8). Atg101 is essential for autophagy and therefore provides another potential nodal point of regulation for complex assembly. These data strongly suggest that mammalian autophagy may be controlled with finesse absent in yeast.
At the center of the autophagy apparatus lie two ubiquitin-like conjugation systems, serving to assemble multiprotein complexes and drive formation of the autophagosome. The first of these is termed the Atg5/12 complex and consists of Atg5, 12, 16 as well as Atg7 and 10. In this system Atg7 acts as an E1-like enzyme, activating Atg12, which is transferred to Atg10 (E2) and conjugated with a lysine residue of Atg5. The Atg5/12 conjugate is then associated with Atg16 and brought to the site of autophagosome formation. Here the Atg5/12/16 complex acts as an E3-like enzyme to mediate the lipid-conjugation of LC3 (Atg8), with the assistance of the Atg3/7 complex. This lipidation step results in the covalent attachment of a phosphatidylethanolamine (PE) moiety to the C-terminus of LC3. The assembly of the Atg5/12/16 complex and lipidation of LC3 is thought to mediate extension of the autophagic membrane, and control vacuole size. Following sealing of the autophagosome the external protein complexes dissociate from the autophagosome and are recycled. The Atg5/12 covalent linkage is reversible, allowing both components to be reused; however LC3 is permanently lipidated and therefore remains embedded within the autophagosomal lumen (3). This property has led to the adoption of fluorescent protein fusions to LC3 as the premier markers of the autophagosome and allowed autophagosomes to be identified and tracked from their formation through to their subsequent fusion with lysosomes. LC3-PE formation is also thought to control autophagosome size (3, 9)and its interaction with the ubiquitin-binding protein P62 may influence cargo selectin and loading (10, 11).
Many of the proteins involved in these core assemblies have been successfully knocked out in mice, including Atg3, 5, 7 and 16L1. In the majority of cases homozygous knockout mice die very shortly after birth, unable to mediate the transition from placental nutrition to intermittent feeding (12–14). However, embryonic fibroblast cell lines derived from these mice have been an excellent resource for further investigation of the roles of individual Atg proteins.
The lipid constitution of the nascent autophagic membrane appears to play a key role in autophagosome function. In general autophagosomes lack intra-membrane proteins, are low in cholesterol and thus form “thin” bilayers. However several lipid kinases are known to be crucial for autophagosome formation, including hVPS34, a type III phosphatidylinositol-3-phosphate generating kinase. It is likely, based upon studies in yeast, that binding to PI3P is involved in regulation of autophagy and might mediate specific subtypes of autophagy (15–17). Similarly to yeast, mammalian membrane traffic is controlled by at least three PI3K complexes, all sharing the Atg6 homologue, Beclin 1 (18). The hVPS34 protein, the regulatory subunit p150 and Beclin 1 form the first of these complexes to be identified (19) (20). Beclin 1 was known to act as a tumor suppressor and interact with Bcl-2, an anti-apoptotic protein. It is now known that Bcl-2/Beclin 1 binding negatively regulates autophagy, providing one of the many links between autophagy and apoptosis (21). In addition to VPS34, p150 and Beclin 1 two positive regulators of autophagy are have also been found associated with this PI3K complex, UVRAG and Ambra1 (22–24). However, homologues of yeast Atg14 were unknown until four recent papers described the independent identification of human Atg14 (18, 25–27). In addition to identifying Atg14, Itakura et al demonstrated that UVRAG and Atg14 do not occur together and form two independent complexes with Beclin 1 and Vps34, results corroborated and extended within the other publications. Matsunaga et al and Zhong et al also both identify the protein Rubicon as a suppressor of autophagosome maturation, via its role in the Rubicon-UVRAG-Beclin 1-Vps34 complex. In contrast it appears that the same complex, but lacking Rubicon, is a promoter of autophagosome maturation as has been previously asserted (19). These three separate PI3K complexes appear to perform similar functions to those of yeast, with the Atg14-containing complex required for autophagy and the UVRAG-only complex largely confined to controlling endocytosis, retrograde transport and autophagosome/lysosome fusion. The Rubicon-containing complex (with or without UVRAG) acts to control the rate of vesicle fusion and thus forms part of a control loop to mediate autophgagic flux. Given the rapid identification of these complexes upon application of biochemical and proteomic approaches, it is possible that other PI3K complexes with specific roles may also be identified.
These PI3K studies have also shed light upon autophagosome initiation and the membrane source for the nascent autophagophore. The Atg14-containing PI3K complex appears to localize with very early membrane puncta, prior to recruitment of other autophagosome markers, such as Atg16L1 (18) and is also associated with the ER membrane (27). Therefore autophagosomes may be formed by the action of this PI3K complex upon ER-derived membrane vesicles (28) (much like the pre-autophagosomal structures in yeast), and the subsequent recruitment of further autophagy complexes. This model would make sense since membrane lipid composition, as has been demonstrated in phagocytosis (29, 30), is key to recruitment of specific proteins and manipulation of membrane dynamics.
It is also known that using ectopic membrane-association signals to target autophagy proteins such as Atg16L1 to non-cognate membrane compartments results in formation of autophagic complexes at these sites (31). It is also known that LC3-PE containing membranes are formed even under conditions where autophagosome assembly is curtailed (e.g absence of Atg13, 14, FIP200, Bectin 1 and Vps34), suggesting that without targeting, spontaneous accumulation of LC3-PE on endogenous membrane surfaces occurs. This dependence upon membrane constitution also extends to other components of the autophagic targeting process. For example Atg16L1 associates with the small GTPase Rab33B, in a GTP-dependant manner (32). GTPase-deficient mutants of Rab33B result in constitutive LC3 lipidation and reduced rates of autophagy. Reciprocally, expression of the Rab33B-binding domain of ATG16L1 attenuated autophagosome formation. These results suggest that Rab33B modulates the activity of autophagosome formation, via its interaction with ATG16L1. It is possible that Rab33B recruits the Atg5/12/16 complex to nascent autophagic membranes to initiate membrane growth. Alternatively, Rab33B may promote the activity of Atg5/12/16 to generate LC3-PE upon Rab33-bound membranes, thus increasing affinity of the core autophagy complex for these membranes and resulting in autophagosome formation. Based upon these results it is likely that there are other Rab proteins as well as membrane fusion and trafficking regulators such as SNAREs involved in handling of autophagosomal membranes, both pre- and post-completion.
As shall be discussed in the following sections, many specific autophagy substrates and substrate receptors appear to bind LC3 itself. This suggests that autophagosome formation might begin with extension of a pre-existing LC3-containing membrane. Alternatively, LC3-binding may serve to recruit LC3 and other Atg proteins to the target substrate and, in concert with as-yet-unknown membrane sources, entire autophagosomes are assembled in situ. In addition to these core complexes the other Atg proteins are presumed to be involved in regulating and controlling the rate of autophagy, and some of these, along with other cellular proteins, are used to mediate the targeting events observed for some cargos. Following completion of the autophagosome other proteins and lipid modifications are required for efficient delivery of autophagic cargos to lysosomes, via promotion of lysosome/endosome fusion (24).
Upon interaction with lysosomes the outer autophagic membrane fuses with the lysosome and the inner membrane is degraded, followed by the autophagosomal contents. Live imaging of this process within cells has shown that this fusion is more complex than initially believed and often partial interchange of membrane and luminal contents occurs between autophagosomes and lysosomes (33). Jahreiss et al also indicate a role for dynein and microtubules in mediated the movement of autophagosomes towards the lysosome-rich area around the microtubule-organizing center. To date the role of microtubules in autophagy has been poorly understood, with conflicting reports upon the necessity for an intact microtubule system to mediate autophagy (34–37). The source of the autophagic membrane and order of autophagosome assembly also remain poorly characterized in mammals, hopefully further live imaging studies will yield conclusive answers to such questions.
Lysosomal storage disorders and IBD
Given that autophagy is a system which delivers cellular contents to the lysosome and apparently minor defects in autophagy can result in predisposition to IBD, it might be assumed that lysosomal disorders might also result in IBD. In the pre-genome-wide association era, it had been observed that one lysosomal storage disorder was associated with IBD. Hermansky-Pudlak syndrome (HPS) is an albinism characterized by platelet abnormalities and accumulation of ceroid lipofuscin in parenchymal cells and urinary sediments. HPS is frequently associated with granulomatous colitis and pulmonary fibrosis, the latter is frequently the cause of mortality. HPS colitis is severe in approximately 15% of patients and is often treatable with steroids and other first-line therapy such as Remicaid, however colectomy may be required (38).
Lipofuscin is a breakdown product of proteins and lipids, thought to be composed of polymerized protein residues, formed by oxidation and peroxidation of lysosomal contents (17306211). Once such material forms, it appears to be refractory to further degradation and continues to accumulate newly-formed lysosmal enzymes, depriving the cell of these vital components. As such lipofuscin-containing cells appear to suffer a general reduction in their autophagic capacity and accumulate defective organelles and aggregates. This defective clearance of cellular material may also lead to further accumulation of lipofuscin, resulting in a positive feedback loop, lysosomal rupture and cell death (39). It is known that lipofuscin accumulates as cells age, however elevation of autophagy levels under conditions of oxidative stress (such as tissue repair or infection) rapidly result in elevated levels of lipofuscin, this material is often called ceroid to distinguish it from the age-related form of lipofuscin. In vitro inhibition of autophagy result in lipfuscin deposition (40), however it is unclear whether loss of autophagic efficacy is responsible for lipofuscin accumulation in HPS. It is likely that initial defects in the lysosome predispose cells to loss of autophagic ability, through the accumulation of defective, lipofuscin-loaded lysosomes. Thus development of colitis may reflect a reduction in productive autophagic rates in the cells of the gut. Mechanisms of IBD pathology would then mimic those seen in other patients with reduced autophagic capability, as discussed in the section “Autophagy and the intestinal epithelium”, below. This area deserves further study and the detailed mechanisms of lysosome regulation and how these impact autophagy discerned.
Autophagy and ubiquitination – partners in protein turnover
The naïve view that proteasome and autophagic degradation were separate arms of protein turnover has been replaced by discoveries of both shared substrates and regulatory interplay between the two systems.
Since 1995, when Cuervo et al demonstrated that proteasomes themselves were degraded by autophagy, it has been known that proteasome and autophagy are intimately linked (41). The major hub of autophagy/proteasome interaction is the ubiquitin-binding protein p62 (SQSTM1), which was observed to co-localise with cytosolic protein aggregates and LC3, suggesting a role in autophagy (42). It is known that these poly-p62 aggregates also contain ubiquitin, form around mutant huntingtin proteins and are degraded by autophagy. Loss of autophagy results in accumulation of such p62 inclusions and these are associated with liver injury and neurodegeneration (42). In the absence of both autophagy and p62, however, aggregate formation is attenuated and some of the subsequent cellular damage is also ameliorated (43). Recent data demonstrate that this LC3 binding is specific and mediated by recognition of LC3 by a conserved sequence within p62. Through this interaction the levels of cytoplasmic p62 are controlled by autophagy and thus autophagy also controls inclusion body formation (11). Kim et al have shown that mono-ubiquitination of diverse protein substrates is sufficient to confer efficient p62-mediated trafficking through the autophagosomal pathway to lysosomes (44). They demonstrated that this process could be blocked by inhibition of autophagy via 3-MA treatment, as well as depletion of Atg12, thus confirming that it is an autophagy-mediated process.
However, p62 is not the only ubiquitin-binding protein which acts as a receptor for autophagic cargo. Kirkin et al identified NBR1 as a second autophagic ubiquitin receptor (45), by screening a human cDNA library for proteins capable of binding yeast Atg8. Similarly to p62, NBR1 forms oligomers and it is capable of forming heteromers with p62 itself. NBR1 is able to both bind LC3 and be degraded by autophagy, independently of p62. However, it appears that p62 and NBR1 cooperate in both aggregate formation and autophagy targeting, since knockdown of either ubiquitin-binding protein resulted in reduced aggregate formation upon puromycin treatment. Therefore there exist at least two ubiquitin-binding, autophagic cargo receptors in human cells, with overlapping roles in substrate selection. It is plausible to speculate that p62/NBR1-binding to ubiqutinated substrates may serve as a general signal for autophagic disposal, although the circumstances under which this takes precedence over proteasomal degradation are unclear. Presumably aggregates are less suitable for proteasome disposal and are primarily degraded via lysosomal delivery via autophagy.
The picture is further clouded by another discovery indicating the complex interplay between the proteasome and autophagy, via p62. Inhibition of autophagy, by depletion of Atg5, 7 or 12 resulted in accumulation of proteasome substrates (46). This increase in non-degraded material was associated with (and could be mimicked by) an increase in the levels of p62, which slowed passage of ubiqitinated targets through the proteasome-mediated degradative pathway. Knockdown of p62 restored the proteasome to full efficiency, indicating that p62-bound proteins are less able to be delivered to the proteasome. The mechanism behind this delay is not known, but masking of ubiquitin by p62 binding, as well as formation of larger ubiquitinated protein/p62-oligomers cannot be ruled out.
Recently, in the model organism Caenorhabditis elegansa novel selective autophagy pathway was identified. P granules are structures associated specifically with germ cells and are removed from non-germ cells both by segregation into germ precursors during development and clearance from somatic cells. This clearance of a specific protein aggregate provides an opportunity to study autophagic cargo selection. Using a genome-wide RNAi strategy Zhang et al (47) determined that P granule clearance from somatic cells is dependant upon the C. elegans Atg8 homologue. Mutants in this, and other components of the autophagy apparatus, phenocopied the RNAi-treated worms. Using a suppression screen in autophagy-mutant worms they identified 11 mutant alleles of the protein SEPA-1 (Suppressor of Ectopic P-granule in Autophagy mutants). Further analysis revealed that SEPA-1 is required for autophagic clearance of P-granules and this action is dependant upon interactions with PGL3 (a P granule protein) and the C. elegans Atg8 homologue. Thus, like p62 and NBR1, SEPA-1 binds both cargo and the autophagy protein Atg8 (LC3). Interestingly the authors suggest that SEPA-1 acts to remove P-granule components prior to their formation of aggregates, underlining the role of basal autophagy in controlling levels of aggregate-prone proteins and suggesting that these proteins may have specific receptors for their delivery to autophagosomes. These studies demonstrate that even though autophagosomes are formed in the cytoplasm in a stochastic fashion, there may operate a heretofore unseen level of cargo selection. Whilst the receptors described so far are largely general, binding to ubiquitin and Atg8/LC3, there may well be uncharacterized cargo-specific receptors, possibly with roles restricted to autophagy subtypes such as mitophagy.
Organelle destruction – selective, or stochastic?
As outlined in the introduction, autophagy serves to remove and recycle many organelles, including mitochondria and peroxisomes. It has been assumed that mitochondria are selectively removed when they become damaged, but it is unknown whether a basal level of non-selective mitophagy also occurs. Recently Nerendra et al identified Parkin as a protein recruited to damaged mitochondria and resulted in autophagic removal of such organelles (48). Parkin possesses a ubiqutination activity, but it is currently unknown whether this is required for selective autophagy of mitochondria. It is also still unclear whether Parkin itself serves as a sensor of mitochondrial damage, or is responding to other factors such as Pink1, an outer membrane kinase with chaperone substrates known to play roles in mitochondrial quality control. Alternatively Parkin may be recruited in its role as an ubiquitin ligase to unfolded proteins on the mitochondrial surface, and thus via ubiquitination of these targets induce autophagy through ubiquitin-binding receptors such as p62 and NBR1. Parkin has been implicated as a candidate for early-onset Parkinson’s disease, since it is mutated in almost 50% of heritable and 10–15% of sporadic early-onset Parkinson’s. In Drosphila models a loss of Parkin or Pink1 results in extensive mitochondrial fusion, a phenotype rescued by overexpression of the fission GTPase Drp1. In mammalian cells subjected to knockdown of Pink1, mitochondria are smaller and more fragmented (49), and cells show enhanced reactive oxygen generation and mitophagy. Knockdown of Atg7 and Atg8/LC3 also yielded a change in mitochondrial morphology, with a loss of extensive fission and an increase in cell death. Expression of Parkin was able to largely ameliorate mitochondrial defects induced by loss of Pink1 and restored mitophagy to rates comparable to control cells. These studies strongly suggest that early-onset neurodegeneration in Parkinson’s is associated with mitochondrial dysfunction. Therefore promotion of selective mitophagy might reduce or delay some of the neural damage seen in this devastating disorder.
The role of mitophagy in maintaining the balance of cellular functions is also seen in models of viral infection. Using vesicular-stomatitis virus infection of Atg5-deficient and replete MEFs Tal et al (50) showed that loss of autophagy is associated with a significant upregulation of reactive oxygen species-dependant RIG-I-like receptor signaling. RIG-I-like receptors are innate immune recognition molecules which recognize single-stranded RNA in the cytoplasm. It is known that dendritic cells rely upon autophagic delivery of ssRNA to TLR7-containing compartments for anti-viral response initiation (as described previously), but this is the first study that implicates autophagy in RIG-I-mediated cytoplasmic ssRNA detection. This enhancement of immune response was shown to rely upon increased levels of reactive-oxygen species (ROS) induced by accumulation of damaged mitochondria. Treatment with ROS-inducing agents resulted in enhanced interferon production following transfection with Poly I:C ligands in both wild-type and Atg5−/− cells. This study provides further evidence that loss of autophagy has diverse immune consequences, here enhancing accumulation of ROS and predisposing cells to elevated levels of innate immune signaling. This might result in increased immunopathology upon exposure to minor viral infections, and the consequences for other modes of immune signaling are still to be determined.
Adaptive immunity is also affected by reductions in autophagy. Microarray analysis of transcription in Atg5+/+ compared to Atg5−/− thymocytes revealed a pronounced mitochondrial signature (51). This Atg5-dependant gene set contained 64 genes with known associations with mitochondrial function (via Gene Ontology). T cells and thymocytes derived from Atg5−/− precursors exhibit a gain in total mitochondrial mass. This fits with the studies described above, where autophagy results in accumulation of mitochondria, both healthy and damaged. In mice, Atg5- or 7-deficient T cell and thymocytes are markedly reduced in numbers and T cells show reduced survival. Mitochondrial mass was also positively correlated with cell death, suggesting a linkage between loss of mitochondrial homeostasis and T cell death. In a recent publication Pua et al (52) demonstrate that this defect is due to loss of autophagic control of mitochondrial number in mature T cells. Upon exit from the thymus T cells move from high to low mitochondrial mass status. In the absence of autophagy (due in this case to Atg7 deficiency) this shift does not occur and therefore cells exhibit increased reactive oxygen stress and an imbalance in expression of pro- and anti-apoptotic signals. Interestingly this developmentally regulated mitochondrial switch is not observed in B lymphocytes, but autophagy is required for survival of pre-B cells and mature B1 B cells (53).
Together these data suggest a role for autophagy in controlling mitochondrial homeostasis through constitutive remodeling of mitochondrial morphology – with mitophagy removing damaged organelles. It is also possible that mitochondria are split by autophagy into fragments, sparing undamaged portions and removing damaged sections. Thus autophagy preserves proper mitochondrial function and regulates the ROS balance to maintain correct functioning of numerous cellular processes, from ROS signaling to innate and adaptive immunity.
Peroxisomes are small, single-membrane-bound organelles which are responsible for oxidation of fatty acids, cholesterol synthesis and ridding the cell of toxic peroxides. In mammals pexophagy, the autophagic destruction of peroxisomes, has been shown to be controlled by ubiquitination, via p62 (44). In addition, the peroxisome protein Pex14p is thought to associate with LC3 and be required for pexophagy upon recovery from nutrient starvation (54). During nutrient-rich conditions Pex14p is bound to Pex5 at the peroxisome membrane, however upon starvation LC3-II is able to bind Pex14p and mediate pexophagy. In the absence of autophagy peroxisome proteins are broken down via the proteasomal pathway, however pexophagy increases the rate of removal of peroxisomal proteins during starvation recovery. Thus pexophagy may serve to mediate rapid turnover of peroxisomes, clearing damaged and potentially toxic proteins from the cell and restoring normal cellular nutrition. Interestingly the interaction between Pex14p and LC3 was reported to be microtubule dependent, the roles of microtubules in autophagy have been controversial and apparently vary according to cell type and cargo (33–37). Perhaps in cells which accumulate larger quantities of peroxisomes microtubules are important for autophagy of these organelles, whilst in other contexts they are more dispensible. In the fission yeast Pichia pastoris pexophagy requires the novel protein ppAtg30 (55), a protein with no currently identified homologue in mammalian organisms. In P. pastoris Atg30 interacts with both Atg11 and 17 and is required for both micro and macro-pexophagy (micropexophagy in yeast occurs when the vacuole engulfs peroxisomes, macropexophagy is akin to the mammalian process and involves formation of a separate membrane, followed by transportation to the vacuole). Peroxisome formation in yeast is extremely robust when grown upon methanol as a sole carbon source and subsequent supplementation with alternative nutrition leads to high levels of pexophagy. It is possible that higher eukaryotes also have an equivalent molecule responsible for marking cargo and controlling selective pexophagy. Alternatively, given that multicellular organisms are rarely required to subsist upon methanol and similar carbon sources, the ubiquitin-mediated mechanisms discussed above may be sufficient.
In recent years there has been a clear link elucidated between ER stress and autophagy, with autophagy thought to participate in recovery from ER stress and induction of the unfolded protein response (UPS). It has also been observed that during ER stress autophagosomes can be found which contain ER-derive proteins and membrane structures, a process termed reticulophagy (56). This suggests that autophagy may directly degrade portions of the ER to maintain protein quality control and eliminate areas of highly-stressed ER. However, the signals by which this selective engulfment takes place have not been fully elucidated to date. It is thought that reticulophagy relies upon at least one of the three ER stress signaling systems present in mammals – the inositol-requiring enzyme 1 (IRE1), a conserved RNA splicing factor required for the transcription of genes involved in the unfolded protein response (57). Other studies have implicated the other two canonical ER stress signaling systems in reticulophagy; the protein kinase regulated by RNA-like ER kinase (PERK) (58), and activating transcription factor 6 (ATF6) systems (59).
Autophagy in Crohns disease – genetic association studies
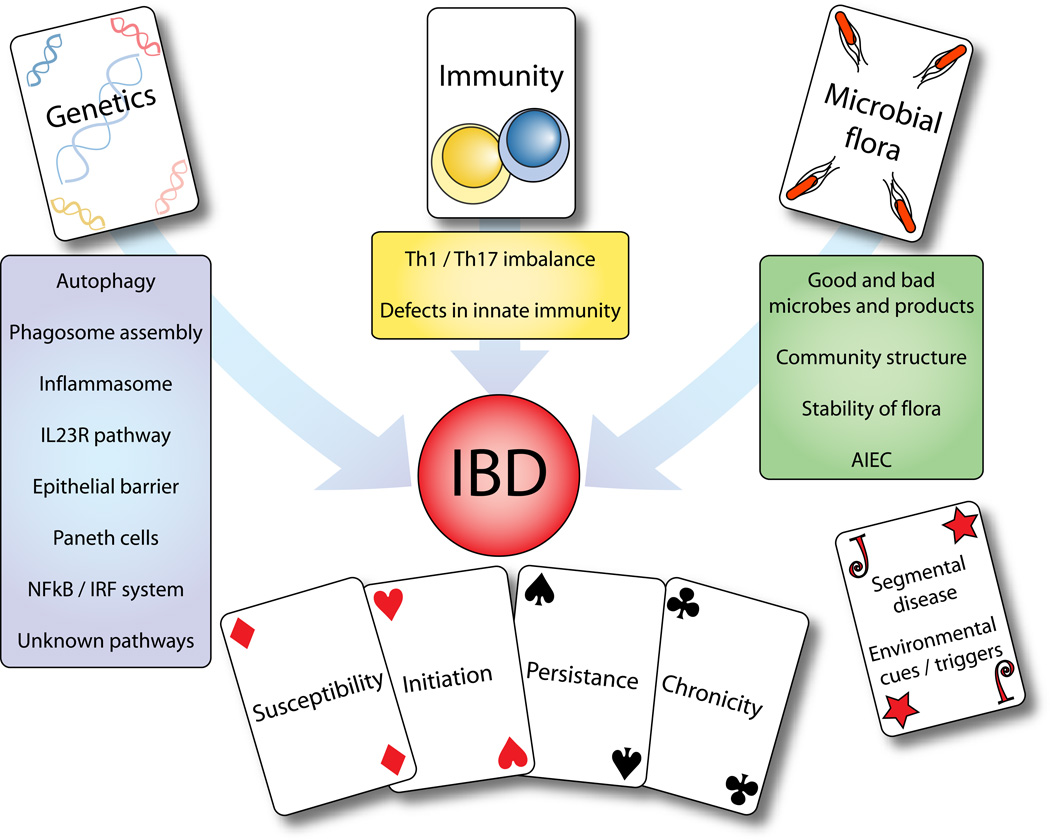
The advent of rapid, cheap and flexible genotyping technologies have opened the possibility of recruiting large cohorts of patients and healthy controls and analyzing their genetics in an unbiased, systematic fashion. The increase in power to find genetic associations given by this approach has enabled a deep analysis of complex, multigenic diseases. In the case of Crohns disease (CD), the number of associated genetic variants has grown from two (Nod2 and the IBD5 locus) to 32 in just five years (60). This explosion is even more impressive given that many of the novel associations are able to be placed with a high degree of confidence to a limited number of single-nucleotide polymorphisms within a genomic region. This enables powerful follow-up studies using deep sequencing, a vast improvement over previous linkage-based analysis where causal variants have not been identified, despite the regional association having been known for nearly a decade (in the case of IBD5). Thus our knowledge of inflammatory bowel disease and CD in particular, has vastly increased and several cohesive themes have emerged as a result. Our current understanding places genetics, innate immunity and microbial interactions as crucial components of IBD susceptibility, however there are many unanswered questions including what factors trigger disease onset and affect the distribution of inflammation within the gut (Figure 1).
Figure 1. Themes in IBD susceptibility.
Our current knowledge of IBD susceptibility yields three main themes: genetics, immunity and microbial flora. Modern genetic techniques have allowed identification of several interactions and pathways, including autophagy. However each of the known genetic variants contributes only a modest amount to overall susceptibility. Immunity has been though as central to IBD for many years, but now the focus is firmly upon innate immunity and triggers of inflammation, and how this relates to skewing of the Th1/Th17 axis of the adaptive immune system. Our knowledge of the microbiome and its interactions with the gut is in its infancy. The microbial flora is likely to be crucial for IBD susceptibility and pathology, both as triggers of initial inflammation as well as the long-term alterations in flora induced by ongoing immune responses. Combined with unknown environmental triggers and other contributors to disease location and pathology, these factors combine to give each individual a unique risk profile.
So far two genes within the autophagy pathway have been identified and replicated as being associated with CD. The presence of two such genes, ATG16L1 and IRGM, in just 32 associations points towards a strong role for autophagy in the pathogenesis of CD.
The associations observed are very different in functional consequence however, the ATG16L1 risk signal corresponds to a single amino acid change in the protein sequence, a threonine to alanine substitution (T300A) (61). In the case of IRGM, no coding variants have been identified, but a large deletion in the promoter region is correlated with the identified risk allele (62), suggesting an expression-level polymorphism.
ATG16L1
Both Atg16l1 and Irgm are essential for the autophagy of intracellular pathogens (61, 63–65), and Atg16l1 is also required for other forms of autophagy. However, the identified risk- and protection-associated Atg16l1 variants appear equally able to mediate basal levels of autophagy, and both effectively rescue starvation / rapamycin induced autophagy following knockdown of endogenous Atg16l1 expression. However in a model of intracellular pathogen infection using Salmonella enterica serovar Typhimurium, the risk-associated Atg16l1 variant is less able to mediate anti-bacterial autophagy and appears less stable (64). Therefore it would appear that Atg16l1 variant stability is altered by both autophagic load and other cellular factors associated with infection. Thus tissues with either a high endogenous autophagic load, or exposed to a high bacterial burden, might be amongst the first to exhibit defects in autophagy due to loss of Atg16l1 stability. Recent data using mice hypomorphic for Atg16l1 has shown that under normal husbandry conditions defects appear largely confined to the gut, where paneth cells appear to be the most affected cell type (66). The phenotype observed includes defective anti-microbial peptide secretion (degranulation) and loss of large apical portions of paneth cells via an apparent secretory defect. A similar reduction in secreted antimicrobial peptides was observed in human biopsies from CD patients, suggesting a common pathogenesis between human disease and the hypomorphic Atg16l1 model.
In another mouse model, this time expressing a truncated Atg16l1 protein, defective in mediating autophagy, Akira and colleagues demonstrated an elevated IL-1B response in macrophages exposed to LPS (67). This enhanced response was related to elevated levels of Caspase-1 (caspase-1 cleaves pro-IL-1B to yield active cytokine), following exposure to LPS or extracellular bacteria. Chimeric Atg16l1-deficient mice were also highly susceptible to DSS colitis, a phenotype partially rescued by administration of anti-IL-1B and IL-18 antibodies. Thus production of pro-inflammatory cytokines by host macrophages in response to LPS appears to be regulated by autophagy. These findings implicate a crucial role for autophagy in regulating the inflammation, especially in the gut where exposure to LPS is both constant, and required for proper healing and homeostasis (68).
These insights reinforce the idea that subtle defects in autophagy are likely to be reflected only in organs where homeostasis imposes large or extremely dynamic loads upon the autophagic machinery. Indeed it is unlikely that patients exhibit severe defects in autophagy, since these would be rapidly lethal.
IRGM
IRGM represents one of two human orthologues of a large family of mammalian interferon-regulated GTPases. In mice there are 20 such IRG proteins expressed and many of those studied to date have been shown to provide host defence against intracellular pathogens (63, 65, 69, 70). In humans the expression of the IRGC gene is thought to be restricted to the testis whilst IRGM, a truncated protein (compared to other mammalian irgs), is widely expressed. IRGM has been shown to be essential for autophagy of the intracellular pathogens S. Typhimurium and Mycobacterium tuberculosis (62, 63) and this activity is associated with expression level (62, 71). In the tuberculosis model, macrophages harboring BCG (a commonly-used variant of M. tuberculosis var. bovis with reduced virulence in humans), were able to clear intracellular bacteria via induction of autophagy in an IRGM-dependant manner (63, 72). This IRGM function was associated with IRGM localization to bacteria-containing compartments and appeared to be common to both human IRGM and mouse IRGM1 in the species-appropriate macrophage cell lines. IRGM-mediated autophagy resulted in mycobacteria-containing vacuoles acquiring lysosomal markers and vacuole acidification, indicating that these bacterial vacuoles were fused with lysosomes to effect anti-bacterial control.
Two studies examining genome-wide associations for CD identified SNPs close to IRGM as being strongly correlated with disease (61, 73). Subsequent detailed analysis of IRGM and the surrounding genomic region identified the functional polymorphisms associated with CD are in perfect linkage with a large (20 kb) deletion of potential regulatory sequence (62). Analysis of allele-specific expression showed a complex regulatory picture, with different heterozygous tissues and cell lines exhibiting varying levels of expression of the two alleles. It is possible that regulation of IRGM is cell-type specific and thus altered regulation induced by the CD-associated deletion may be reflected by functional deficits in specific cell types. Alternatively, IRGM is predicted to exhibit alternative splicing, with at least five splice variants. The protein products of the splice variants have predicted molecular weights of between 19 and 24 kD and their expression at the protein level has not been documented. In most cell types studied to date, expression of IRGM transcripts is very low and detection of the endogenous protein has been difficult. We also know from studies of knockout mice that defects in immunity induced by loss of IRGM1 can be largely rescued by deletion of IRGM3 (70). Therefore there may be circumstances where high-level IRGM expression compromises host resistance in the face of an infectious challenge.
The IRG gene family has a rich evolutionary history, whilst most non-primate mammals studied have undergone significant gene duplication and diversification events to generate up to 21 interferon-regulated IRG genes, humans have only two. Neither of the human genes appears to be regulated by interferons and do not possess canonical interferon-responsive promoter elements. In a recent paper Beckpen et al undertook to examine the evolutionary history of the IRGM locus in the primate lineage (74). They revealed a fascinating story of gene loss and resurrection under selective pressures and over deep evolutionary time. Prosimians, such as the two sequenced lemur species (Microcebus murinus and Lemur catta) have three IRG genes, organized in a tandem gene family as seen in the mouse. Analysis of Old world and New World monkey species has revealed that IRGM became a non-functional pseudogene in the common ancestor of these monkey species around 35–40 million years ago. This loss of IRGM was due to integration of an Alu repeat sequence, disrupting the ORF. However, expression of a truncated ORF reappears in humans and African Great Apes. This restoration is accompanied by insertion of a retroviral element into the IRGM ORF, this results in a start site within the ERV and addition of a retrovirally-derived 5’ UTR. There is also preliminary evidence that the deletion polymorphism identified by Mcarrol et al (62) results in alterations to the relative levels of IRGM splice variants, thus opening the door for further study of the role of splicing and CD susceptibility. The evidence accumulated by Beckpen and colleagues demonstrate that IRGM is subject to significant and possibly conflicting evolutionary pressures. Orangutans and gibbons appear to possess both functional and pseudogenic IRGM alleles, whilst humans have fully resurrected the functional allele to express a truncated IRGM transcript (possibly with cell-type specificity and specific splicing). It is tempting to speculate that differential pressures such as the high prevalence of tuberculosis in the human population might have selected for IRGM resurrection, although further studies will be needed before such theories can be tested.
Identifying the complete autophagic interactome – chemical and genetic screening
Historically the discovery of novel biological pathways or processes has led to a drive towards finding all the components both directly involved and impinging upon such processes, both by candidate-selection and screening approaches. There is a general consensus that autophagy in higher, multicellular eukaryotes is likely to exhibit increased complexity of operation and regulation than in yeast. Therefore there have been a number of attempts to utilize screening approaches to identify novel regulatory players and pathways as well as increase the available toolset for both modifying and monitoring autophagy.
Firstly we will consider the use of small molecules as probes to identify pathways involved in the regulation of autophagy. It has been known for many years that small molecules such as rapamycin and 3-methyladenine have profound effects upon the rate of autophagy, elevating or reducing the rate of autophagosome formation, respectively. The mechanism of autophagy induction by rapamycin, through its effect upon mTOR (a crucial integrator of cellular nutritional state), has been extensively studied. Indeed, the mTOR pathway has been mapped out in detail, mostly independently of the autophagy field, largely due to interest in its role in cell growth and insulin signaling. This suggests that finding the targets of autophagy-altering small molecules might reveal additional regulatory pathways and the molecules within these novel networks.
With the current maturity if high-throughput technologies it has become possible to conduct such small molecule screens rapidly and sensitively and several recent publications demonstrate the power of such approaches. Zhang et al utilized a microscopy-based screen to identify eight novel compounds with potent autophagy-inducing effects (75). Seven of these compounds were FDA-approved drugs (albeit for apparently unrelated human diseases), and exhibited the ability to enhance the degradation of proteins carrying expanded polyglutamine repeats. Accumulation of polyglutamine-rich proteins is a hallmark of Huntington’s disease and therefore the ability to effect clearance of such molecules might yield clinical benefit.
Rubinszteins et al utilized a different approach, identifying in a yeast model enhancers or inhibitors of rapamycin-induced cell growth arrest. From a set of 32 primary screen hits, they were able to identify 13 compounds delaying clearance of mutant alpha-synuclein in a mammalian cell model, as well as four compounds which enhanced protein clearance. Further testing revealed three of these compounds to be able to enhance autophagy in several mammalian cell models as well as in a Drosophila model of neurodegeneration.
Rubinsztein has also performed a similar screen to that of Zhang et alhowever in this case the primary readout was the clearance of alpha-synuclein from a neuronal cell line (76). In this case the compounds selected for validation were three calcium channel antagonists, a potassium channel opener and a G(i) signaling activator. Using these drugs they identified an mTOR-independent cycle controlling autophagy. This cycle initiates with cAMP; elevated cAMP levels result in increased calpain activity and reduced autophagy. Calpain also cleaves G(s)alpha, resulting in its activation and completing the cycle, since G(s)alpha itself regulates cAMP levels.
These studies demonstrate the power of high-throughput chemical biology to identify novel pathways and regulators of cellular processes, in this case autophagy. In a very short time the number of autophagy-modulating compounds identified has exploded, from trehalose, rapamycin and 3-methyladenine to a wide range of drugs and small molecules. Encouragingly many of these compounds have therapeutic potential and exploit a diverse range of cellular targets, greatly increasing our knowledge of autophagy regulation.
Contemporaneously with these small molecule-based screens, others have developed novel tools for use in identifying autophagy-related components via genetic screens. The use of automated microscopy to monitor the cellular distribution of GFP-LC3 has been used by several groups, and a recent paper from He et al demonstrates the power of the approach. Using a 1050 clone cDNA human library the authors identified three genes whose overexpression resulted in elevated autophagosome formation (77). Interestingly all three were transmembrane proteins, and follow-up of a single candidate revealed that TM9SF1 was required for optimal induction of starvation-mediated autophagy. TM9SF1 is thought to be a ubiquitously-expressed transmembrane protein and shows a high level of sequence conservation with its yeast orthologue, suggesting a conserved role in an important cellular process.
This theme of conservation of gene function can be exploited to find novel components of the human autophagy apparatus based upon their known roles in yeast. The yeast protein interactome has been exceptionally amenable to investigation, primarily due to the ability to utilize two-hybrid methods with “endogenous” proteins. This has resulted in a number of large, well-defined datasets which can be mined to construct protein-protein interaction networks. We used this approach to generate a yeast protein interaction network centered around the “core” autophagy apparatus, capturing many of the characterized interaction described in the first section of this review. We then “humanized” this network by mapping the interactors to their human orthologues. By this approach we identified 14 human proteins whose involvement in autophagy was previously uncharacterized, several of which had putative roles in membrane organization or dynamics (78). Detailed examination of one of these candidates, FNBP1L, an ATG3 interactor, showed that it played a crucial role in anti-bacterial autophagy, but was dispensable for classical autophagy induced by starvation or rapamycin. FNBP1L contains an F-BAR domain, thought to be able to sense and/or effect membrane bending. In addition, the binding of FNBP1L and ATG3 led us to propose a model whereby FNBP1L acts as a scaffold for ATG3, holding the ATG3/7 complex at the growing autophagosome. Since autophagosome size is dependant upon the supply of LC3-PE created by Atg3 (14, 79), it is likely that anti-bacterial autophagy requires more LC3-PE than classical vesicle formation. By tethering ATG3/7 close to the cargo FNBP1L would allow local generation of high levels of lipidated LC3, and drive formation of the large anti-bacterial autophagosome.
So far these screens have exploited either specific autophagic substrates e.g. alpha synuclein, S. Typhimurium, or used automated microscopy to quantify punctate LC3 structures. Both of these methods have considerable drawbacks when it comes to finding general regulators of autophagy. Using specific substrates or cargos will yield only components involved in handling those cargos, and as the FNBP1L story illustrates these may not be generalisable to other autophagic contexts. Of course the specificity of these components is also of great interest and cargo-specific screens are likely to yield important insight, especially into the mystery of cargo selection. However a broader approach would allow capture of all involved components, and could be extended to cargo-specific secondary screens.
Microscopy analysis is much slower and more laborious than a simple enzymatic readout (e.g. luciferase or B-galactosidase) and requires sophisticated software analysis. Therefore, it would be of value to obtain a general readout of autophagic activity amenable to high-throughput analysis. Recently Ketteler et al described just such a sensor (80, 81). Using a luciferase tethered to actin by the LC3 protein, they were able to construct a biosensor which releases luciferase into the culture supernatant upon LC3 cleavage by ATG4B. ATG4B is the endogenous protease which serves to cleave pro-LC3, prior to its conversion to LC3-PE. Therefore elevated levels of LC3 cleavage are correlated to luciferase levels in the growth medium. The stability and high activity of the Gaussia luciferase used allows the use of extensive automation, without the need for microscopic analysis. Kettler et al demonstrated proof-of-concept by treatment of cell lines with shRNAs to either ATG4B or AKT, an kinase upstream of mTOR. Knockdown of ATG4B resulted in loss of luciferase activity, as predicted through loss of LC3-cleavage. AKT knockdown yielded increased levels of autophagy-mediated luciferase, an effect of similar magnitude to that of rapamycin treatment.
This tool is likely to prove extremely valuable, since luciferase-based screens have a proven track record in other settings, due to their sensitivity and reproducibility (82).
Autophagy and microbes
Mucosal surfaces represent the primary interface between the human body and the microbial world. Following birth our mucosa are rapidly colonized by microbes in incredible quantity and diversity (83), and these organisms are kept at bay through constant immuno-vigilance. Despite such surveillance there are numerous pathogens capable of infecting otherwise healthy hosts and clearance of such invaders requires the initiation of powerful inflammatory cascades. It has recently become clear that autophagy is a cell-autonomous response to infection by many microbes and also plays a role in controlling the innate and adaptive immune responses to infection. In this section we will consider some of the diverse range of microbes which have evolved to infect via mucosal surfaces and their relationships to the host autophagic response. Table 2 shows a selection of mucosal pathogens for which a role of autophagy in their pathogenesis has been shown.
Table 2.
Mucosal pathogens and roles of autophagy
| Pathogen | Infection route |
Disease | Role of autophagy | Putative triggers or signals |
Known autophagy evasion / subversion strategy |
|---|---|---|---|---|---|
| Salmonella enterica serovar Typhimurium | Oral | Food poisoning | Control of replication in epithelial cells | Type III secretion-mediated vacuolar damage | Intracellular shutdown of type III secretion system |
| Shigella flexneri | Oral | Dysentery | Control of intracellular replication | Binding of ATG5 to the bacterial protein VirG | Masking of VirG via binding of IcsB and subsequent actin motility |
| Mycobacterium tuberculosis | Inhalation | TB | Clearance of bacterium from macrophages | Location of IRGM to M. tuberculosis-containing vacuoles | Unclear, although endogenous levels of IRGM appear insufficient to mediate effective clearance |
| Listeria monocytogenes | Oral | Listeriosis | Killing of intracellular listeria in infected macrophages | Listeriolysin-damaged vacuoles | Actin motility and phospholipase activity, as well as vacuolar lysis via lysteriolysin O |
| Legionella pneumophila | Inhalation | Legionnaire’s disease | Killing of phagocytosed bacteria by macrophages | Unknown type IV-secreted proteins, sensing by murine Birc1e/Naip5 | Remodeling of phagosome and subsequent fusion with ER compartment |
| Poliovirus | Oral | Polio | Autophagic membrane provides viral replication site | Unknown | Autophagy appears required for viral replication |
| Chlamydia trachomatis | Sexually transmitted | Urethiritis, trachoma, pelvic inflammatory disease | Autophagy provides protection in a MEF model of infection | Accumulation of Irga6 protein promotes autophagocytosis of bacterial inclusions | Irg protein induction is IFN-g dependant, evasion of IFN induction may play a role |
| Vibrio cholerae | Oral | Cholera | Autophagy protects cells against secreted cholera cytolysin | N/A | N/A |
| Vibrio parahemolyticus | Oral | Acute gastroenteritis | Induction of autophagy leads to cell death | Uncharacterized type III-secreted effectors | Cell lysis releases nutrients for replication |
| Porphyromonas gingivalis | Oral | Periodontitis | Internalized bacteria enter autophagosomes | Unknown | Bacteria appear to survive within autophagosomes, in the absence of lysosomal fusion |
| Burkholderia pseudomallei | Inhalation | Melliodosis | Autophagy induction appears to restrict replication and promote clearance | Unknown | Evasion of autophagy is dependant upon synthesis of the Type III-secreted protein BopA |
Autophagy has been observed to serve as an anti-microbial process in many model infections. A diverse sampling of infections in which autophagy has been implicated are outlined, demonstrating both what is known about autophagy during host-microbial interaction and how much is still to be determined.
In the majority of cases studied so far autophagy appears to be a protective, anti-microbial response which serves to compromise the niche of many intracellular pathogens. However, there are cases where autophagy is subverted to increase the replication rate of both bacteria and viruses or the autophagosome is arrested to yield a novel replication niche, e.g. poliovirus. The majority of the described interactions initiate in the gastrointestinal tract, a unique site under constant exposure to bacteria and their products. It has been known for many years that the gut represents a largely tolerant environment, and that immune responses to many highly immuno-stimulatory molecules are heavily dampened (84). Inflammatory bowel disease has often been characterized as a failure of such dampening or “loss of tolerance” to the normal gut flora and antigens, resulting in inappropriate immune activity. What roles might autophagy play in this context?
In many of the autophagy anti-microbial models studied to date autophagy appears to target pathogens within damaged vacuoles, either as pathogens attempt to escape into the cytoplasm e.g. Listeria monocytogenesor fail to maintain their vacuolar niche e.g. Salmonella Typhimurium. The signals by which the autophagic apparatus detects such vacuolar damage are unknown, but several possibilities exist: recognition of ubiquitinated proteins upon the microbial surface; direct binding of autophagy adaptors/scaffolds to membrane leaflets of specific lipid compositions; sensing of microbial-derived molecules (pathogen-associated molecular patterns) by innate immune factors and subsequent recruitment of the autophagic apparatus. In mice it is known that the interferon-induced Irg protein family play a role in autophagic recognition of pathogen targets, however, as discussed in the IRGM section of this review, the picture in humans is less clear. Evidence from the S. Typhimurium model of infection suggests that bacteria are ubiquitinated prior to capture within autophagosomes (85). However, it is not known whether this is an absolute requirement for autophagy, or merely represents a failure of vacuolar integrity. We have been able to localize p62 to intracellular S. Typhimurium and therefore consider it plausible that binding of p62 to ubiquitinated bacteria results in recruitment of the autophagic apparatus to the site (Huett and Kuballa, unpublished data). Careful experiments using live imaging may reveal more about the requirements for this and other molecules in these models. The p62-triggered model is attractive since this approximates the chain of events seen in mitophagy, where damaged mitochondria are specifically removed and degraded.
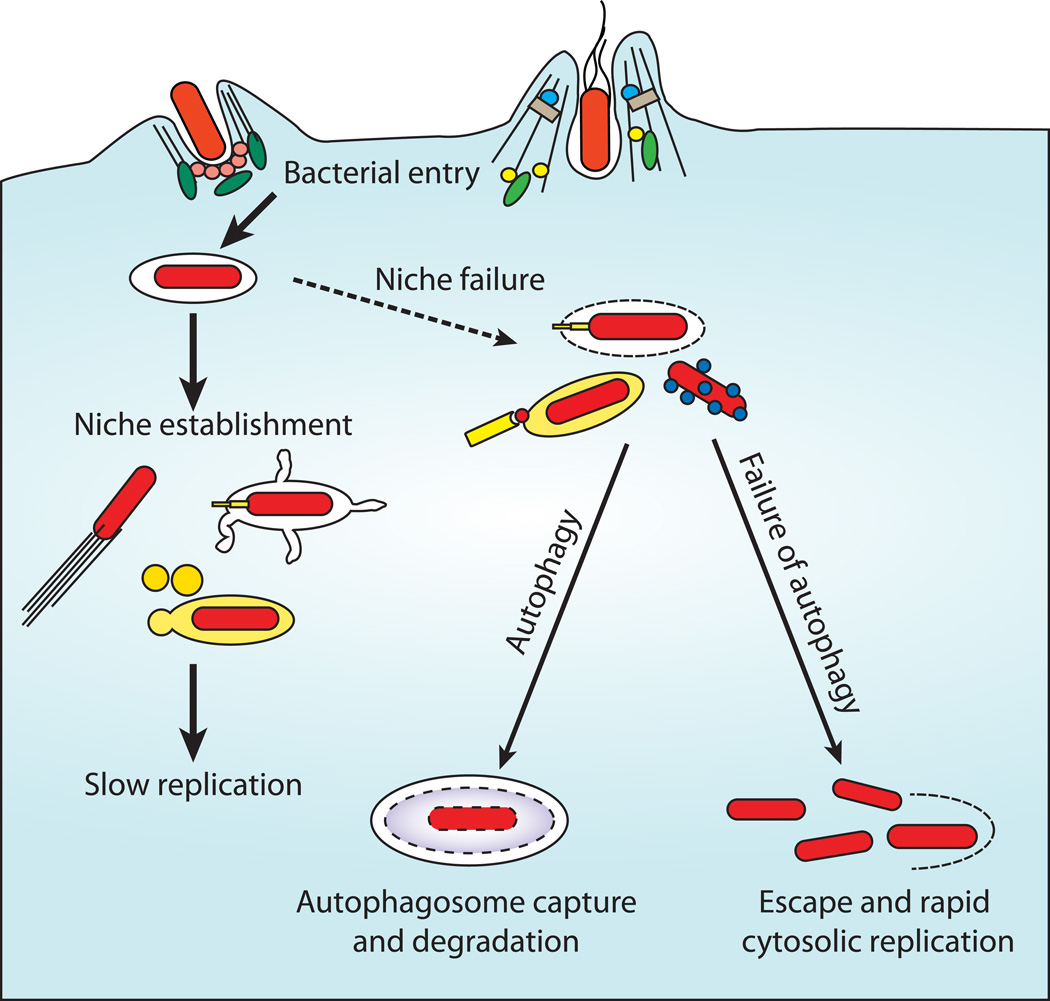
Whilst much work in this field has concentrated upon intracellular bacterial pathogens within phagocytes, these pathogens are often able to grow much more rapidly within the more permissive environment of the epithelial cell cytosol. Autophagy represents a cellular defense mechanism able to eliminate such pathogens, or prevent their escape into the cytosol (Figure 2), compromising the intracellular niche and restricting replication to within the vacuole. In the case of S. Typhimurium, it appears that autophagy targets bacteria in the process of vacuolar lysis, this would be bactericidal in macrophages, but in epithelial cells allows rapid replication once free of the restrictions of the Salmonella-containing vacuole. Shigella flexneriendeavors to escape autophagy by masking an autophagic recognition motif upon VirG via the binding of IcsB. IcsB binds to VirG in a location allowing the function of VirG in actin polymerization to be preserved, whilst blocking binding of Atg5 and subsequent autophagic capture (86). Without IcsB, cytoplasmic replication of Shigella plateaus after 4 hours and approximately 40% of internalized bacteria are enclosed within autophagosomes.
Figure 2. Autophagy as cytoplasmic defence.
Following entry into epithelial cells or other non-phagocytic cells, many bacteria are able to replicate in two niches. The initial niche is within the induced entry vacuole (e.g. Salmonella Typhimurium) or within the cytoplasm itself (e.g. Shigella flexneri). In the case of Salmonella this environment supports only slow replication, and Shigella must establish motility to evade autophagy and induce cell to cell spread. Escape from this initial niche into the cytoplasm, or loss of niche integrity results in one of two outcomes. Pathogens may be recognized (e.g. by NOD-like receptors) marked (e.g. by ubiquitination) and destroyed, by autophagy or other mechanisms; or they may establish rapid replication within the cytosol. When autophagy is unable to restrict bacteria to a vacuolar niche bacterial replication is elevated, yielding increased spread of bacteria and more severe disease.
Thus autophagy represents another battleground in the evolutionary arms race between host and pathogen. Host cells have evolved methods by which they restrict the availability of permissive niches, whilst pathogens attempt to evade or subvert such systems and establish replicating populations in host tissues.
Innate immunity, the role of autophagy
We have already discussed the role that autophagy plays in restricting the cytoplasmic niche for many pathogens. However, there are other innate immune processes that are enhanced and controlled by autophagy. The formation of an autophagic membrane around a pathogen and subsequent lysosomal fusion results in a concentrated pool of pathogen-derive molecules, many of which are likely to be ligands for immune receptors. These PAMPs must be presented to TLRs, NODs and other innate immune receptors to facilitate induction of inflammatory mediators and a full-blown inflammatory response. The restriction of such molecules within a membrane compartment allows delivery of the contents to receptor-containing compartments in a controlled and concentrated form.
In addition, directly anti-microbial products of autophagy such as ubiquitin, ubiquicidin and ribosomal polypeptides can also be delivered to pathogens within autophagosomes, prior to lysosomal fusion (87–89).
Recent publications have demonstrated that many TLR-ligands can result in induction of autophagy in stimulated cell lines. This is likely to both render the stimulated cells less permissive to pathogen infection (90), and in the case of immune effector cells provide nutrition to fuel de-novo synthesis of pro-inflammatory molecules and anti-microbial pathways, such as reactive-oxygen production. So far TLR-induced autophagy had been observed upon addition of ligands for TLR2, TLR3, TLR4, TLR7 and TLR8 (reviewed in (91)). Although the cytoplasmic RNA receptors form the RIG-I family, as well as NOD-like receptors are both likely to detect cytoplasmic pathogens, there is no direct evidence to date that either pathway has an effect upon autophagy induction.
Autophagy and the inflammasome
The inflammasome is a multi-protein complex which serves to generate pro-inflammatory molecules such as IL-1B or IL-18. The composition of the inflammasome is variable and stimulus dependant, with a variety of proteins assembled around a central core complex. This assembly is mediated via CARD-CARD and pyrin-pyrin domain protein-protein interactions. So far three inflammasomes have been identified; all share the ASC adaptor protein, caspase-1 and a linking CARD or pyrin-containing NLR protein. These complexes are usually named after the linking protein, e.g. NLRC4 (IPAF), NLRP1 or NLRP3 (NALP3) and are thought to respond to differing stimuli. NLRC4 is thought to sense intracellular flagellin (in a TLR5-independent manner) and mediates pyroptosis (pro-inflammatory programmed cell death) in response to S. Typhimurium infection. Other ligands for the NLRC4 inflammasome also exist, since aflagellate pathogens also induce robust activation via this complex, possibly via additional sensor proteins such as Naip5 (ref Nunez shigella paper). NLRP1-containing inflammasomes respond to MDP, a bacterial cell wall component and the ligand for NOD2. MDP appears to induce oligomerisation of NLRP1 and activation of the inflammasome – NLRP1 inflammasome activity is not strictly ASC-dependent, but is reduced in the absence of ASC. The NLRP3 (Nalp3) inflammasome appears to be the most promiscuous and is activated by a variety of bacterial ligands, as well as other endogenous compounds. It is known that NLRP3 inflammasomes can be induced by LPS, MDP, bacterial and double-stranded RNAs as well as uric acid crystals, ATP, pore-forming toxins and asbestos fibers. The presence of extracellular ATP likely represents a signal of nearby cellular lysis and is mediated via the P2X7R receptor. This may be important in the gut where normal cell shedding may become disrupted or accelerated by infection and/or inflammation.
In the context of CD and autophagy, inflammasome activation appears to be down-regulated by autophagy, in that ATG16L1- or Atg7-deficient macrophages exhibited enhanced IL-1B responses to LPS exposure (67). Chimeric mice generated from ATG16L1-deficient fetal liver cells injected into irradiated hosts showed a dramatic susceptibility to DSS-induced colitis. However, these mice did not develop spontaneous colitis and DSS-mediated injury could be partially rescued by administration of anti-IL-1B or anti-IL-18 antibodies. It has therefore been proposed that autophagy may serve to sequester and degrade the inflammasome, thus reducing the output of IL-1B. Alternatively, it is possible that autophagy increases the degradation rate of inflammasome-activating components, such as LPS and other microbial ligands.
A further wrinkle to the inflammasome / autophagy tale has been added with the identification of regulation of autophagy by inflammasome components in a S. flexneri infection model. Using infections of macrophages lacking Ipaf or ASC, Suzuki et al demonstrated that Ipaf is able to activate the inflammasome in the absence of flagellin. In Shigella-infected macrophages autophagy was enhanced by the absence of caspase-1 or Ipaf, but not ASC. In this model elevated autophagy in the absence of caspase-1 or Ipaf appears able to prevent or delay the pyroptotic death of macrophages infected with S. Typhimurium or S. flexneri. This is consistent with other non-infection models where autophagy is a survival response to stress. These findings indicate that autophagy and inflammasome / caspase activation are reciprocally regulated and that pyroptosis is induced through inhibition of autophagy, leading to rapid cell death and induction of inflammation. This would serve to rapidly rid hosts of infected cells and ensure robust inflammatory responses. However, if the balance tips too far, e.g. reduced ATG16L1 function, then pathological inflammation will result.
Education and activation of the adaptive immune system
Autophagy also plays an important part in the adaptive immune response, both in immune system development and immune education and activation. We have discussed the role of autophagy in controlling the mitochondrial status of T cells and how this relates to their development and survival. However it has become apparent that autophagy is responsible for more than this in the T cell compartment. Firstly autophagy appears to be crucial for T cell repertoire development in the thymus (92). It has been known for some time that thymic epithelial cells express a variety of otherwise cell-type restricted proteins. This “promiscuous” or ectopic expression allows presentation of these self antigens upon MHC II molecules to eliminate self-reactive T cells via negative selection. It has come to light that the thymic epithelium also has a high level of constitutive autophagy, in addition to a robust starvation-induced response (92). This constant level of autophagy appears to endow the thymic epithelium with enhanced antigen-presentation abilities, increasing the diversity of self-antigens presented to T cells and driving negative selection. In athymic mice transplanted with Atg5−/− thymi a significant autoimmunity developed, with severe colitis, uterine atrophy and lymphoadenopathy. This phenotype was associated with the CD4+ T cell repertoire, since most symptoms could be generated by transfer of CD4+ T cells from Atg5−/−chimeric mice. In addition positive selection is also affected, with several monoclonal MHCII-restricted transgenic TCR-expressing mice, showing altered CD4+ T cell generation within engrafted Atg5−/− thymic tissues. However, these effects were quite subtle, probably due to the relative flexibility of positive selection with regard to the spectrum of MHC-protein complexes available.
B cells also appear to have a requirement for Atg5 and Atg5-deficient B cells show a compromised transition from pro- to pre-B cells (53). This reduction in mature B cells was accompanied by cell death during B cell development in the bone marrow, thus autophagy is likely required for B cell survival during development. Whilst a population of peripheral B cells does survive in the absence of Atg5, the peritoneal B1 B cell compartment is almost completely ablated. This is a result both of deficiencies in B cell development as well as a requirement for Atg5 to maintain B1a B cells in the periphery. Thus Atg5 is differentially required for the development of B cell subsets and their maintenance.
Autophagy and antigen presentation
It is well known that T cell responses are largely MHC-restricted and that this results in differential presentation of antigen based upon its source. For example, CD4+ T cells are MHCII-restricted and thus view antigen derived from the lysosomal compartment, whilst MHCI-restricted CD8+ T cells are presented with antigen loaded in the ER, derived via the proteasomal degradation system. However, it has been shown that the natural MHCII-bound peptide repertoire contains a diverse array of cytoplasmic and nuclear antigens (93–95), approaching 30% of total MHC-bound peptides. Some of these peptides result from known autophagy components, e.g. LC3 or substrates, GAPDH (96, 97). A series of elegant studies from the Munz laboratory and others have demonstrated that the vesicular compartments where MHCII peptide loading occurs receive constant input from autophagosomes (98). This cytosolic peptide uptake is dependant upon autophagy, since chemical inhibitors and knockdown of autophagy components result in loss of such MHCII presentation (97, 99, 100). Indeed, Munz was able to show that fusion of a model antigen to LC3 and was directed to such compartments, resulting in enhanced immune presentation of the antigen (98). Both viral and bacterially-derived antigens have been shown to be presented on MHCII via this pathway, demonstrating its importance as a host defence mechanism (101) (102).
Autophagy and the intestinal epithelium
The intestinal epithelium presents several unique features relevant to autophagy – the process of constant epithelial renewal and shedding of apical epithelial cells, and the secretion of antimicrobial peptides by specialized Paneth cells. In its role as an absorptive organ the intestine has a special relationship with nutrients, following a meal the epithelium is loaded with a vast array of nutrient molecules which must be taken up and redistributed. This is then followed by periods of relative lower nutrient abundance. Until recently little was known of what effect these periods of feast and famine might have upon the regulation of autophagy in the epithelium itself, although it was of interest since several candidate genes for IBD are themselves nutrient transporters (e.g. SLC22A4 and SLC22A5) (60, 103).
It has become clear that amino acid availability greatly influences the rate of autophagy within intestinal epithelial cells and that amino acid levels are sensed by sophisticated transporter-based systems. Sakiyama et al reported that glutamine increased the levels of autophagy in both basal conditions and following heat or oxidative stress, and this increase was related to cell survival (104). They also observed that glutamine starvation induced increased caspase-3 and poly (ADP ribose) polymerase activity following stress, these increases were reduced by mTOR or p38 MAP kinase inhibition. These findings suggest that autophagy protects cells undergoing glutamine restriction from apoptosis.
Recently, Nicklin et al have shown that mTOR signaling is mediated by glutamine uptake via SLC1A5, followed by efflux from the cell (105). This glutamine efflux is required for uptake of leucine, since the leucine transporter SLC7A5/SLC3A2 is an obligate exchanger and thus requires an intracellular substrate to allow leucine import. Ingress of leucine into the cell results in activation of mTORC1, therefore uptake of glutamine is a prerequisite for essential amino acid signaling to mTORC1. Therefore glutamine appears to be the hub around which nutritional and growth factor signaling is organized in intestinal epithelial cells. The central role of amino acids in intestinal homeostasis is further underlined by the roles played by arginine, glutamine and leucine in mucosal cell migration and restitution.
As part of barrier maintenance the epithelium is constantly shed and renewed, forming an “epithelial escalator” from near the base of the crypt to the villus tip. Here cells are shed from the gut surface, a process which must be controlled to both limit nutrient and fluid loss and prevent bacterial ingress. Therefore it is likely that prior to shedding cells attempt to recover and recycle as many nutrients as possible. Although studies of autophagy have not been performed on these cells, it is plausible that cells consume their stores of amino acids and other nutrients prior to being shed.
The loss of cells from the villus must take place within a carefully organized series of events to prevent loss of epithelial integrity; therefore junctions between viable epithelial cells must be maintained to prevent osmotic leakage and bacterial invasion. Again it is not known whether autophagy is required for this process, but there are plausible reasons to believe that it may play a role. As has been discussed in the previous section, focal adhesion interacting proteins have been implicated in autophagy (106, 107) and focal adhesions have been observed to be lost during nutrient starvation of cultured cells. In addition, models of detachment-induced cell death have shown induction of autophagy following detachment from the extracellular matrix, inhibition of which resulted in increased levels of apoptotic cell death (108, 109). Therefore defects in autophagy may alter the state of shed epithelial cells, as well as affect the permeability of the epithelium they leave behind.
Shed epithelial cells may take on special immunological significance during gut inflammation. It is known that during development the clearance of cell corpses is dependant upon autophagy, without which cells are unable to release lysophosphatidylcholine or present phosphatidylserine to repectively trigger recruitment of, and engulfment by, phagocytes (110). This failure to promptly engulf and clear dead and dying cells is strongly linked to a number of autoimmune disorders, including models of arthritis and lupus (111–114). During infection and subsequent inflammation of the gut, prematurely shed or dying cells may be required to be cleared by phagocytes. This too is likely to be dependant upon defined membrane signals, and thus possibly affected by reduced levels of autophagy. Combined with the increased leakage of soluble factors from the gut lumen during inflammation and tissue disruption, this may lead to enhanced auto-antigen exposure and increased risk of autoimmunity. In addition, a combination of anti-pathogen autophagy and autophagy-mediated cell corpse signaling following cell death may result in more rapid immune detection of pathogens. Infected cells with intact autophagy are likely to both present more pathogen-derived antigens to immune cells, as well as speed their take-up by phagocytes upon their death. This is likely to result in a reduction of pathogens capable of spreading infection and increase the speed of adaptive immune responses, both leading to a more prompt resolution of inflammation.
Related to the subject of cell shedding there has been significant gut abnormality associated with autophagic defects in mouse models. Mice expressing a hypomorphic variant of ATG16L1, an essential autophagy gene, show a distinctive Paneth cell defect, characterized by defective degranulation (66, 115). This defect is accompanied by a variety of gene-expression changes, including many related to mucosal homeostasis and healing, as well as a strong pro-inflammatory signature. Similar phenotypes were identified in mice lacking Atg5 or Atg7 expression and patients with CD, confirming both that the defect is autophagy related and relevant to human disease. In the mouse model it appears that Paneth cell degranulation is largely lost and the apical portion of the Paneth cell is often seen to break away from the basal portion. This results in loss of Paneth cells and a paucity of antimicrobial peptides and other secreted proteins in the luminal environment. Such a loss is likely to result in an alteration in the frequency and character of the bacterial / epithelial interactions, as the level of antimicrobial peptides in the crypt is reduced. It is likely that loss of defensins and other active peptides result in increased intrusion of microbial flora into the crypt, closer to the self-renewing stem cell compartment (116). Indeed, reduction of defensin expression appears to be a strategy employed by several intestinal pathogens (117, 118). This profound alteration in the gut environment is likely to significantly alter the overall microbiome of the gut and not merely the level of bacteria close to the crypts. The interconnected and relatively stable nature of complex microbial communities means that such differences in gut flora are likely to be life-long and may directly affect susceptibility to infection (119, 120). In the future, experiments examining the extent and role of autophagy in the guts of germfree, mono-associated or defined-flora-reconstituted mice will be important to determine the interplay between these factors.
Autophagy is known to be required for optimal antigen cross-presentation upon MHCII, both in thymic development and viral infections (ref (92) (98). However antigen presentation in the gut is also performed by the specialized M-cell, which continuously samples luminal antigen and delivers these to macrophages and other antigen-presenting cells. Recent data has shown that such sampling takes place not just in lymphoid tissues such as Peyer’s patches, but also in isolated M-cells within the crypt epithelia and such sites are occupied by large numbers of follicular dendritic cells (121). In addition it is known that M cells are preferentially targeted by many pathogens and their toxins as portals to the systemic compartment (122–126). Thus M cells are likely to be crucial in both determination of which antigens are presented to immune cells, and controlling the invasion of pathogens attempting to effect entry into the gut tissue. Indeed, it is known that M-cell-mediated immune responses differ markedly compared to those driven by dendritic cells. M-cell antigen presentation is required to generate secretory IgA, whilst these cells are not needed for systemic IgG antibody production in a S. Typhimurium model (127) (128). M-cells and intact Peyer’s patches are also required to generate innate secretory antibodies (129). These innate antibodies are usually characterized as low-affinity, multi-valent molecules capable of binding a variety of self and non-self antigens. Innate antibodies are thought to be important agents of gut tolerance through their ability to remove commensal and self-derived antigens from the gut environment. In addition, they provide immediate, albeit less potent, defence against pathogens whilst high-affinity, specific antibodies are being produced (130, 131). Given the known role of autophagy in antigen presentation and defense against intracellular pathogens, it would seem reasonable to propose that defects in autophagy might also affect the ability of M-cells and Peyer’s patches to fulfill their roles in immune recognition and antigen display. This might predispose affected individuals to infections, as well as reduce or alter the innate antibody repertoire, exposing the gut and systemic immune system to an elevated and more diverse antigen burden.
Enteroendocrine (neuroendocrine) cells represent another specialized gut cell type which has been associated with CD and other inflammatory disorders. Following identification of a CD-associated SNP within the genomic region adjacent to the PHOX2B gene, we examined the location of PHOX2B expression in the gut by immunohistochemistry. Cells staining positive for PHOX2B protein were morphologically identified as enteroendocrine cells. These cells are thought to mediate control of innate immune responses and gut motility through their secretion of hormones and other factors (132) and participate in the “tasting” of luminal contents. A recent report also identified functional TLR signaling within enteroendocrine cells and TLR stimulation resulted in calcium flux and secretion (133). Whilst intestinal epithelial cells are sensitive to ER stress (134, 135), secretory cells, such as Paneth cells and goblet cells, appear to be acutely sensitive to ER stress (66, 115, 136). This may be induced by secretory blockade leading to accumulation of protein within the ER compartment; this may hold true for neuroendocrine cells, given their secretory nature. With the potential role for the autophagy apparatus in controlling both ER stress and exocytosis, it is possible that defects in autophagy disproportionately affect the function of all gastrointestinal secretory cells, e.g. Paneth, enteroendocrine and goblet cells. It will be of interest to ascertain whether these cells exhibit high levels of autophagy and if they are affected in IBD patients.
Non-autophagic roles for autophagy proteins
In addition to the importance, conservation and diversity of autophagic processes, some autophagy proteins appear to play additional roles, both in response to infection and during homeostasis. For example Zhao et al describe an autophagy-independent function for the core autophagy protein Atg5 in resistance to Toxoplasma gondii (137). As with many autophagy-based resistance mechanisms, this function appears dependant upon assembly of protein complexes around the pathogen-containing vacuole. In this case, such complexes result in vacuolar membrane lysis induced by Atg5-dependant recruitment of the interferon-induced GTPase Irga6. It is interesting to compare this to resistance seen in models of C. trachomatis infection, where Irga6 recruitment appears to redirect bacterial inclusions into the autophagic pathway and subsequently results in lysosome fusion and bacterial killing (69). Al-Zeer et al demonstrate that Irga6 is required for optimal IFN-g induced autophagy, suggesting that the interaction with Atg5 is required to trigger this innate response in interferon-activated cells. In both infections killing is Atg5 dependant, however in the case of T. gondii-infected macrophages, no autophagic features surrounding parasites were observed, in contrast to Al-Zeer et al who were able to observe GFP-LC3 surrounding bacterial inclusions. However, since Zhao et al did not perform staining against LC3 it is possible that these results are complementary and that both Atg5 and LC3 accumulate upon intracellular pathogen-containing vacuoles in the absence of full-blown autophagy. It is interesting to note that LC3 has also been observed upon phagocytic membranes and a role in lysosome fusion imputed (138). Therefore Zhao et al suggest that the role of Atg5 in T. gondii infection is similar to that seen in phagocytosis of beads coated with TLR ligand (139), where Atg5 serves to recruit key proteins to the nascent membrane compartment. This may be a phenomenon generalisable to other intracellular infections, including C. trachomatisand may represent an alternative “sub-autophagic” route to direct cargos to the lysosomal compartment dependant upon IFN-g and induced GTPases. There is a growing body evidence suggesting that autophagic proteins play a role in phagosome assembly (138, 139), and given the similarities between the two processes it is not surprising that much of the membrane handling may be conserved.
In plants and worms autophagy is also largely conserved from yeast and the use of genetics has yielded significant insight and a variety of powerful tools, including many mutants. Arabidopsis thaliana mutants in Atg6, but not other autophagy genes tested, exhibit a defect in pollen germination - such an Atg6-specific defect points towards a non-autophagic role of this protein in pollen formation (140). Similarly, Atg6-deficient C. elegans have defects in both autophagy and endocytosis, perhaps hinting at a role for the autophagy-related PI3K complex in endocytic traffic (141). Also of interest is that mice lacking Beclin 1 (mammalian Atg6) die upon entry into the second week of embryogenesis, whilst most other autophagy mutants survive until birth (142). All of these findings are suggestive of a role outside of autophagy for Beclin1 and its homologues, further study of mutants in diverse species might yield interesting data upon the roles of Atg6 / Beclin 1.
Aside from the core autophagic apparatus there are other proteins with emerging roles in autophagy which exhibit other cellular functions. One such example is FNBP1L (Toca-1) which is known to be essential for cdc42-mediated actin dynamics (143–145). We recently identified this as a component required for anti-bacterial autophagy (78). Interestingly FNBP1L association with Atg3 depends upon the HR1 domain of FNBP1L, this is the same domain responsible for binding of cdc42. Therefore, just as FNBP1L and FBP17 may be responsible for generation of actin polymerization upon curved membrane surfaces (Takano et al EMBO 2008), FNBP1L may be required to generate autophagosomes on and around bacteria-containing vacuoles. Indeed FBP17 (a related F-BAR containing protein) has been shown to mediate phagocytic cup formation in macrophages, and recruitment is thought to be dependant upon enrichment of phosphatidylinositol-4,5-bisphosphate. Phagocytosis has some similarity to autophagosome formation and, as has been elegantly shown for phagocytosis by Grinstein and others (29, 30), autophagsome formation is likely to rely upon both binding of specific proteins as well as generation of lipid modifications on the growing, bending membrane. Indeed it is known that autophagosome membranes are generally cholesterol-poor and lacking in intramembrane proteins, suggesting that they may exhibit specific properties favoring binding of proteins required for their growth. Initiation and completion of a classical autophagosome in the cytoplasm in a stochastic fashion may therefore be self-generated by the core autophagic machinery; however larger cargoes require additional membrane growth. Therefore it is possible that the anti-bacterial autophagosome begins at a single focus, triggered by an as-yet-uncharacterized signal. This initial, curved membrane may subsequently be stabilized and extended by the actions of FNBP1L to completely engulf the desired cargo. Such a mechanism has been proposed by others to explain the requirement for Bif-1 (an N-BAR containing protein) in classical autophagosome formation (146, 147). This use of different BAR domains makes sense in the context of differing vesicle sizes, since N-BAR proteins generate narrower membrane tubules in vitro and increased membrane curvatures than F-BAR proteins (148).
It has also been suggested that Atg1/ULK1 may have autophagy-independent roles in cell adhesion, since focal adhesion components have been indentified as required for autophagic function. Atg1 has been associated with neuronal axon development in both C. elegans and mammals and overexpression of ULK1 or 2 in tissue culture triggers morphological changes and loss of adhesion (106, 149–151). In addition in both a D. melanogaster model and mammalian cells, paxillin was observed to be required for autophagy via an Atg1/ULK interaction (107). These findings suggest that ULK proteins may regulate formation and placement of non-autophagic protein complexes, particularly those involved in membrane-cytoskeletal interactions.
Defense-in-depth, it’s a numbers game
Over the past 100 years our knowledge of innate and adaptive immunity has exploded, from the first descriptions of phagocytes and immune defense molecules to the integrated systems biology of today. What has become apparent is that host defense in mammals is a diverse, multi-layered phenomenon, with each layer serving to reduce the probability of an infectious insult resulting in a productive infection. Some of these layers are easily observed, for example the barrier function of the skin and mucosa. Others are less obvious, such as anti-microbial peptides and sensors of pathogen-associated molecular patterns. In a naïve individual these responses are required in order to control or delay the onset of infection until a full adaptive response can be mounted. However, no single mechanism functions alone to entirely control a given infection. Thus the body operates a defense-in-depth strategy, with each mechanism reducing the probability that an infection can penetrate beyond the defense or maintain a replicative niche.
This concept of pathogenesis as probability was first demonstrated by Meynell and Stocker in 1957 (152) and has largely been borne out in further analysis of animal models (153, 154). In the Stocker model the probability of any given organism within a clonal inoculum progressing to a replicative infection is independent of the other organisms within the inoculum. Thus an ID50 is a purely mathematical phenomenon, rather than a requirement that an inoculum exceed a physical number of organisms to initiate an infection. Dosage of a single organism could be sufficient to generate an infection, albeit in a tiny proportion of instances.
By combining these two ideas; defence in depth and the probability model of infections, it is possible to generate novel insights into infections and their progression. Recent work utilizing molecular genetics to tag individual bacterial clones within an intra-venous delivered population of S. Typhimurium has shown that during infection of mice bacterial replication occurs alongside host-mediated killing (154). In successful infections pathogen replication during establishment of a systemic infection just barely outpaces the killing rate. Since initial numbers of invading organisms are small, modest gains in killing efficiency or niche restriction result in greatly improved outcomes for the host, including resolution of infections whilst disease remains sub-clinical.
In the gut, where bacterial exposure is constant and occurs over a vast surface area, itself under constant renewal, these concepts take on an unrivalled importance. It is crucial for gut homeostasis that microbes are immediately confined and removed when they gain entry into intestinal tissues, both to avoid replication and spread and to limit the induction of problematic inflammation. Pathogens, commensals and their products must be rapidly handled and degraded to limit the extent of immune recognition and activation.
In the context of IBD, minor defects in autophagy may predispose individuals to disease in many ways, summarized in Figure 3. Increased stress upon secretory cells results in loss or depletion of the mucus barrier and anti-microbial peptides, increasing epithelial exposure to luminal flora. In intestinal crypts, where anti-microbial peptide concentrations are usually highest, loss of peptide secretion results in flora entering the base of crypts (Figure 3A). The presence of elevated numbers of auto-reactive or flora-reactive T cells, due to improper negative selection during thymic development, results in inflammatory responses (Figure 3B). This may be ongoing, or initiated by acute exposure to microbial products and antigens upon injury. In addition, there is a loss of innate antibody production de to a paucity of B1-B cells, further decreasing epithelial barrier defence. This initial inflammation promotes epithelial turnover, however in the absence of fully functional autophagy junctional regulation may be compromised, resulting in improper coordination of cell shedding (Figure 3C). Exposure of the basolateral surfaces of epithelial cells, as well as depletion of mucus and the epithelial layer itself, all predispose tissues to infection with classical pathogens and “normal” gut flora. It is known that many proteins utilized by bacteria to effect entry into epithelial cells are restricted to the basolateral surface (155, 156). Thus loss of epithelial integrity is likely to result in increased invasion and attachment of bacteria. The defects in mucus, antimicrobial peptide and antibody production also expose M-cells to greater numbers of bacteria and potential pathogens. Underlying lymphoid tissues are therefore exposed to higher amounts of pro-inflammatory ligands and signals (Figure 3D). Under all these circumstances epithelial cells, as well as professional phagocytes, have to respond to higher burdens of bacterial invasion/attachment and epithelial breakthrough (Figure 3E). This results an increased requirement for autophagic elimination of bacteria which enter both phagocytes and the cytoplasm of permissive cell types such as epithelial cells. If the autophagic system is unable to keep up with the bacterial challenge, then these cells may become infected with bacteria and perpetuate the inflammatory cycle.
Figure 3. The diverse roles of autophagy in IBD pathology.
Autophagy may play several special roles in the gut related to epithelial barrier function, immunity and bacterial infection in addition to normal cellular homeostasis. (A) Under normal conditions intestinal crypts are kept free of the majority of microbial flora by Paneth cell secretion of anti-microbial peptides. Loss or reduction of such secretion results in an increase in bacteria within the crypts and alterations in overall flora composition. Paneth cell secretion defects have been observed in CD patients with ATG16L1 polymorphisms. (B) Autophagy is required for normal T cell development and plays a role in negative selection of T cells during thymic development. Defects in autophagy may result in increased auto-reactive T cells within the gut, as well as a reduced B1 cell numbers. T cells may therefore be triggered to induce inflammation by self-antigen or increased exposure to microbial products. Loss of innate antibodies from B1 cells will also yield lowered resistance to bacterial colonization. (C) Several lines of evidence point towards a link between autophagy and regulation of cell-cell junctions. Maintenance of junctions is important to prevent leakage of gut contents through the epithelium. Disruptions in cell junctions may directly render the epithelium leakier, or result in loss of integrity during cell shedding and replacement, exposing sub-epithelial layers and basolateral cell surfaces to microbes. (D) The role of autophagy in M cell antigen uptake and presentation is largely unknown. However M cells are preferred portals of a number of infectious agents. Thus it is likely that without fully competent autophagy M cells are more prone to infection and less able to present antigens to immune cells. This will result in more infections, increased inflammation and altered immune responses to gut flora. (E) Epithelial cells and phagocytes rely upon autophagy to control invading bacteria and to present cytosolic antigens upon MHCII. Therefore autophagic defects will result in less capable bacterial handling and antigen delivery, increasing the risk of infection and slowing initiation of immune responses.
In conclusion we believe that autophagy is likely to play a key role in the process of gastrointestinal homeostasis, serving to remove bacteria that make it into tissues and degrade any non-self immunostimulatory molecules to maintain intestinal tolerance. For these reasons autophagy may be under a particularly high load in the tissues of the gastrointestinal tract, especially in areas where barrier function has been compromised by previous injury. Perhaps this is why even minor defects in autophagy are associated with inflammatory bowel disease, in the absence of overt symptoms in other organ systems.
Acknowledgments
We should like to apologize to all of our colleagues within the autophagy field whose work we were unable to cite due to space constraints.
RJX and AH are supported by the following grants from the NIH: AI062773, DK83756 and DK043351.
References
- 1.Matsuura A, Tsukada M, Wada Y, et al. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nature cell biology. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. The Journal of biological chemistry. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 5.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 Association with the ULK1-Atg13-FIP200 Complex Required for Autophagy. Molecular biology of the cell. 2009 doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. Molecular biology of the cell. 2009 doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5 doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 9.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Molecular biology of the cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pankiv S, Hoyvarde Clausen T, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of Ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 11.Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. The Journal of biological chemistry. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 12.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sou YS, Waguri S, Iwata J, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Molecular biology of the cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dove SK, Piper RC, McEwen RK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. The EMBO journal. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stromhaug PE, Reggiori F, Guan J, et al. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Molecular biology of the cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obara K, Sekito T, Niimi K, et al. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. The Journal of biological chemistry. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itakura E, Kishi C, Inoue K, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Molecular biology of the cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 20.Kihara A, Kabeya Y, Ohsumi Y, et al. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO reports. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. The Journal of biological chemistry. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 23.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature cell biology. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 24.Liang C, Lee JS, Inn KS, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nature cell biology. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nature cell biology. 2009 doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Q, Fan W, Chen K, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature cell biology. 2009 doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 28.Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of cell biology. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamen LA, Levinsohn J, Swanson JA. Differential association of phosphatidylinositol 3-kinase, SHIP-1, and PTEN with forming phagosomes. Molecular biology of the cell. 2007;18:2463–2472. doi: 10.1091/mbc.E07-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee WL, Mason D, Schreiber AD, et al. Quantitative analysis of membrane remodeling at the phagocytic cup. Molecular biology of the cell. 2007;18:2883–2892. doi: 10.1091/mbc.E06-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita N, Itoh T, Fukuda M, et al. The Atg16L Complex Specifies the Site of LC3 Lipidation for Membrane Biogenesis in Autophagy. Molecular biology of the cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh T, Fujita N, Kanno E, et al. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Molecular biology of the cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic (Copenhagen, Denmark) 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fass E, Shvets E, Degani I, et al. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. The Journal of biological chemistry. 2006;281:36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- 35.Iwata A, Riley BE, Johnston JA, et al. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. The Journal of biological chemistry. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 36.Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell structure and function. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- 37.Kochl R, Hu XW, Chan EY, et al. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic (Copenhagen, Denmark) 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 38.Gahl WA. GeneReviews at GeneTests: Medical Genetics Information Resource (database online) Seattle: University of Washington; Hermansky-Pudlak Syndrome. Updated November 27, 2007. [Google Scholar]
- 39.Zhao M, Antunes F, Eaton JW, et al. Lysosomal enzymes promote mitochondrial oxidant production, cytochrome c release and apoptosis. European journal of biochemistry / FEBS. 2003;270:3778–3786. doi: 10.1046/j.1432-1033.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 40.Stroikin Y, Dalen H, Loof S, et al. Inhibition of autophagy with 3-methyladenine results in impaired turnover of lysosomes and accumulation of lipofuscin-like material. European journal of cell biology. 2004;83:583–590. doi: 10.1078/0171-9335-00433. [DOI] [PubMed] [Google Scholar]
- 41.Cuervo AM, Palmer A, Rivett AJ, et al. Degradation of proteasomes by lysosomes in rat liver. European journal of biochemistry / FEBS. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- 42.Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. The Journal of cell biology. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 44.Kim PK, Hailey DW, Mullen RT, et al. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Molecular cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 46.Korolchuk VI, Mansilla A, Menzies FM, et al. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Molecular cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Yan L, Zhou Z, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 48.Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagda RK, Cherra SJ, 3rd, Kulich SM, et al. Loss of pink1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. The Journal of biological chemistry. 2009 doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tal MC, Sasai M, Lee HK, et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephenson LM, Miller BC, Ng A, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5 doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pua HH, Guo J, Komatsu M, et al. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 53.Miller BC, Zhao Z, Stephenson LM, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 54.Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Experimental cell research. 2008;314:3531–3541. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 55.Farre JC, Manjithaya R, Mathewson RD, et al. PpAtg30 tags peroxisomes for turnover by selective autophagy. Developmental cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klionsky DJ, Cuervo AM, Dunn WA, Jr, et al. How shall I eat thee? Autophagy. 2007;3:413–416. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- 57.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annual review of cell and developmental biology. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 58.Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell death and differentiation. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 59.Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Molecular and cellular biology. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature genetics. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature genetics. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nature genetics. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh SB, Davis AS, Taylor GA, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 64.Kuballa P, Huett A, Rioux JD, et al. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henry SC, Daniell X, Indaram M, et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) J Immunol. 2007;179:6963–6972. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 66.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 68.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Al-Zeer MA, Al-Younes HM, Braun PR, et al. IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS ONE. 2009;4:e4588. doi: 10.1371/journal.pone.0004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry SC, Daniell XG, Burroughs AR, et al. Balance of Irgm protein activities determines IFN-{gamma}-induced host defense. Journal of leukocyte biology. 2009 doi: 10.1189/jlb.1008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huett A, McCarroll SA, Daly MJ, et al. On the level: IRGM gene function is all about expression. Autophagy. 2009;5:96–99. doi: 10.4161/auto.5.1.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gutierrez MG, Master SS, Singh SB, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 73.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nature genetics. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bekpen C, Marques-Bonet T, Alkan C, et al. Death and resurrection of the human IRGM gene. PLoS genetics. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L, Yu J, Pan H, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams A, Sarkar S, Cuddon P, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nature chemical biology. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He P, Peng Z, Luo Y, et al. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy. 2009;5:52–60. doi: 10.4161/auto.5.1.7247. [DOI] [PubMed] [Google Scholar]
- 78.Huett A, Ng A, Cao Z, et al. A Novel Yeast-Human Network Analysis Reveals an Essential Role for FNBP1L in Antibacterial Autophagy. J Immunol. 2009 doi: 10.4049/jimmunol.0803050. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanada T, Satomi Y, Takao T, et al. The amino-terminal region of Atg3 is essential for association with phosphatidylethanolamine in Atg8 lipidation. FEBS letters. 2009 doi: 10.1016/j.febslet.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 80.Ketteler R, Seed B. Quantitation of autophagy by luciferase release assay. Autophagy. 2008;4:801–806. doi: 10.4161/auto.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ketteler R, Sun Z, Kovacs KF, et al. A pathway sensor for genome-wide screens of intracellular proteolytic cleavage. Genome biology. 2008;9:R64. doi: 10.1186/gb-2008-9-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay and drug development technologies. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 83.Palmer C, Bik EM, Digiulio DB, et al. Development of the Human Infant Intestinal Microbiota. PLoS biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rescigno M, Lopatin U, Chieppa M. Interactions among dendritic cells, macrophages, and epithelial cells in the gut: implications for immune tolerance. Current opinion in immunology. 2008;20:669–675. doi: 10.1016/j.coi.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 85.Birmingham CL, Smith AC, Bakowski MA, et al. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. The Journal of biological chemistry. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 86.Ogawa M, Yoshimori T, Suzuki T, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 87.Scherz-Shouval R, Shvets E, Fass E, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. The EMBO journal. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kieffer AE, Goumon Y, Ruh O, et al. The N- and C-terminal fragments of ubiquitin are important for the antimicrobial activities. Faseb J. 2003;17:776–778. doi: 10.1096/fj.02-0699fje. [DOI] [PubMed] [Google Scholar]
- 89.Howell SJ, Wilk D, Yadav SP, et al. Antimicrobial polypeptides of the human colonic epithelium. Peptides. 2003;24:1763–1770. doi: 10.1016/j.peptides.2003.07.028. [DOI] [PubMed] [Google Scholar]
- 90.Delgado MA, Elmaoued RA, Davis AS, et al. Toll-like receptors control autophagy. The EMBO journal. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delgado M, Singh S, De Haro S, et al. Autophagy and pattern recognition receptors in innate immunity. Immunological reviews. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nedjic J, Aichinger M, Emmerich J, et al. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 93.Chicz RM, Urban RG, Gorga JC, et al. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. The Journal of experimental medicine. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dongre AR, Kovats S, deRoos P, et al. In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. European journal of immunology. 2001;31:1485–1494. doi: 10.1002/1521-4141(200105)31:5<1485::AID-IMMU1485>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 95.Dengjel J, Schoor O, Fischer R, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aniento F, Roche E, Cuervo AM, et al. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. The Journal of biological chemistry. 1993;268:10463–10470. [PubMed] [Google Scholar]
- 97.Fengsrud M, Raiborg C, Berg TO, et al. Autophagosome-associated variant isoforms of cytosolic enzymes. The Biochemical journal. 2000;352(Pt 3):773–781. [PMC free article] [PubMed] [Google Scholar]
- 98.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dorfel D, Appel S, Grunebach F, et al. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 2005;105:3199–3205. doi: 10.1182/blood-2004-09-3556. [DOI] [PubMed] [Google Scholar]
- 101.Munz C, Bickham KL, Subklewe M, et al. Human CD4(+) T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1. The Journal of experimental medicine. 2000;191:1649–1660. doi: 10.1084/jem.191.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jagannath C, Lindsey DR, Dhandayuthapani S, et al. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nature medicine. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 103.Peltekova VD, Wintle RF, Rubin LA, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nature genetics. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 104.Sakiyama T, Musch MW, Ropeleski MJ, et al. Glutamine increases autophagy under Basal and stressed conditions in intestinal epithelial cells. Gastroenterology. 2009;136:924–932. doi: 10.1053/j.gastro.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. The Journal of cell biology. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen GC, Lee JY, Tang HW, et al. Genetic interactions between Drosophila melanogaster Atg1 and paxillin reveal a role for paxillin in autophagosome formation. Autophagy. 2008;4:37–45. doi: 10.4161/auto.5141. [DOI] [PubMed] [Google Scholar]
- 108.Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy. 2008;4:351–353. doi: 10.4161/auto.5523. [DOI] [PubMed] [Google Scholar]
- 109.Fung C, Lock R, Gao S, et al. Induction of autophagy during extracellular matrix detachment promotes cell survival. Molecular biology of the cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qu X, Zou Z, Sun Q, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 111.Bijl M, Reefman E, Limburg PC, et al. Inflammatory clearance of apoptotic cells after UVB challenge. Autoimmunity. 2007;40:244–248. doi: 10.1080/08916930701357125. [DOI] [PubMed] [Google Scholar]
- 112.Norsworthy PJ, Fossati-Jimack L, Cortes-Hernandez J, et al. Murine CD93 (C1qRp) contributes to the removal of apoptotic cells in vivo but is not required for C1q-mediated enhancement of phagocytosis. J Immunol. 2004;172:3406–3414. doi: 10.4049/jimmunol.172.6.3406. [DOI] [PubMed] [Google Scholar]
- 113.Fossati-Jimack L, Cortes-Hernandez J, Norsworthy PJ, et al. C1q deficiency promotes the production of transgenic-derived IgM and IgG3 autoantibodies in anti-DNA knock-in transgenic mice. Molecular immunology. 2008;45:787–795. doi: 10.1016/j.molimm.2007.06.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Potter PK, Cortes-Hernandez J, Quartier P, et al. Lupus-prone mice have an abnormal response to thioglycolate and an impaired clearance of apoptotic cells. J Immunol. 2003;170:3223–3232. doi: 10.4049/jimmunol.170.6.3223. [DOI] [PubMed] [Google Scholar]
- 115.Cadwell K, Patel KK, Komatsu M, et al. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–252. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vaishnava S, Behrendt CL, Ismail AS, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Salzman NH, Chou MM, de Jong H, et al. Enteric salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infection and immunity. 2003;71:1109–1115. doi: 10.1128/IAI.71.3.1109-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sperandio B, Regnault B, Guo J, et al. Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. The Journal of experimental medicine. 2008;205:1121–1132. doi: 10.1084/jem.20071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lawley TD, Bouley DM, Hoy YE, et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infection and immunity. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Glaysher BR, Mabbott NA. Isolated lymphoid follicle maturation induces the development of follicular dendritic cells. Immunology. 2007;120:336–344. doi: 10.1111/j.1365-2567.2006.02508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Anosova NG, Chabot S, Shreedhar V, et al. Cholera toxin E. coli heat-labile toxin, and non-toxic derivatives induce dendritic cell migration into the follicle-associated epithelium of Peyer's patches. Mucosal immunology. 2008;1:59–67. doi: 10.1038/mi.2007.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Keita AV, Salim SY, Jiang T, et al. Increased uptake of non-pathogenic E. coli via the follicle-associated epithelium in longstanding ileal Crohn's disease. The Journal of pathology. 2008;215:135–144. doi: 10.1002/path.2337. [DOI] [PubMed] [Google Scholar]
- 124.Martinez-Argudo I, Jepson MA. Salmonella translocates across an in vitro M cell model independently of SPI-1 and SPI-2. Microbiology (Reading, England) 2008;154:3887–3894. doi: 10.1099/mic.0.2008/021162-0. [DOI] [PubMed] [Google Scholar]
- 125.Martinez-Argudo I, Sands C, Jepson MA. Translocation of enteropathogenic Escherichia coli across an in vitro M cell model is regulated by its type III secretion system. Cellular microbiology. 2007;9:1538–1546. doi: 10.1111/j.1462-5822.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 126.Ouzilou L, Caliot E, Pelletier I, et al. Poliovirus transcytosis through M-like cells. The Journal of general virology. 2002;83:2177–2182. doi: 10.1099/0022-1317-83-9-2177. [DOI] [PubMed] [Google Scholar]
- 127.Martinoli C, Chiavelli A, Rescigno M. Entry route of Salmonella typhimurium directs the type of induced immune response. Immunity. 2007;27:975–984. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 128.Hashizume T, Togawa A, Nochi T, et al. Peyer's patches are required for intestinal immunoglobulin A responses to Salmonella spp. Infection and immunity. 2008;76:927–934. doi: 10.1128/IAI.01145-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wijburg OL, Uren TK, Simpfendorfer K, et al. Innate secretory antibodies protect against natural Salmonella typhimurium infection. The Journal of experimental medicine. 2006;203:21–26. doi: 10.1084/jem.20052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hangartner L, Zellweger RM, Giobbi M, et al. Nonneutralizing antibodies binding to the surface glycoprotein of lymphocytic choriomeningitis virus reduce early virus spread. The Journal of experimental medicine. 2006;203:2033–2042. doi: 10.1084/jem.20051557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McCoy KD, Stoel M, Stettler R, et al. Polyclonal and specific antibodies mediate protective immunity against enteric helminth infection. Cell host & microbe. 2008;4:362–373. doi: 10.1016/j.chom.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 132.Di Comite G, Grazia Sabbadini M, Corti A, et al. Conversation galante: how the immune and the neuroendocrine systems talk to each other. Autoimmunity reviews. 2007;7:23–29. doi: 10.1016/j.autrev.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 133.Bogunovic M, Dave SH, Tilstra JS, et al. Enteroendocrine cells express functional Toll-like receptors. American journal of physiology. 2007;292:G1770–G1783. doi: 10.1152/ajpgi.00249.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shkoda A, Ruiz PA, Daniel H, et al. Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterology. 2007;132:190–207. doi: 10.1053/j.gastro.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 136.Heazlewood CK, Cook MC, Eri R, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS medicine. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao Z, Fux B, Goodwin M, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell host & microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shui W, Sheu L, Liu J, et al. Membrane proteomics of phagosomes suggests a connection to autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16952–16957. doi: 10.1073/pnas.0809218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 140.Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant physiology. 2007;143:1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Takacs-Vellai K, Vellai T, Puoti A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 142.Yue Z, Jin S, Yang C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ho HY, Rohatgi R, Lebensohn AM, et al. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 144.Kakimoto T, Katoh H, Negishi M. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. The Journal of biological chemistry. 2006;281:29042–29053. doi: 10.1074/jbc.M604025200. [DOI] [PubMed] [Google Scholar]
- 145.Leung Y, Ally S, Goldberg MB. Bacterial actin assembly requires toca-1 to relieve N-wasp autoinhibition. Cell host & microbe. 2008;3:39–47. doi: 10.1016/j.chom.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nature cell biology. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/Endophilin B1: a candidate for crescent driving force in autophagy. Cell death and differentiation. 2009 doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Henne WM, Kent HM, Ford MG, et al. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 149.Ogura K, Wicky C, Magnenat L, et al. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes & development. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 150.Tomoda T, Kim JH, Zhan C, et al. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes & development. 2004;18:541–558. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhou X, Babu JR, da Silva S, et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meynell GG, Stocker BA. Some hypotheses on the aetiology of fatal infections in partially resistant hosts and their application to mice challenged with Salmonella paratyphi-B or Salmonella typhimurium by intraperitoneal injection. Journal of general microbiology. 1957;16:38–58. doi: 10.1099/00221287-16-1-38. [DOI] [PubMed] [Google Scholar]
- 153.Brown SP, Cornell SJ, Sheppard M, et al. Intracellular demography and the dynamics of Salmonella enterica infections. PLoS biology. 2006;4:e349. doi: 10.1371/journal.pbio.0040349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grant AJ, Restif O, McKinley TJ, et al. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS biology. 2008;6:e74. doi: 10.1371/journal.pbio.0060074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bishop A, House D, Perkins T, et al. Interaction of Salmonella enterica serovar Typhi with cultured epithelial cells: roles of surface structures in adhesion and invasion. Microbiology (Reading, England) 2008;154:1914–1926. doi: 10.1099/mic.0.2008/016998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pentecost M, Otto G, Theriot JA, et al. Listeria monocytogenes invades the epithelial junctions at sites of cell extrusion. PLoS pathogens. 2006;2:e3. doi: 10.1371/journal.ppat.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]