Abstract
Microfabricated elastomeric scaffolds with 3D structural patterns are created by semi-automated layer-by-layer assembly of planar polymer sheets with through-pores. The meso-scale interconnected pore architectures governed by the relative alignment of layers are shown to direct cell and muscle-like fiber orientation in both skeletal and cardiac muscle, enabling scale up of tissue constructs towards clinically relevant dimensions.
Keywords: polyglycerol sebacate (PGS), microfabrication, alignment, heart, muscle
Mammalian tissue is a complex three-dimensional (3D) environment of cells and extracellular matrix in which structural order can be found over different length scales. Particularly for tissues and organs whose function depends on the alignment and directionality of cells and larger multi-cellular structures, control over architectural patterns and mechanical anisotropy are critical in developing functional tissue for regenerative medicine applications. In this work, we describe an approach to bridge the gap between the wealth of information and progress that has been made in controlling cell orientation on two-dimensional (2D) patterns and the less characterized effects of 3D structural cues in scaled-up tissue constructs. We developed a semi-automated layer-by-layer assembly method, using microelectromechanical systems (MEMS) fabrication and packaging technologies, to assemble planar elastomer sheets with through-pores into multi-layered scaffolds with tunable 3D structural patterns. With precise control over the micro-scale pore structure of individual layers and meso-scale pore interconnectivity patterns of multi-layered scaffolds, we were able to guide the alignment of cells into multi-cellular bundles approaching macro-scale dimensions using mouse myoblasts (C2C12) and neonatal rat heart cells. This design paradigm for scaling up polymer scaffolds is not only useful for furthering an understanding of cellular interactions with geometric and topographic cues in their 3D microenvironment, but also as a practical method to produce scaled-up, functional tissue constructs which is adaptable to a higher-throughput processing flow more suitable for larger scale manufacturing than previous MEMS fabrication approaches.
Controlling cell function is of central importance in engineering functional substitutes for organs and tissues.[1] Polymer scaffolds or matrices are often used in conjunction with cells to provide physical support and guidance, deliver specific growth factors and biochemical cues, and to offer protection from mechanical and other stresses.[2–3] Porous polymer scaffolds with stochastic or disordered structures at the micro-scale are among the oldest and most successful templates used for tissue engineering,[4–5] due in large part to the accessibility of techniques required to fabricate them, such as textile technologies[6] and porogen leaching.[7–8] Development of these random structures is still very relevant for applications such as microvascular networks[9–10] and as templates for host remodeling,[11] but as many tissues and organs are characterized by hierarchical and multi-scale 3D organization,[12] the scale-up of tissue engineered devices for increased clinical relevance and in vitro diagnostic applications will require control over tissue architecture. Maintaining function at the cellular level is particularly challenging for tissues and organs where structure and directionality at hierarchical scales provide the basis for tissue function, such as the heart, in which interwoven sheets of myocardial syncytia and collagen are found in a laminar architecture that varies throughout the thickness of the ventricular wall,[13–14] and the brain, in which cerebral fiber pathways form a recti-linear grid continuous with the principal axes of development.[15] Replicating these types of multi-scale arrangements will be crucial to building large pieces of functional tissue.
Different approaches have been used to mimic key features of this intricate environment, such as application of chemical and physical signals[16–17] and recreating native tissue structures by cell printing or seeding within pre-fabricated scaffolds. Various methods to print or place elements of tissue, such as different cell types, hydrogels, growth factors, and polymers in patterned proximity show promise for the development of complex tissue types,[18–23] but are not yet capable of replicating all the structural interactions which guide cells at the micro-scale. The geometric and architectural cues critical for guiding cell alignment and orientation are most often studied using various 2D or so-called 2.5D models, where lithographic and other microfabrication methods have been used to systematically evaluate parameters such as feature shape and size that influence cell alignment.[24–31] Extension of these studies to 3D has until recently been difficult due to a lack of technology to fabricate structures with control over architectural geometries in a third dimension. Solid freeform fabrication/additive manufacturing (SFF/AM) can be used to leverage specific material properties to spatially direct 3D structures,[32] and has been used to create scaffolds with micro-scale features for a number of cell systems.[33–34] These techniques, however, rely on optical, chemical, and/or thermal manipulation of a material are therefore limited in their applicability, and can also be relatively slow as fabrication of the entire scaffold occurs in one extended processing step.
Here we propose a novel method to assemble porous elastomeric sheets with controlled architectural order in 3D. Specifically, a semi-automated layer-by-layer assembly method is used to stack planar sheets of poly(glycerol sebacate) (PGS) with pre-defined and fabricated through-pores into multi-layered structures with unique pore geometries and interconnected architectures as a result of the relative alignment patterns of the different layers. The pore-interconnectivity patterns have a characteristic length scale on the order of hundreds of micrometers, defined here as the mesoscale, a transition between the microscale (1 – 1000 µm) and the microscale (1 – 1000 mm). Our fabrication method (Fig. 1a) relies heavily on bioMEMS technologies, including standard photolithography and etching to create re-usable silicon wafers for micro-molding of individual polymer sheets with through-pores, followed by a new procedure to align, stack, and bond the layers using an automated, precision alignment device. While various bioMEMS approaches incorporating some form of layer-by-layer assembly of pre-fabricated sheets have previously been used to create 3D porous cell culture scaffolds[35–39] and 3D microvascular networks,[40–41] these stacking methods relied on manual alignment. Some attempts have been made to improve stacking of soft polymers to enable more complex structures [42], but stacking processes are generally cited as being too slow[10] or requiring highly specialized equipment, [43] limiting their scalability. However, a similar stacking process is used routinely for circuit board assembly and 3D integrated circuit (IC) packages in the electronics packaging industry.[44–46] These standard assembly techniques are scalable from prototype research and development up to production scale capabilities. This 3D IC assembly process is typically used for silicon based, high modulus integrated circuit components. We have adapted the process to handle our flexible and porous biodegradable elastomer sheets.
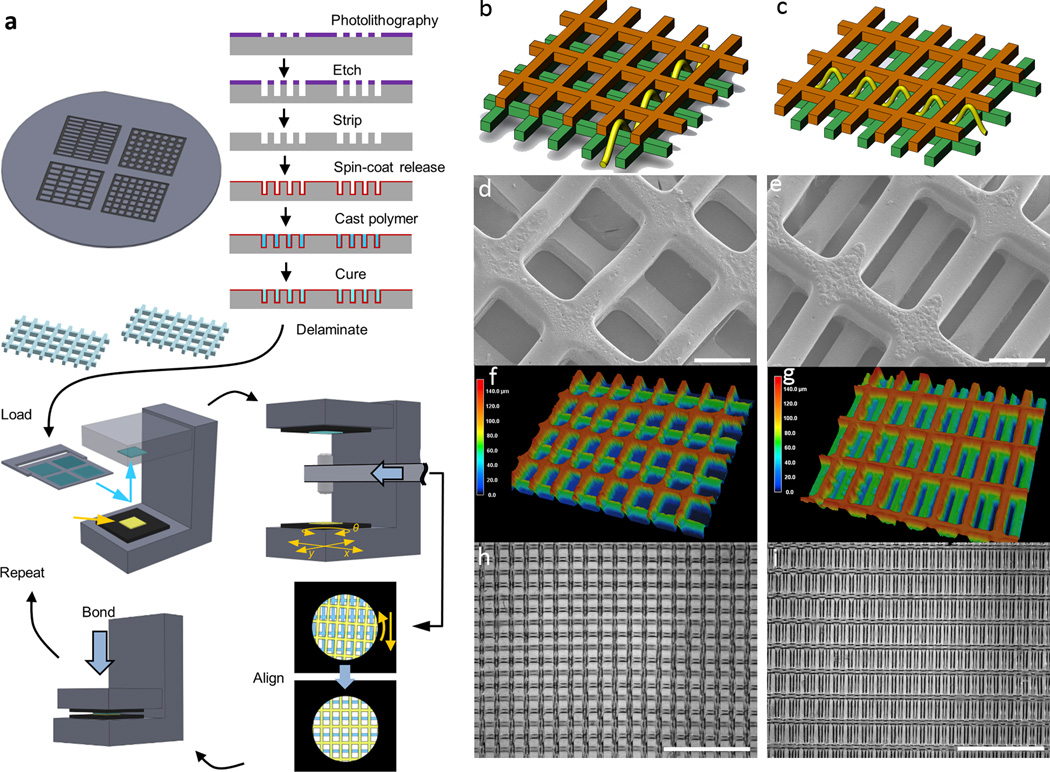
Figure 1.
Fabrication of multi-layer scaffolds with unique 3D structural patterns. (a) Process flow for creating multi-layer scaffolds. To make polymer sheets with throughpores, Si wafers were etched with specific patterns to serve as reusable molds, on which poly(glyercol sebacate) (PGS) was cast, cured, and delaminated by dissolving a sacrificial layer of maltose. Multi-layer scaffolds were assembled using a semi-automatic process to align and bond the sheets into multi-layer structures. (b)-(i) Demonstration of two specific two-layered (2L) alignment patterns, short strut offset (SSoff) (b,d,f,h) and long strut offset (LSoff)(c,e,g,i), for PGS polymer sheets, each sheet with rectangular pores 250 µm×125 µm and thickness of 70 µm. (b-c) Schematic drawings with yellow line indicating pore interconnectivity pattern, (d-e) SEM showing internal pore structures, (f-g) 3D laser scanning micrographs indicating features in the third dimension via color bar, (h-i) bright-field micrographs indicating multi-layered alignment over a macro-scale on the order of several mm. Scale bars are (d,e) 100 µm (h,i) 1 mm.
For a clear demonstration of the importance of the 3D structural patterns that are created based on the relative alignment of multiple layers, here we focus on two specific architectures created from PGS sheets with the same rectangular pore pattern (125 µm×250 µm inner pore dimensions with 50 µm strut width, 70 µm thick): short strut offset (SSoff) and long strut offset (LSoff) two layer (2L) scaffolds (Fig. 1b-i). The alignment of specific out-of-plane microfeatures was used to create two distinct pore interconnectivity patterns from the same 2D pore shape, as schematically shown by sinusoidal arrows in Fig. 1b and Fig. 1c. These periodic structural patterns were demonstrated by micrography (Fig. 1d-i) and realized over macro-scale distances approaching the centimeter scale (Fig. 1h-i).
To test whether 3D architectural cues could be used to align cells and multi-cellular tissues, we first cultured a murine skeletal myoblast cell line (C2C12) on scaffolds with these two patterns. We found striking alignment of both cells and multi-cellular muscle-like bundles extending the width of the scaffolds (5 mm) (Fig. 2a-b). Histological cross sections (Fig. 2c-f) and confocal images (Fig. 2g-j) confirmed that cells elongated in a sinusoidal path extending over and under the offset PGS struts in both patterns, to form interwoven muscle-like bundles. Further, these bundles were aligned in divergent directions relative to the 2D rectangular pore shape, where bundles in the SSoff pattern were aligned in the PD (parallel to the long rectangle axis), but bundles in the LSoff pattern were aligned in the XD (perpendicular to the long rectangle axis) (Fig. 2a-b,g-j). Quantification of alignment at the cellular level was performed on confocal slices of filamentous actin (F-actin) stained constructs in the middle of the muscle-like bundles at the interface of the two scaffold layers using a fast Fourier transform (FFT)-based algorithm (Fig. 2k, Fig. S1, Supporting Information). This again indicated perpendicular cellular orientations for LSoff versus SSoff patterns, and further that the orientation index, a measure of how well the cells were aligned in the predominant direction, was almost identical for the two patterns (Fig. 2l). The control over cell orientation was due primarily to the meso-scale pore interconnectivity pattern, in turn dictated by the relative alignment of the two scaffold layers. The scaffolds provided micro-scale structural cues (on the order of 50-to−100 µm), including widths of the struts and the gaps between offset struts, that cells used for attachment and elongation,[35] but the directionality of elongation was dictated by the meso-scale pathways (on the order of hundreds of µm) through the scaffolds that were created from periodic patterns of micro-features in the specific 3D architecture. This effect of 3D meso-scale architecture created by periodic micro-features was demonstrably more important than 2D pore shape.
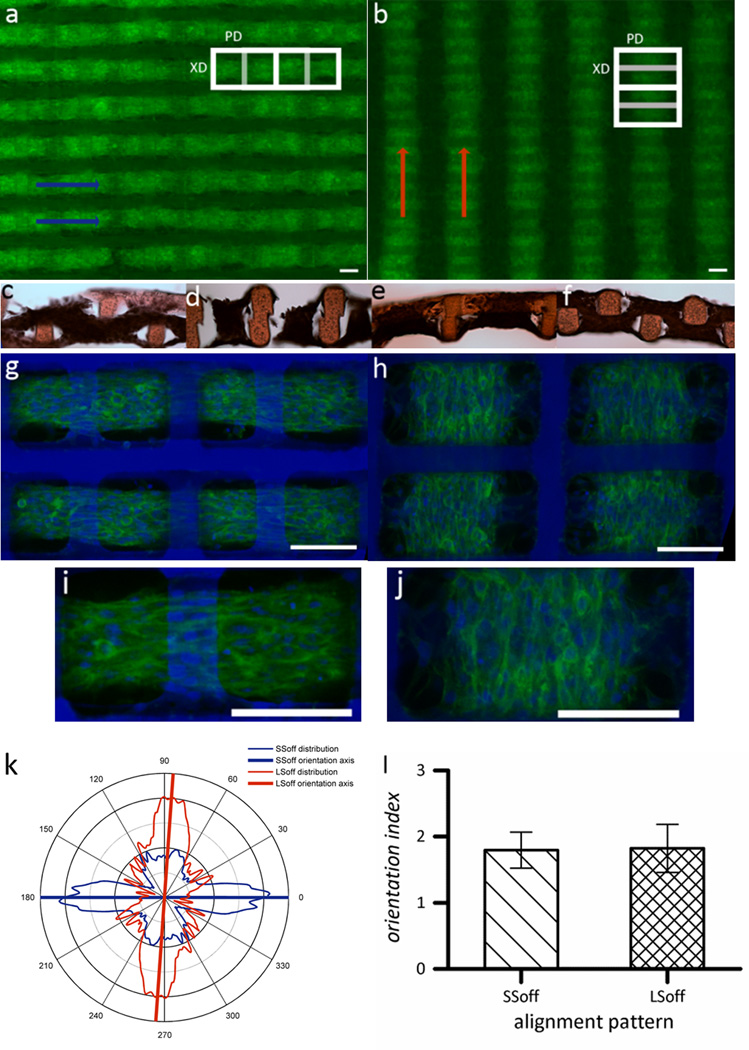
Figure 2.
Multi-scale alignment of C2C12 myoblasts cultured on 2L PGS scaffolds with different 3D structural patterns. (a,c,d,g,i) SSoff pattern (b,e,f,h,j) LSoff pattern. (a-b) Wide view fluorescent micrographs of F-actin stained tissue constructs, where macroalignment of muscle tissue bundles is indicated by arrows. (c-f) Cross sections stained with hematoxylin and eosin indicated tissue weaving throughout both scaffold layers (c) PD-section of SSoff (d) XD-section of SSoff (e) PD-section of LSoff (f) XD-section of LSoff. (g-j) Confocal micrographs of tissue constructs stained for F-actin (green) and counterstained for nuclei (blue) taken at the interface of the two scaffold layers demonstrating meso-scale pore interconnectivity patterns which guided tissue architecture and cell alignment. (k) Graphical representation of cellular alignment quantified via FFT analysis of n=6 circular masks cropped from confocal micrographs (see Supplementary Figure S1), with the axis lines representing the predominant alignment direction. (l) Orientation index (average of n=6) indicating that both distributions are equally ‘well-aligned,’ calculated by normalizing the percentage of cells aligned within ± 20° of the mean to the percentage of cells aligned within ±20° of the mean if cells were randomly oriented. Scale bars in all images are 100 µm.
Similar results of aligned tissue bundles interwoven with PGS scaffold were seen with neonatal rat heart cells after 5 days in culture (Fig. 3a-f). Cardiac function and differentiation were indicated by contractility in response to electrical field stimulation (Vid. S1–S2, Supporting Information), staining of cells and striated myofibers with sarcomeric α-actinin (Fig. 3b,f), and ultrastructural demonstration of myofilaments with clearly defined sarcomeres and z-lines, abundant mitochondria, and well-formed junctional complexes (Fig. 3g, h). The muscle-like tissue bundles shown in Fig. 3 may be comprised of cardiac myocytes, fibroblasts, smooth muscle cells, and endothelial cells,[17] since the scaffolds were seeded with a mixed population of heart cells. It is likely that the cardiac fibroblasts provided the structural support and directionality (Fig. 3e) for the tissue bundles[47] and also served as electrical conduits to form a syncytium between clusters of distributed cardiac myocytes.[48] Cardiac myocytes appeared as a mixture of more mature elongated and striated cells together with immature, rounded cells (Fig. 3f), where their elongation was presumably guided by the direction of the tissue bundles. In previous studies, scaffolds assembled by manual stacking did not exhibit sufficient 3D architectural order to enable the consistent, extensive alignment of muscle cells or striated cardiac myocytes found here.[35–36]
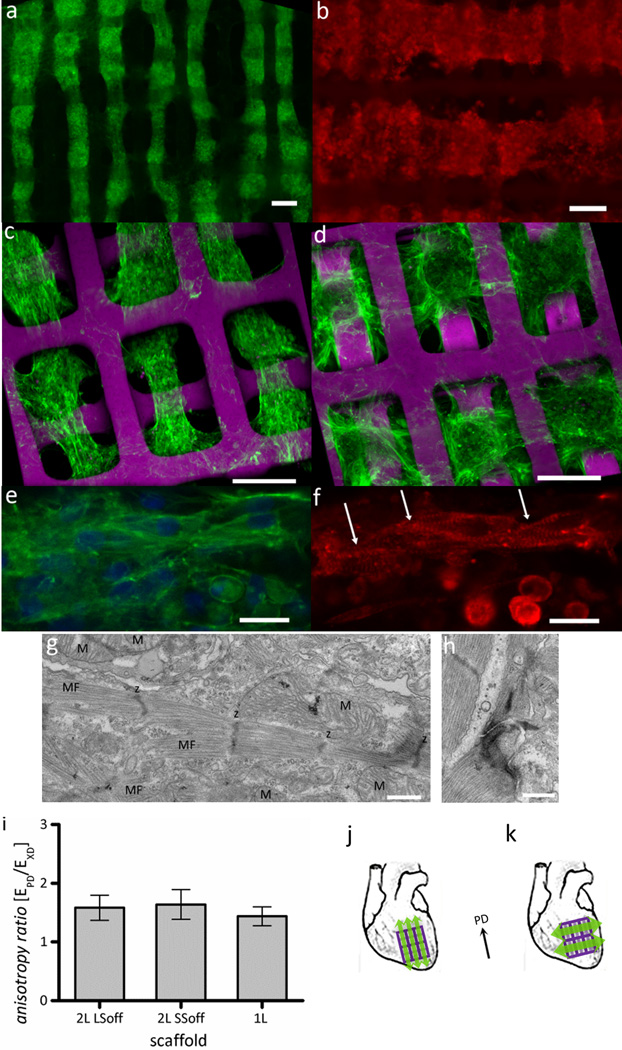
Figure 3.
Alignment and differentiation of neonatal rat heart cells cultured on 2L PGS scaffolds with different 3D structural patterns. (a,c) SSoff, (b,d)LSoff, shown at different magnifications and imaged by (a-b) fluorescence or (c-d) confocal microscopy (a,c-e) of F-actin stained (green) and nuclear DNA counter-stained or (b,f) sarcomeric α-actinin stained (red); arrows indicate striations in cardiac myocytes. (g,h) TEM images showing (g) abundant mitochondria (M) positioned between myofilaments (MF) with clearly defined sarcomeres and z-lines (z) and (h) a well-formed junctional complex. (i) Mechanical testing confirmed that mechanical anisotropy was independent of pore interconnectivity pattern for the 2L scaffolds (LSoff versus SSoff) and independent of the number of layers (2L versus 1L scaffolds). (j-k) As cell alignment and mechanical anisotropy can be effectively decoupled, tissue constructs can be implanted with muscle contraction from cell alignment (green arrows) and mechanical support (PD, black arrow) from PGS scaffolds in the (j) same or perpendicular (k) directions. Scale bars are (a-d) 100 µm (e,f) 10 µm (g,h) 500 nm.
The use of rectangular pores in the basic 1L scaffold design was motivated by their 2D mechanical anistropy, which was previously demonstrated to be similar to native adult rat ventricular myocardium.[30, 36] Tensile mechanical testing verified the mechanical anisotropy of 1L scaffolds was governed by the 2D pore shape as previously suggested and demonstrated [30–31, 35–36, 49] and that mechanical anisotropy of 2L scaffolds was maintained for the two specific pore interconnectivity patterns (LSoff versus SSoff) (Fig. 3i, Table 1 Supporting Information). As a result, multi-layered tissue constructs could be developed for implantation that provide anisotropic mechanical support in the longitudinal direction of the left ventricle, which has been demonstrated to improve systolic function[50–51] and contractility in either the longitudinal or circumferential direction (Fig. 3k,l), in a two-pronged approach to aid in recovery post- myocardial infarction. One of the key implications of this scaffold design is that while 3D structural cues can be used to guide cell and muscle fiber alignment, thereby enabling directional contractile force generation, 2D pore shape can be independently used to determine construct mechanical anisotropy and passive mechanical restraint.
Ultimately, to be of clinical relevance, tissue constructs must be scaled up to the macro-scale not only in length and width, but also in thickness. This presents unique challenges not limited to the practical problem of how to create thicker (i.e. many layered) constructs with controlled 3D architectures in a manner suitable for higher production volumes, but also the shifting of critical design paradigms that occur when crossing scales. Specifically, we have demonstrated that in moving the scaffold design space from 2D or 2.5D (i.e. a single layer with topological features or through-pores) to 3D (multi-layer structures with a third relevant dimension), the dominant factor guiding cell alignment is no longer in-plane features such as pore shape but instead the architectural order that includes a third dimension, which in this case is the characteristic meso-scale pore interconnectivity patterns created by periodic microfeatures. Towards fabricating thicker scaffolds, the semi-automated stacking method described here was easily used to add additional layers to the scaffolds, including three layered scaffolds that supported contractile cardiac tissue seeded throughout (Fig. 4a-b, Video S3 Supporting Information), four layered scaffolds (Fig. 4c-d), and so on.
Figure 4.
Scale-up of multi-layer scaffolds for implantation of cardiac tissue constructs. (a) Bright-field image of a three-layered (3L) scaffolds with rectangular pores with “staircase” alignment pattern (b) Confocal cross-sections of heart cells culture on the 3Lscaffold shown in (a), stained for F-actin (green) and counterstained for nuclear DNA (purple), demonstrating seeding throughout all layers (c,d) SEM images of four-layered scaffolds in a short-strut offset alignment pattern, with number indicating individual layers (e,f) SEM images showing (e) longitudinal and (f) cross section views of a five-layered tissue construct including two layered scaffolds on either side of a central layer with channels instead of through-pores, which can be used for perfusion of nutrients and/or endothelialization. Channels are indicated by asterisks. Scale bars are (a) 350 µm (b-f) 100 µm.
However, scaffolds cannot be indefinitely scaled up in this manner, as tissue more than a few hundred µm thick will require a supporting channel network to enhance transport, analogous to the transport function of the vasculature.[43] Semi-automated layer-by-layer assembly could also be used to incorporate microfluidic flow channels which could serve as templates for vascularization in vivo or simply perfusion support in vitro. One such possible design was fabricated using this stacking method, in which a layer of microfluidic channels each with a cross-section ~ 100 µm×100 µm was assembled in between two 2L offset rectangular scaffolds (Fig. 4 e-f). Regardless of the merits of this specific design, whose further characterization is the subject of ongoing work, it demonstrates the flexibility of the stacking approach to reliably assemble various layers into a large device. A higher throughput manufacturing process could easily be envisioned based on similar semi-automated stacking technology, where individual layers could be fabricated in parallel using wafer molds as described here, or with other microfabrication technologies such as laser microablation or even various SFF/AM techniques, as to be applicable to a wide range of polymers.
In this work, we furthered a bioMEMS based approach for creating micro-scale pores in elastomeric polymer scaffolds, by using precision assembly technology to extend these 2.5D scaffolds to 3D with control over the architectural order in this additional dimension. The semi-automated method we developed to align, stack, and bond multiple porous polymer sheets is useful in that enables a more efficient scale up of scaffolds both in size and in throughput, but perhaps more importantly, it opens up an additional design space in scaffold fabrication. This 3D architectural order, which we controlled here by precise patterning of out-of-plane micro-scale features to create distinct meso-scale structural characteristics, was demonstrated to be of critical importance in guiding cell elongation and tissue architecture, superseding the effects of the 2D features alone. This conclusion has profound implications when scaling up scaffolds to create tissue constructs that are relevant to clinical and in vitro diagnostic applications, in that design criteria used to control cells and tissue in 2D must be considered in the context of the 3D structure it imparts upon scale-up. In addition, the method we developed will enable the systematic evaluation of relationships and synergies between various micro-scale 2D features and meso-scale 3D periodic patterns in efforts to further optimize tissue structure and function. Finally, this design paradigm of guiding tissue orientation via controllable, meso-scale pore interconnectivity patterns has an immediate impact on developing implantable devices to augment or replace tissue function, as it provides a clear path forward towards more closely replicating the complex weavings, layerings, and anisotropies found in native tissues such as the heart.
Experimental
Scaffold fabrication and characterization
Silicon wafer molds were etched and prepared for use with a sacrificial layer of maltose as previously described [35]. PGS was synthesized by reacting a 1:1 molar ratio of glycerol and sebacic acid under heat and vacuum as previously described [35, 52]. To make one layered sheets, PGS was dissolved in ethanol (25% w/v), cast into 4 cm2 etched regions of the wafer mold, pre-cured on a hotplate at 110°C for 30 min, and vacuum cured at 165°C for 6.5 h at 10–15 mTorr. After curing, the wafers were soaked in dI-H2O at 80°C for 16–24 h and PGS sheets were delaminated off the wafers. To make multi-layered scaffolds, PGS sheets were transferred to poly(tetrafluoroethylene) (Teflon) backings, and loaded into an automated device bonder (model FC150 with alignment resolution to better than 0.5 µm, Smart Equipment Technology, Saint Jeoire, France). We programmed the tool to execute an automated bonding cycle that included scaffold pick up and positioning, before allowing the operator to align the scaffolds to the desired pattern using a dual-objective microscope and control of x, y, and θ on a mechanical stage. Layers were automatically brought together, and upon contact bonded at 100 g, 160°C for 2 min. For additional layers the cycle was repeated. Mechanical properties of the scaffolds were measured using an ELF3200 tester (Bose, Framingham, MA) as previously described [30–31, 35]. In brief, scaffold mechanical anisotropy was assessed by measuring effective stiffnesses under uniaxial tension in two orthogonal directions, the long rectangular pore axis (EPD) and the short rectangular pore axis (EXD), and calculating the associated anisotropy ratio (EPD/EXD). Samples were imaged by scanning electron microscopy (SEM) after sputter-coating with gold-palladium (108auto sputter coater, Cressington Scientific Instruments) using a Hitachi S3500 SEM (Hitachi High Technologies America). Other samples were imaged using a VK-X100 3D laser scanning microscope (Keyence America) for 3D topography or a Zeiss Axiovert 200M inverted microscope (Zeiss) at 1X–5X magnification for wide field-of-view confirmation of alignment.
Tissue construct preparation and maintenance
Multi-layered scaffolds (5mm diameter disks) were soaked in complete culture media (Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (P/S) (Invitrogen)) for 5 days prior to cell seeding, with media replaced after 2 days. Scaffolds were placed inside 6mm ID×8mm OD×3 mm thick poly(dimethylsiloxane) gaskets on non-cell culture treated 6-well plates for cell seeding. C2C12 myoblasts were obtained from American Type Culture Collection (ATCC), maintained in complete culture media, and seeded at 2500 cells mm−2. C2C12 constructs were maintained in complete culture media for 3 days, followed by 1.5 days in differentiation media (DMEM supplemented with 2% horse serum and 1% P/S). Heart cells were isolated from 1–2 day old neonatal rats following an Institutional Animal Care and Use Committee-approved protocol as previously described [30, 35–36], and seeded at 1–2×105 cells mm−2. Heart cell constructs were maintained in supplemented complete culture media (DMEM with 10% FBS, 1% P/S, and 50 ng mL−1 of insulin-like growth factor I) for 5 days with a complete media replacement 2 days after seeding, and were assessed for contractility at the end of the culture period. Samples to be analyzed by immunohistochemistry or histology were fixed (10% neutral buffered formalin overnight at 4°C). Samples to be analyzed by transmission electron microscopy (TEM) were fixed overnight (2.5% glutaraldehyde and 3% paraformaldehyde with 5% sucrose in 0.1M sodium cacodylate buffer (pH 7.4)) and postfixed (1% OsO4 in veronal acetate buffer).
Construct Analysis
The response of heart cell constructs to electrical stimulation was assessed using a temperature controlled test-chamber with carbon electrodes connected to an electrical pulse generator (S88X, Astro-Med Inc. West Warwick, RI) which was used to apply monophasic pulses at a 1-Hz frequency, as previously described [53], and video was captured on a Zeiss Axiovert microscope. To visualize filamentous actin (F-actin), constructs were rinsed, permeabilized (1 h, 0.1% Triton-X in phosphate buffered saline (PBS)), and stained with fluorescein isothiocyanate (FITC)-phalloidin conjugate (3 h, 1:40, Invitrogen) followed by DRAQ5 nuclear counterstain (1:200, Axxora LLC). For differentiated marker assessment, sarcomeric α-actinin primary antibody (1:100, Sigma) for cardiac muscle was used in conjunction with FITC-phalladoin and an additional blocking step (1 h, 0.05% Triton-X, 0.1% bovine serum albumin in PBS), followed by Texas-red conjugated secondary antibody (1 h, 1:200, Invitrogen) and counterstaining with DRAQ5. After staining, samples were examined with either a Zeiss Axiovert microscope for low-magnification images or a Nikon 1AR Spectral Scanning Confocal Microscope for detailed 3D reconstructions. For assessment of cell penetration into the scaffold, samples were embedded in Tissue Tek O.C.T. Compound (Sakura Finetek), frozen, cross-sectioned to 50 µm, stained with hematoxylin and eosin (H&E), and imaged on a Zeiss Axiovert microscope. Samples fixed for TEM were stained en bloc (overnight, 5.5% uranyl acetate in veronal acetate buffer, pH 6.0 (UA)), dehydrated, embedded in Spurr’s resin, cross-sectioned to 50 nm using a microtome (Reichert Ultracut E) with diamond knife (Diatome), stained (2% UA followed by 0.1% lead citrate), and examined using an EM410 TEM instrument (Philips, Eindhoven, The Netherlands) at 80kV.
Quantification of cell orientation
Cell orientation within 2L scaffolds was analyzed from images corresponding to the interface of the two layers from confocal stacks of F-actin stained tissue constructs. Images were cropped in circular, intra-pore regions to reduce edge and scaffold effects, respectively, and analyzed using an FFT-based algorithm in MATLAB adapted from several previous approaches [30, 33, 54]. The FFT power spectrum was integrated over a 360° rotation using the center beam of a ‘fanbeam’ projection [33], which was normalized, and averaged over n=6 pores for each scaffold. The orientation index was defined as the fraction of cells or fibers aligned within 20° of the peak angle, normalized to the fraction of randomly oriented fibers that would lie within this range [54].
Supplementary Material
Acknowledgments
This work was funded by the National Heart, Lung and Blood Institute (NHLBI), Award 1-R01-HL107503 (LEF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or NIH. We thank R. Morrison, J. Hsaio, and M. Bancu for help with MEMS Fab, E. Kim for help with drawings, L. Lu for help with frozen sections, and E. Vasile and N. Watson for help with microscopy.
Footnotes
Author Disclosure Statement: No competing financial interests exist. A patent application has been filed with the United States Patent and Trademark Office
((Supporting Information is available online from Wiley InterScience or from the author)).
Contributor Information
Martin E. Kolewe, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Hyoungshin Park, Microsystems Development and Microfabrication Process Engineering Groups, Charles Stark Draper Laboratory, Cambridge, MA 02139, USA.
Caprice Gray, Microsystems Development and Microfabrication Process Engineering Groups, Charles Stark Draper Laboratory, Cambridge, MA 02139, USA.
Xiaofeng Ye, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Robert Langer, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Lisa E. Freed, Email: lfreed@mit.edu, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Microsystems Development and Microfabrication Process Engineering Groups, Charles Stark Draper Laboratory, Cambridge, MA 02139, USA.
References
- 1.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Park H, Cannizzaro C, Vunjak-Novakovic G, Langer R, Vacanti CA, Farokhzad OC. Tissue Eng. 2007;13:1867. doi: 10.1089/ten.2006.0198. [DOI] [PubMed] [Google Scholar]
- 3.Freed LE, Engelmayr GC, Borenstein JT, Moutos FT, Guilak F. Adv. Mater. 2009;21:3410. doi: 10.1002/adma.200900303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed LE, Marquis JC, Nohria A, Emmanual J, Mikos AG, Langer R. J. Biomed. Mater. Res. 1993;27:11. doi: 10.1002/jbm.820270104. [DOI] [PubMed] [Google Scholar]
- 5.Radisic M, Park H, Chen F, Salazar-Lazzaro JE, Wang YD, Dennis R, Langer R, Freed LE, Vunjak-Novakovic G. Tissue Eng. 2006;12:2077. doi: 10.1089/ten.2006.12.2077. [DOI] [PubMed] [Google Scholar]
- 6.Moutos FT, Freed LE, Guilak F. Nat. Mater. 2007;6:162. doi: 10.1038/nmat1822. [DOI] [PubMed] [Google Scholar]
- 7.Gao J, Crapo PM, Wang YD. Tissue Eng. 2006;12:917. doi: 10.1089/ten.2006.12.917. [DOI] [PubMed] [Google Scholar]
- 8.Chen VJ, Ma PX. Biomaterials. 2004;25:2065. doi: 10.1016/j.biomaterials.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 9.Bellan LM, Kniazeva T, Kim ES, Epshteyn AA, Cropek DM, Langer R, Borenstein JT. Adv. Healthcare Mater. 2012;1:164. doi: 10.1002/adhm.201100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellan LM, Pearsall M, Cropek DM, Langer R. Adv. Mater. 2012;24:5187. doi: 10.1002/adma.201200810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, Allen RA, Wang YD. Nat. Med. 2012;18:1148. doi: 10.1038/nm.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Cuddihy MJ, Kotov NA. Tissue Eng. Part B Rev. 2008;14:61. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- 13.Legrice IJ, Smaill BH, Chai LZ, Edgar SG, Gavin JB, Hunter PJ. Amer. J. Phys.-Heart Circ. Phys. 1995;269:H571. doi: 10.1152/ajpheart.1995.269.2.H571. [DOI] [PubMed] [Google Scholar]
- 14.Pope AJ, Sands GB, Smaill BH, LeGrice IJ. Amer. J. Phys.-Heart Circ. Phys. 2008;295:H1243. doi: 10.1152/ajpheart.00484.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wedeen VJ, Rosene DL, Wang RP, Dai GP, Mortazavi F, Hagmann P, Kaas JH, Tseng WYI. Science. 2012;335:1628. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu LLY, Montgomery M, Liang Y, Liu HJ, Radisic M. Proc. Nat. Acad. Sci. USA. 2012;109:E3414. doi: 10.1073/pnas.1210580109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naito H, Melnychenko I, Didie M, Schneiderbanger K, Schubert P, Rosenkranz S, Eschenhagen T, Zimmermann WH. Circulation. 2006;114:I72. doi: 10.1161/CIRCULATIONAHA.105.001560. [DOI] [PubMed] [Google Scholar]
- 18.Khademhosseini A, Suh KY, Yang JM, Eng G, Yeh J, Levenberg S, Langer R. Biomaterials. 2004;25:3583. doi: 10.1016/j.biomaterials.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Culver JC, Hoffmann JC, Poche RA, Slater JH, West JL, Dickinson ME. Adv. Mater. 2012;24:2344. doi: 10.1002/adma.201200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du YA, Lo E, Ali S, Khademhosseini A. Proc. Nat. Acad. Sci. USA. 2008;105:9522. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, Parlakgul G, Bernstein J, Xing W, Boyden ES, Demirci U. Adv. Mater. 2013;25:1192. doi: 10.1002/adma.201203261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norotte C, Marga FS, Niklason LE, Forgacs G. Biomaterials. 2009;30:5910. doi: 10.1016/j.biomaterials.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu F, Wu CAM, Rengarajan V, Finley TD, Keles HO, Sung YR, Li BQ, Gurkan UA, Demirci U. Adv. Mater. 2011;23:4254. doi: 10.1002/adma.201101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Proc. Nat. Acad. Sci. USA. 2010;107:565. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang BY, Xiao Y, Hsieh A, Thavandiran N, Radisic M. Nanotechnology. 2011;22:10. doi: 10.1088/0957-4484/22/49/494003. [DOI] [PubMed] [Google Scholar]
- 26.Hume SL, Hoyt SM, Walker JS, Sridhar BV, Ashley JF, Bowman CN, Bryant SJ. Acta Biomaterialia. 2012;8:2193. doi: 10.1016/j.actbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Feinberg AW, Alford PW, Jin HW, Ripplinger CM, Werdich AA, Sheehy SP, Grosberg A, Parker KK. Biomaterials. 2012;33:5732. doi: 10.1016/j.biomaterials.2012.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bettinger CJ, Orrick B, Misra A, Langer R, Borenstein JT. Biomaterials. 2006;27:2558. doi: 10.1016/j.biomaterials.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 29.Bian WN, Bursac N. Biomaterials. 2009;30:1401. doi: 10.1016/j.biomaterials.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelmayr GC, Cheng MY, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat. Mater. 2008;7:1003. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillemette MD, Park H, Hsiao JC, Jain SR, Larson BL, Langer R, Freed LE. Macromol. Biosci. 2010;10:1330. doi: 10.1002/mabi.201000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melchels FPW, Domingos MAN, Klein TJ, Malda J, Bartolo PJ, Hutmacher DW. Prog. Polymer Sci. 2012;37:1079. [Google Scholar]
- 33.Hanson Shepherd JN, Parker ST, Shepherd RF, Gillette MU, Lewis JA, Nuzzo RG. Adv. Funct. Mater. 2011;21:47. doi: 10.1002/adfm.201001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melissinaki V, Gill AA, Ortega I, Vamvakaki M, Ranella A, Haycock JW, Fotakis C, Farsari M, Claeyssens F. Biofabrication. 2011;3:12. doi: 10.1088/1758-5082/3/4/045005. [DOI] [PubMed] [Google Scholar]
- 35.Neal RA, Jean A, Park H, Wu PB, Hsiao J, Engelmayr GC, Jr., Langer R, Freed LE. Tissue Eng. 2013;19:793. doi: 10.1089/ten.tea.2012.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park H, Larson BL, Guillemette MD, Jain SR, Hua C, Engelmayr GC, Freed LE. Biomaterials. 2011;32:1856. doi: 10.1016/j.biomaterials.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folch A, Mezzour S, Du, ring M, Hurtado O, Toner M, Mu, ller R. Biomed. Microdev. 2000;2:207. [Google Scholar]
- 38.Ryu W, Min SW, Hammerick KE, Vyakarnam M, Greco RS, Prinz FB, Fasching RJ. Biomaterials. 2007;28:1174. doi: 10.1016/j.biomaterials.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Ryu W, Fasching RJ, Vyakarnam M, Greco RS, Prinz FB. J. MEMS. 2006;15:1457. [Google Scholar]
- 40.King KR, Wang CCJ, Kaazempur-Mofrad MR, Vacanti JP, Borenstein JT. Adv. Mater. 2004;16:2007. [Google Scholar]
- 41.Bettinger CJ, Weinberg EJ, Kulig KM, Vacanti JP, Wang YD, Borenstein JT, Langer R. Adv. Mater. 2006;18:165. doi: 10.1002/adma.200500438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marentis TC, Vacanti JP, Hsiao JC, Borenstein JT. J. MEMS. 2009;18:531. [Google Scholar]
- 43.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat. Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knickerbocker JU, Andry PS, Dang B, Horton RR, Interrante MJ, Patel CS, Polastre RJ, Sakuma K, Sirdeshmukh R, Sprogis EJ, Sri-Jayantha SM, Stephens AM, Topol AW, Tsang CK, Webb BC, Wright SL. IBM J. Res. Dev. 2008;52:553. [Google Scholar]
- 45.Al-Sarawi SF, Abbott D, Franzon PD. IEEE Trans. Components Pack. Manf. Tech. Part B-Adv. Pack. 1998;21:2. [Google Scholar]
- 46.Das RN, Egitto FD, Lauffer J, Bonitz B, Wilson B, Marconi F, Poliks MD, Markovich VR. Ieee, 2012 IEEE 62nd Elec. Comp. Tech. Conf. 2012:1333. [Google Scholar]
- 47.Radisic M, Park H, Martens TP, Salazar-Lazaro JE, Geng WL, Wang YD, Langer R, Freed LE, Vunjak-Novakovic G. J. Biomed. Mater. Res. Part A. 2008;86A:713. doi: 10.1002/jbm.a.31578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaudesius G, Miragoli M, Thomas SP, Rohr S. Circulation Res. 2003;93:421. doi: 10.1161/01.RES.0000089258.40661.0C. [DOI] [PubMed] [Google Scholar]
- 49.Jean A, Engelmayr GC. J. Biomech. 2010;43:3035. doi: 10.1016/j.jbiomech.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fomovsky GM, Macadangdang JR, Ailawadi G, Holmes JW. J. Cardio. Trans. Res. 2011;4:82. doi: 10.1007/s12265-010-9241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fomovsky GM, Clark SA, Parker KM, Ailawadi G, Holmes JW. Circulation-Heart Fail. 2012;5:515. doi: 10.1161/CIRCHEARTFAILURE.111.965731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang YD, Ameer GA, Sheppard BJ, Langer R. Nat. Biotech. 2002;20:602. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 53.Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, Freed LE, Vunjak-Novakovic G. Proc. Nat. Acad. Sci. USA. 2004;101:18129. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng CP, Hinz B, Swartz MA. J. Cell Sci. 2005;118:4731. doi: 10.1242/jcs.02605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.