Abstract
Development of exosome based semi-synthetic nanovesicles for diagnostic and therapeutic purposes requires novel approaches to load exosomes with cargo. Electroporation has previously been used to load exosomes with RNA. However, investigations into exosome colloidal stability following electroporation have not been considered. Herein, we report the development of a unique trehalose pulse media (TPM) that minimizes exosome aggregation following electroporation. Dynamic light scattering (DLS) and RNA absorbance were employed to determine the extent of exosome aggregation and electroextraction post electroporation in TPM compared to common PBS pulse media or sucrose pulse media (SPM). Use of TPM to disaggregate melanoma exosomes post electroporation was dependent on both exosome concentration and electric field strength. TPM maximized exosome dispersal post electroporation for both homogenous B16 melanoma and heterogeneous human serum derived populations of exosomes. Moreover, TPM enabled heavy cargo loading of melanoma exosomes with 5 nm superparamagnetic iron oxide nanoparticles (SPION5) while maintaining original exosome size and minimizing exosome aggregation as evidenced by transmission electron microscopy. Loading exosomes with SPION5 increased exosome density on sucrose gradients. This provides a simple, label free means to enrich exogenously modified exosomes and introduces the potential for MRI driven theranostic exosome investigations in vivo.
Keywords: exosomes, electroporation, superparamagnetic, iron oxide, trehalose
INTRODUCTORY STATEMENT
Cells communicate with their microenvironment through multiple mechanisms. One mechanism of relaying information relies on the production and secretion of messengers known as exosomes [1]. Exosomes are naturally occurring biological nanovesicles 30–200 nm in size [2; 3; 4; 5]. They are formed constitutively by the inward budding of multivesicular body (MVB) membranes. MVB’s fuse with the internal cell plasma membrane and release their exosome vesicles extracellulary. The nanoscale size of exosomes facilitates their penetration and interaction with distant tissue microenvironments [6]. For example, melanoma exosomes participate in the inflammatory responses of sentinel lymph nodes by establishing pre-metastatic niches that attract metastatic tumor cells [7]. Tumor exosomes also facilitate tumor immune evasion by direct suppression of immune cell activation [3; 4; 8].
One primary aim of exosome research is the development of exosome based semi-synthetic nanocarriers to detect and treat disease microenvironments [1]. The design of exosomal nanocarriers requires exogenous manipulation of exosome contents and/or biophysical properties (e.g., electrokinetic mobility, density, size, etc.) while simultaneously avoiding exosome aggregation. Exosome nanovesicles essentially exist as natural colloidal suspensions in most media (buffer, culture media, serum, and plasma). For many years, the nanotechnology industry has recognized that one of the hurdles in creating efficacious nanomedicines is overcoming the tendency of colloidal particles to aggregate or flocculate in suspension. Accordingly, ensuring homogenous dispersal of nanovesicles or nanoparticles is necessary for preserving functionality in vivo and for enhancing stability during storage.
To generate semi-synthetic nanovesicles from exosomes, one must establish methods to move cargo across the exosome membrane bilayer. A traditional approach for transferring cargo across cellular membranes is electroporation. In general, electroporation occurs when the voltage potential across a cell membrane exposed to an electric field is high enough to cause spontaneous pore formation in an attempt to neutralize the voltage difference. Electroporation has been used previously to load exosomes with RNA cargo [9; 10] which is logical given that exosomes transport RNA naturally [11; 12]. Moreover, the use of electroporation for exosome studies minimizes perturbation of sensitive exosome components such as ligands and receptors. Electroporation relies on transient pore formation in the absence of chemical modification. Despite the potential to generate heat through electrical resistance, electroporation produces no thermal damage to membrane components because electroporation exposure times are in the millisecond range resulting in minimal temperature rises in pulse media at ~ 1°C per pulse [13].
However, while electroporation is traditionally considered as a means to introduce DNA, RNA, enzymes, drugs or biochemical reagents into cells [13], it can also be used to trigger fusion between cells to form hybrid cells as in the case of generating hybridomas for antibody production [14] or liposomes to study the electrofusion process [15]. Natural electrofusion occurs in living cells at the intracellular organelle level to facilitate the formation of vesicles during the exocytic process [16].
While electrofusion may be either a desirable or an inconsequential side effect in cellular applications it is conceivable that the electroporation process may exacerbate the undesirable potential for exosomes to aggregate. To ameliorate this possibility, we hypothesized that the use of a membrane stabilizer during the electroporation process was warranted. Based on past experience with synthetic liposome stabilizers, we reasoned that use of the safe and biocompatible sugar additive trehalose to simple low impedance PBS pulse field media would enable exosome electroporation while minimizing aggregation. Trehalose is generally accepted as being the best disaccharide membrane protectant for preserving liposomes and their contents during freeze drying [17]. Moreover, a previous study demonstrated that inclusion of 50 mM trehalose in pulse field media greatly improves survival and transfection efficiency of mammalian cells post electroporation [18]. In combination, these membrane stabilizing properties of trehalose make it an ideal candidate for exosome based preparations and electroporation studies.
Herein we demonstrate for the first time that a simple pulse media containing 50 mM trehalose in PBS (TPM) is sufficient to minimize exosome aggregation following the electroporation process. Further, use of TPM simultaneously allows for loading of exosomes with 5 nm superparamagnetic iron oxide nanoparticle (SPION5) test cargo. SPIONs are biocompatible, safe and effective magnetic resonance imaging agents in vivo [13]. The development of a process to minimize exosome aggregation while loading exosomes with SPIONs signifies a major advancement in facilitating the reality of exosome based semi-synthetic nanocarriers.
MATERIALS AND METHODS
Materials and cell culture
Mouse B16-F10 melanoma cells were purchased from ATCC and maintained in culture with 90% DMEM and 10% heat inactivated fetal bovine serum at 37ºC and 5% CO2.
Isolation of exosomes
B16-F10 melanoma cell cultures were grown to 70% confluence in three 300 cm2 flask. Culture media was removed and cells washed in PBS. Cells were cultured for 48 hrs in the presence of conditioned media. Conditioned culture media was prepared by subjecting normal culture media to overnight ultracentrifugation at 110,000 x g to remove bovine exosomes [19]. B16 melanoma exosomes were collected from 48 hr culture in conditioned media through standard differential centrifugation steps using a 70 Ti rotor as previously described [19]. Briefly, culture media was diluted 1:1 in PBS or 50 mM trehalose in PBS or 50 mM sucrose in PBS, spun and supernatants collected from 300 x g for 10 min, 2000 x g for 10 min, to remove residual cells and debris, 10,000 x g for 30 min to remove microparticles [20], and 100,000 x g for 2 h (in the presence or absence of 1.0 μM DiI (InVitrogen, CA)). For isolation of human serum exosomes, pooled human serum (Innovative Research Inc., Novi MI) was diluted 1:10 in PBS or 50 mM trehalose PBS, subjected to centrifugation and supernatants collected at 300 x g for 10 min, 2000 x g for 10 min, and 10,000 x g for 30 min. Post 10,000 x g, supernatants were filtered through a 0.22 micron filter (MIDSCI™, St. Louis MO) to further reduce non-exosome vesicles followed by ultracentrifugation at 100,000 x g for 2 h to collect exosomes. Exosome pellets were resuspended in 1 ml of PBS, 50 mM trehalose PBS or 50 mM sucrose PBS and stored at −80ºC until use. Protein content was measured via BCA absorbance (Pierce).
Electroporation of Exosomes and Sizing by DLS
Exosomes, 50 μg protein, were re-suspended in 0.75 ml of PBS, 50 mM trehalose PBS or 50 mM sucrose PBS pulse media and kept at 4ºC while waiting for electroporation or sizing by dynamic light scattering. To load exosomes with 5 nm superparamagnetic iron oxide nanoparticles (SPION5, Ocean Nanotech), 50 mM trehalose PBS pulse media containing 0.25 μg/ml SPION5 iron was used. Suspended exosomes were electroporated in 4 mm path length electroporation cuvettes using a BTX Harvard Apparatus ECM 399 system (Holliston MA). A single pulse was applied to each exosome sample using the high voltage setting. The pulse length for each sample regardless of the type of pulse media utilized was measured by the electroporation device to be < 1 ms. Following electroporation, 75 μl of each exosome sample was set aside to assess RNA release indicative of successful electroextraction as a result of pore formation. Exosome samples were then diluted in 1 ml of pulse media and exosome effective diameter size assessed by dynamic light scattering (DLS, Brookhaven Instruments). In the case of SPION5 loaded exosomes, exosome sizing by DLS was performed using the intensity versus size mode to eliminate the potential for under sizing brought about by the contribution of free of 5 nm SPIONs to readings according to manufacturer application notes [21].
Measuring Exosome RNA release
RNA packaged in exosomes is protected by a lipid membrane bilayer containing numerous proteins. RNA has a maximal absorbance of 260 nm (purine and pyrimidine nucleotides) whereas maximal protein absorbance is 280 nm (tryptophan and tyrosine amino acids). However, protein can absorb light submaximal at 260 nm [22]. It follows that if RNA is released from exosomes it is unbuffered by the 260 nm absorption capacity of exosome membrane proteins. An absorbance spectrophotometer will thus detect more free exosome RNA at 260 nm proportional to the amount electroextracted and the total RNA detected will increase. Thus, following electroporation of each 750 μl exosome sample, 75 μl was placed in an absorbance cuvette and RNA measured by absorbance at 260 nm using an eppendorf BioPhotometer (Eppendorf North America Inc., Westbury, N.Y.) indicative of successful electroextraction as a result of pore formation. The amount of RNA detected in exosome samples, which are conventionally stored in PBS at −80°C, was used as the normalization point of comparison to electroporated samples +/− trehalose pulse media, set to 100% and averaged for individual exosome collections given the wide batch to batch variation of baseline exosome RNA storage levels.
In gradient fluorescent detection of exosomes
To assess the density of exosomes loaded with SPION5, exosomes were incubated passively or post electroporation for an additional 20 minutes at 4ºC in 0.75 ml of 50 mM trehalose pulse media containing 0.25 μg/ml SPION5 iron. Exosomes were subsequently washed and re-isolated by ultracentrifugation at 100,000 x g for 2 h in 50 mM trehalose PBS containing 0.4 μM DiI to fluorescently label exosomes and remove extravesicular iron. According to the manufacturer's technical data, the ultrasmall nanosize of SPION5 prevents the particles from being pelleted using exosome ultracentrifugation speeds. Isolation of SPION5 requires an average RCF ~ 376,523 x g to pellet. This property makes SPION5 an ideal test cargo loading agent for exosome electroporation studies. Flotation of exosomes (150 μg protein) on a continuous sucrose gradient (2.0 – 0.25 M sucrose, 20 mM HEPES/NaOH, pH 7.4) was performed as previously described but using an SW 41 rotor [23]. The gradient was produced using a Gradient Master (Biocomp Instruments, Fredericton, NB, Canada) and was spun for >15 hours at 100,000g. Post centrifugation, 1 ml fractions were collected from the bottom up. The density of each fraction was calculated using a refractometer [19]. 200 μl of each fraction was added to a black 96 well plate and DiI exosome fluorescence detected using a Xenogen IVIS fluorescent imager (Caliper Life Sciences).
Measuring Exosome SPION5 content
SPION5 loading of exosomes was measured using inductively coupled plasma optical emission spectroscopy (ICP-OES). Briefly, 100 μl of sucrose gradient exosome peak fractions 3,4 and 5 detected by in gradient fluorescence were placed in pre-acid rinsed tubes (Teflon coated etc.). To each sample, Optima grade nitric acid (HNO3) 0.5mL, hydrochloric acid (HCL) 0. 2mL, and hydrogen peroxide (H2O2) 0.1 mL was added and the mixture was mixed by repetitive pipetting. Samples were placed in a microwave for approximately two hours with power ramping over 20 min to 1000 W, then held for 10 min. to allow for cool down. The digested samples were then diluted to 10 ml in DI water which is an equivalent volume to that of iron calibration standards. The concentration of iron in the calibration standards was 0.5 ppm, 1 ppm and 2 ppm, respectively. Samples included 5 ppm yttrium as an internal standard.
Electron Microscopy
Purified exosome pellets were fixed with 2.5% glutaraldehyde in PBS for 30 minutes on ice. After rinsing, the pellet was sequentially stained in osmium tetroxide, tannic acid, and uranyl acetate; then dehydrated and embedded in Polybed 812. Tissue was thin sectioned on a Reichert-Jung Ultracut and viewed on a Jeol (JEM 1400) Electron Microscope. Images were recorded with an AMT V601 digital camera. E.M. reagents were purchased from Electron Microscopy Sciences.
Statistics
Statistical analyses were performed using JMP Version 10 Statistical Software (SAS Institute). One factor ANOVA was used when either pulse media type or electric field strength varied but not both. For analyses where both pulse media type and electric field varied, 2-factor ANOVA was utilized. Following ANOVA, Tukey HSD analysis was used to determine statistical significance for multiple comparisons within a data set. P values < 0.05 were considered statistically significant.
RESULTS
Evaluation of Trehalose Pulse Field Media for Exosome Electroporation
In the initial set of experiments we sought to determine the extent of exosome aggregation following electroporation in the presence of TPM. For all experiments we optimized the electroporation process for 50 μg of B16 melanoma exosomes based on protein content. We selected this batch size based on our previous experience efficiently isolating, labeling and tracking this quantity of B16 melanoma exosomes in vivo [7]. Moreover, B16 melanoma exosomes are an ideal model system for testing electroporation parameters because they are syngeneic with wild type C57/BL6 mice and enable future exosomal semi-synthetic investigations in vivo.
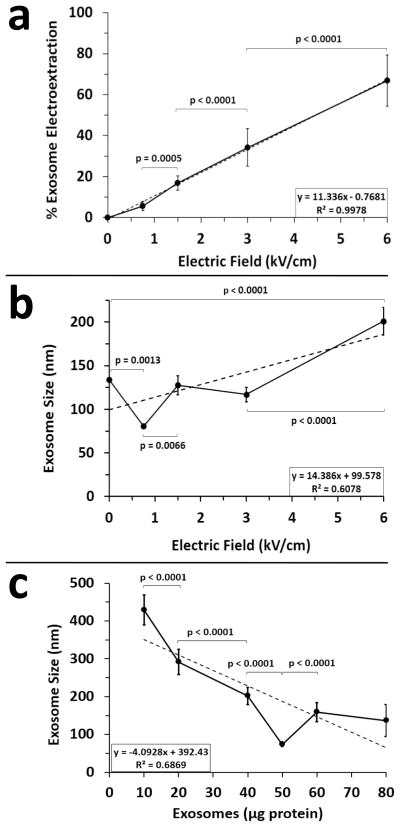
We first assessed the influence of TPM on exosome pore formation at select electric field strengths. During any electroporation event within an electric force field, there is two-way traffic. External contents move into cells or vesicles while internal contents move out in a process known as electroextraction. Electroextraction has been used previously to study cell contents [24]. Fortuitously, exosomes contain an exemplary electroextraction marker, RNA, the release of which is indicative of pore formation during the electroporation process. Therefore, we reasoned that exosome RNA content could be used to assess pore formation. As shown, increasing the electric field strength applied to a fixed 50 μg exosome sample size resulted in a significant linear increase in RNA extraction (Fig. 1a). This is to be expected given that increasing the strength of the electric field increases the magnitude of the electric force applied to the exosomes thus extracting more content.
Figure 1.
Exosome electroporation in the presence of trehalose pulse media. Post storage at −80° C, B16 melanoma exosomes were subjected to electroporation at 4°C using selected electric field strengths. (a) The amount of RNA electroextracted from 50 μg of exosomes was determined by RNA absorbance and reported as percent electroextraction. Background RNA absorbance in the absence of electroporation (0 kV/cm) was set to 0%; (n = 15 for 3 experiments and 5 measurements per experiment); error bars = s.d. (b) B16 melanoma exosome (50 μg) size was measured by DLS as a function of varying electric field strengths; (n = 27 for 3 experiments and 9 measurements per experiment); error bars = s.e.m. (c) B16 melanoma exosome size was measured using a constant electric field strength of 0.75 kV/cm while varying exosome concentration (micrograms of protein per 0.75 ml of trehalose pulse media); (n = 10 for 3 pooled exosome isolations); error bars = s.d. Best fit dotted lines denote linear regression analyses indicated by R2 values. Connecting bars denote statistically significant differences; p values < 0.05 were considered significant.
In the subsequent experiment, DLS was used to gage post electroporation exosome aggregation. As shown, we demonstrated a significant increase in exosome size post pellet isolation at 100,000 x g at an electric field strength of 6 kV/cm in contrast to no electroporation (0 kV/cm) (Fig. 1b). No change in exosome aggregation was observed at field strengths of 3 or 1.5 kV/cm. However, exosome dispersal significantly increased at a field strength of 0.75 kV/cm.
Interestingly, while a strong linear relationship for electroextraction versus electric field was observed for a fixed 50 μg exosome sample size (R2 = 0.99, Fig. 1a), this was not the case for size versus electric field (R2 = 0.61, Fig. 1b) because an electric field of 0.75 kV/cm uniquely dispersed the exosomes. We therefore hypothesized that an electric field of 0.75 kV/cm generates the required force to disperse a 50 μg batch of aggregated exosomes without further increasing clumping. To test this hypothesis, we varied exosome batch size while keeping the electric field constant at 0.75 kV/cm. As shown, exosome size is significantly higher if exosome batch size is decreased or increased relative to 50 μg while keeping the electric field constant at 0.75 kV/cm (R2 = 0.69, Fig. 1c). To summarize, electroporation of a 50 μg batch of melanoma exosomes in TPM at an electric field of 0.75 kV/cm will generate pores in exosomes as evidenced by RNA electroextraction while simultaneously maximizing exosome dispersal.
Assessment of PBS versus Trehalose PBS Pulse Field Media for Electroporation of Exosomes
Given the previous results, we next sought to determine whether electroporation mediated exosome dispersal required the presence of trehalose. Using electric fields of 0, 0.75 and 1.5 kV/cm, we compared exosome size and electroextraction of 50 μg batches of melanoma exosomes in the presence of PBS versus TPM. As shown, there is no difference in size between post exosome pellet isolation at 100,000 x g at an electric field strength of 0 kV/cm (Fig. 2a). However, significant differences exist between PBS and TPM using electric fields of 0.75 and 1.5 kV/cm. Moreover, there is a trend toward increasing size with increasing electric field for PBS. In contrast, exosome size at 0.75 kV/cm is significantly less than 0 or 1.5 kV/cm for TPM.
Figure 2.
Comparing the effects of PBS and trehalose PBS pulse media on exosome size and electroextraction. B16 melanoma exosomes were electroporated in the presence of PBS or trehalose PBS pulse media using electric field strengths of 0, 0.75 and 1.5 kV/cm. (a) Exosome size (n = 30 for 3 experiments and 10 measurements per experiment, error bars = s.e.m.) and (b) RNA electroextraction (n = 15 for 3 experiments and 10 measurements per experiment, error bars = s.d.) were determined by DLS and RNA absorbance post electroporation. For background RNA absorbance in the absence of electroporation, electroextraction in PBS at 0 kV/cm was normalized to 100%. Connecting bars denote statistically significant differences; p values < 0.05 were considered significant.
We next assessed differences in exosome electroextraction at 0, 0.75 and 1.5 kV/cm using PBS versus TPM. For purposes of comparison, exosome electroextraction was normalized to PBS background RNA at 0 kV/cm. As shown, electroextraction in PBS versus TPM was significantly greater at 0, 0.75 and 1.5 kV/cm. Using PBS, a linear trend for increasing RNA extraction with concomitant electric field strength was observed (Fig. 2b). Such a trend was more subtle for TPM. In summary, aggregation and electroextraction of exosomes in TPM is less than PBS at 0.75 and 1.5 kV/cm. Moreover, while PBS and TPM produced similar sizing by DLS post cold storage at 0 kV/cm, TPM increases exosome cold storage stability in that less background RNA is present after thawing from −80°C.
Assessment of Sucrose versus Trehalose PBS Pulse Field Media for Electroporation of Exosomes
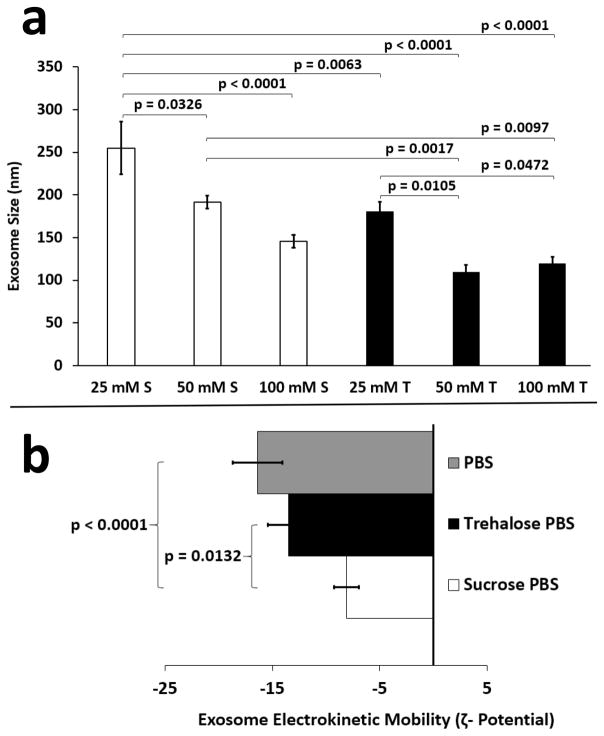
Given that TPM maximized exosome dispersal at 0.75 kV/cm, we investigated whether the same result could be achieved by substituting sucrose for trehalose. Similar to trehalose, sucrose is routinely used as a protein stabilizer [25] and cryoprotectant [17]. The ability of sucrose to maximize retention of solutes or drugs in ~ 100 nm vesicles during freezing is comparable to trehalose [17]. Thus, sucrose should exert similar stabilizing effects on exosomes. To maximally elucidate a subtle differential effect between sucrose and trehalose on exosome aggregation size, individual batches of exosomes were divided into different concentrations of TPM and sucrose pulse media (SPM) for isolation, storage and electroporation. This served to minimize the potential influence of batch to batch exosome size variation on the results. As shown, comparison of 25, 50 and 100 mM sucrose in PBS versus 25, 50, and 100 mM trehalose in PBS revealed that trehalose was superior overall to sucrose in reducing exosome aggregation post electroporation at 0.75 kV/cm (Fig. 3a).
Figure 3.
Comparing the effects of sucrose and trehalose pulse media on exosome size and zeta potential. B16 melanoma exosomes (50 μg) were electroporated in the presence of different concentrations of sucrose PBS (25, 50, 100 mM) or trehalose PBS (25, 50, 100 mM) pulse media using an electric field strength of 0.75 kV/cm. (a) Exosome size (n = 30 for 3 experiments and 10 measurements per experiment, error bars = s.e.m.) was determined by DLS. S = sucrose; T = trehalose (b) Exosome electrokinetic mobility was determined using a ZetaPlus Zeta Potential analyzer in the presence of PBS, 50 mM sucrose PBS, or 50 mM trehalose PBS pulse media (n = 50 for 4 experiments, error bars = s.e.m.). Connecting bars denote statistically significant differences; p values < 0.05 were considered significant.
Interestingly, there was no apparent statistical difference in post electroporation exosome size between 100 mM sucrose, 50 and 100 mM trehalose (Fig. 3a) suggesting that any one of these buffers might be useful for preventing exosome aggregation. However, the overall trend demonstrates that TPM increases exosome colloidal stability better than SPM. When formulating nanoparticles or nanovesicles, colloidal stability is determined by assessing particle electrokinetic mobility (zeta (ζ) potential). A zeta potential >/= + 30 or − 30 mV is considered a very stable colloidal suspension which resist particle aggregation or flocculation over time [26]. To further investigate exosome colloidal stability, we assessed exosome kinetic mobility in PBS, TPM and SPM. As shown, there is no difference in ζ-potential between PBS (−16 mV) and TPM (−14 mV) (Fig. 3b). However, use of either PBS or TPM resulted in a significantly more negative ζ-potential for exosomes than did SPM (−8 mV). In summary, these data demonstrate that exosomes have inherently low colloidal stability but TPM is preferable to SPM for maximizing exosome dispersal after electroporation at 0.75 kV/cm. Moreover, TPM is preferable to SPM for preserving exosome colloidal stability, which attenuates aggregation.
Use of TPM to Minimize Aggregation of Human Serum Exosomes Following Electroporation
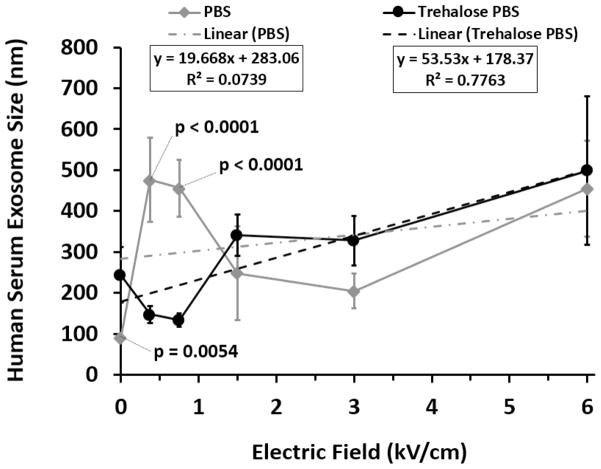
Having optimized the TPM electroporation process for a homogenous population of cultured melanoma exosomes, we next investigated whether this same approach would work for a heterogeneous population of normal human serum exosomes. Human serum exosomes were isolated, stored and electroporated in PBS or TPM. For purposes of comparison, serum exosomes isolated in PBS and not subjected to electroporation served as the baseline exosome reference size (PBS curve point 0, 0) (Fig. 4). As demonstrated, electroporation of human serum derived exosomes in TPM at an electric field strength of 0.75 kV/cm resulted in maximal re-dispersal of exosomes as evidenced by restoration of size near the 100 nm PBS reference point (Fig. 4). In contrast, use of PBS pulse media increased exosome clumping above the 100 nm reference baseline at all electric field strengths tested. Moreover, similar to melanoma exosomes (Fig. 1b), the relationship between electric field strength and human serum exosome size post electroporation in TPM was linear (R2 = 0.78, Fig. 4). However, pronounced nonlinearity was observed post electroporation in PBS (R2 = 0.07, Fig. 4). The coefficients of determination indicate excellent linear responsiveness of exosome size to electric field strength for trehalose data, but significant residual variation in the PBS data such that other additional factors must govern the size outcomes that are at this point unpredictable. The results confirm the suitability of TPM for use in electroporation of exosomes derived from human serum.
Figure 4.
Comparing the effects of PBS and trehalose pulse media on human serum exosome size. After storage at −80°C, exosomes isolated from pooled human serum (Innovative Research Inc.) were subjected to electroporation in the presence of PBS or trehalose PBS pulse media at 4°C at selected electric field strengths. Exosome size was measured by DLS (n = 10 measurements; error bars = s.d.) Best fit dotted lines denote linear regression analyses indicated by R2 values. Significant differences between PBS and trehalose pulse media at selected electric field strengths are denoted by p values; p values < 0.05 were considered significant.
Loading Exosomes with Exogenous Cargo using Trehalose Pulse Field Media
Having demonstrated the ability of TPM to maximize exosome dispersal and maintain colloidal stability post electroporation, our final experiment was to evaluate whether TPM would allow exosomes to be loaded with cargo. Arguably, increased density rather than size and content is the most distinguishing biophysical characteristic of exosomes versus other biological vesicles [1]. We reasoned that successful validation of exosome cargo loading could be achieved by loading exosomes with a test cargo that would increase exosome density.
Electroporation has been successfully used in previous studies for loading SPIONS into cells and tracking them in vivo [15; 16; 17]. Cells with diameters > 2000 nm are easily loaded with 100 nm SPIONs. A quick calculation reveals that the ratio of cell to SPION loading diameter is at least 20:1. Applying this ratio to the exosome size scale led us to hypothesize that we could load 100 nm exosomes with 5 nm SPIONs (SPION5).
Having demonstrated that an electric field strength of 0.75 kV/cm in TPM is optimal for simultaneously permeabilizing and dispersing 50 μg batches of exosomes, we proceeded to load melanoma exosomes with SPION5 (Fig. 5). As visualized by TEM, exosomes were no more electrofused or aggregated non-electroporation versus post electroporation loading with SPION5 (Fig. 5a). However, individual morphological changes throughout the electroporation exosome population could be observed. Approximately one third of post electroporation exosomes might best be described as electroporation transition intermediates (Fig. 5b). While most exosomes resumed their default spheroidal shape post electroporation (Fig. 5b1), a number of exosomes appeared to be caught in different stages of electroextraction whereby dark “electron dense” endogenous exosome contents were either being emptied (Fig. 5b2–3) or mostly replaced with exogenous granular SPION5 particles (Fig. 5b5–6). Some exosomes appeared both partially emptied of endogenous cargo and filled with exogenous SPION5s (Fig. 5b4). Size assessment by DLS further supported TEM results in that no difference in exosome size following passive incubation with SPION5 versus electroporation at 0.75 kV/cm in TPM was observed indicating no appreciable influence of SPION5 on post electroporation exosome aggregation in TPM (Fig. 5c).
Figure 5.
Loading exosomes with SPION5. B16 melanoma exosomes were loaded with SPION5 passively or by electroporation in the presence of trehalose pulse media at 0.75 kV/cm. (a) B16 melanoma exosomes visualized by TEM in the absence of electroporation or post electroporation with SPION5; the black lower left corner scale bar = 100 nm (b) Selected stages of SPION5 loading or unloading of exosome contents are observed by TEM after electroporation. (c) SPION5 loaded exosome size was assessed with DLS (n = 6; error bars = s.d.) (d) DiI labeled SPION5 loaded exosomes were isolated on a representative continuous sucrose gradient, fractions corresponding to peak exosome densities collected, densities determined and exosomes detected using Xenogen IVIS. (e) SPION5 loaded exosome iron content was assessed for exosome peak fractions using ICP-OES (n =3; error bars = s.d.). EP = electroporated; F = fraction
It follows that SPION5 loading of exosomes might be expected to increase exosome density. To assess this possibility, exosomes passively incubated with SPION5 and exosomes loaded with SPION5 using electroporation at 0.75 kV/cm were purified on sucrose density gradients. As expected, iron loading of exosomes by electroporation shifted peak melanoma exosome density from 1.14 g/ml as we previously reported [6] to 1.180 g/ml (Fig. 5d) which is near the upper end of published exosome densities ranging from 1.08 – 1.22 g/ml [19; 20; 23; 27; 28; 29]. Interestingly, passive incubation of SPION5 with exosome also increased peak melanoma exosome density to 1.177 g/ml. In general, a subtle shift toward increased exosome density was observed in peak fractions 3–5 for electroporation versus passive SPION5 loading.
Assessment of iron content in exosome peak fractions 3–5 by ICP-OES revealed a significant difference in exosome iron content between passive and electroporated exosomes (Fig. 5e). More iron was detected in EP loaded exosomes in the greater density fraction 3. In contrast, more iron was detected in passive loaded exosomes in the lesser density fraction 5. No difference was observed between passive and electroporation loading for fraction 4. In summary, the use of TPM during exosome electroporation enables heavy cargo loading of exosomes with SPION5 while maintaining original exosome size and minimizing exosome aggregation as evidenced by TEM and DLS.
DISCUSSION
These data demonstrate the first proof of concept that a simple biocompatible pulse media containing 50 mM trehalose in PBS can be used to maximize exosome dispersal following the electroporation process at 0.75 kV/cm for both homogeneous populations of melanoma exosomes and heterogeneous populations of human serum derived exosomes. Interestingly, throughout these studies we have observed batch to batch variation in B16 exosome size averages post electroporation at 0.75 kV/cm. This size variation observation likely reflects the knowledge that an individual source cell can produce a heterogeneous population of exosomes [30]. The implication is that large exosomes cannot become smaller post electroporation and vice versa as evidenced by the reported wide standard deviation in size despite high numbers of replicate measurements. For example, averaging all of the size data for 10 independent collections revealed a post dispersal size range of 89 +/− 30 nm (n = 97). However, this size range is consistent with our current TEM data (Fig. 5a) and previously reported findings for purified and maximally dispersed B16 exosomes [6] demonstrating that the TPM electroporation process achieves maximal exosome dispersal regardless of the innate pre-electroporation exosome aggregation state.
Use of TPM during exosome electroporation is dependent on both exosome concentration and electric field strength. For the purpose of our future translational experiments we optimized the procedure for processing 50 μg batches of exosomes given that we find this quantity to be efficiently isolated and tracked in vivo [7]. Processing of alternative batch sizes and exosome subtypes will undoubtedly require new optimization parameters. Nevertheless, the optimization approaches we used for melanoma and human serum exosomes are suitable for any such investigation.
Larger membrane structures such as cells are most effectively permeabilized using electric fields of approximately 1kV/cm whereas smaller vesicular structures may require higher field strengths [13]. However, higher field strengths potentiate events that can produce aggregation. Our data demonstrate that an electric field of 0.75 kV/cm provides the optimal balance for simultaneously loading and dispersing 50 μg batches of exosomes in TPM. Use of TPM at an electric field of 0.75 kV/cm generated pores in exosomes as evidenced by RNA electroextraction while simultaneously maximizing exosome dispersal. Moreover, while PBS and TPM produced similar sizing of exosomes as demonstrated by DLS post frozen storage at −80°C, TPM proved to be superior to PBS at increasing exosome cold storage stability as evidenced by decreased RNA background. Additionally, TPM is superior to SPM in maximizing exosome colloidal stability after storage at −80°C and post electroporation at 0.75 kV/cm.
Our data further demonstrate that the relationship between electric field force and melanoma exosome electroextraction or size was linear. However, while we obtained a coefficient of determination of R2 ~ 1.0 for electroextraction of exosomes in TPM (Fig. 1a), the coefficients for melanoma or human serum exosome size in TPM were R2 ~ 0.7 and 0.8 respectively (Fig.1c, 4). Combined, this suggest that only 20–30% of the variance in the data cannot be accounted for by electric field force alone and may be mediated in part by TPM effects.
The data further suggest the potential for higher concentrations of trehalose (100 mM) but not lower (25 mM) to be equally useful for decreasing post exosome electroporation aggregation. However, when evaluating higher concentrations, it is important to consider downstream exosome applications. Conceivably, if the concentration of trehalose chosen for exosome processing is too high, it could interfere with exosome isolation by equilibrating the density of isolation media to that of exosomes thus impeding exosome pelleting during ultracentrifugation. Moreover, exosome loading with cargo might be impeded at very high concentrations of trehalose given the increased viscosity of the buffer. Moreover, while trehalose is relatively inexpensive, it is nevertheless economically advantageous to use the lowest effective concentration of trehalose for isolating, storing and loading exosomes with cargo. Previous studies indicate that 50 mM trehalose is non-toxic to cells [18] and thus may be a safe concentration for in vivo applications.
Finally, the use of TPM during exosome electroporation enables heavy cargo loading of exosomes with 5 nm SPIONs while maintaining original exosome size and reducing exosome aggregation. Loading exosomes with SPION5 increases their density. This allows SPION5 modified exosomes to be potentially resolved from unmodified exosomes using sucrose density gradients. The interesting observation that SPION5 passively associates with exosomes in the absence of electroporation can likely be explained by one or two or a combination of mechanisms. One explanation is that exosomes contain some membrane holes which are an unfortunate side effect of ice crystals formed during frozen storage despite the cryoprotectant properties of TPM and this allows for uptake of some SPION5. In such a scenario, the addition of electroporation would increase the number or size of holes allowing for even more SPION5 uptake. Alternatively, what is more likely, given that exosome internalization of SPION5 in the absence of electroporation would require rather large > 5 nm holes is that the SPION5s are adsorbed to the exosome surface. The SPION5s express carboxylic acid functional groups. This maximizes their colloidal stability in PBS as evidenced by a very high zeta potential of approximately - 60 mV as reported by the manufacturer. However, previous studies demonstrate that iron oxide nanoparticles containing functional carboxylic acid groups can become coated with a corona of proteins when incubated with serum [31] which is known by exosome researchers to contain appreciable numbers of exosomes. It follows that the reverse could be true in that exosomes may adsorb carboxylic acid functionalized SPION5s on their surface. The effect might be further facilitated by the inherently low colloidal stability of exosomes in buffer (Fig. 3 ). The result would be to increase exosome density on a sucrose gradient, but not quite to the extent of exosomes loaded through electroporation with SPION5 as we observed (Fig. 5d) which should contain internal and external SPION5.
Future investigations will be required to determine the maximal extent to which exosomes can be loaded externally or internally with SPION5 while maintaining exosome dispersal and whether both forms of SPION5 loading might be useful depending on the application. For example, it might be advantageous to contain SPION5 in the exosomes with other compounds for the purposes of theranostic applications as described below. Alternatively, the ability to label the surface of exosomes with SPION5 without modifying endogenous exosome contents might provide for experimentation and tracking of native exosome functions in vivo using MRI analogous to our previous investigations where melanoma exosomes carried fluorescent cargo to sentinel lymph nodes [7]. Beyond SPION5 loading, additional studies will be necessary to determine whether our TPM based electroporation parameters are suitable in their current form or require additional tuning for loading other types of cargo including much smaller molecules such as RNA which has been successfully loaded into exosomes by other investigators using alternative electroporation methods [9; 10].
Our findings can best be interpreted in the context of biologic stabilization properties unique to trehalose. Trehalose is a unique disaccharide formed from a 1, 1 linkage of two D-glucose molecules. Trehalose naturally protects the cells of many organisms from dehydration or freezing [32]. For example, trehalose allows sawfly larvae to survive at −40°C and for the plant Selaginella lepidophylla to be completely dried out as if dead and yet return to normal activity upon rehydration. The sugar is naturally found in many food sources including honey, yeast, mushrooms and crustaceans. In consumer food products, trehalose is used as an additive for taste and preservation. It is a safe, non-toxic energy source for human consumption given the presence of the enzyme trehalase in the small intestine and renal proximal tubules [33].
Because of its high stability across many temperature and pH ranges, low viscosity and minimal interaction with proteinaceous molecules, trehalose is used in many medical and pharmaceutical applications [18]. Trehalose is used in the preservation of organs during transport for surgical procedures. It also protects the viability of mammalian cells and tissues during cryostorage and maintains the structural integrity of liposomes during freeze drying and rehydration.
In contrast to other sugars, trehalose can easily transition between dihydrate and anhydrate forms thus preventing ice nucleation during freezing and subsequent membrane damage by resulting ice crystals [32]. Moreover, the rotational flexibility of the glucose-glucose bond in trehalose allows it to conform and stabilize polar macromolecular head groups much more effectively than other sugars [32]. These unique structural properties allow trehalose to successfully control water stability and order around membranes. Trehalose is also essentially chemically inert with respect to its interactions with membrane proteins. Accordingly, the unique physical properties of trehalose make it an ideal exosome storage and stabilization agent during electroporation.
Development of exogenous exosome loading strategies that minimize exosome aggregation is essential for basic, translational and clinical exosome research. Development of TPM for exosome electroporation represents one such strategy. TPM facilitates both diagnostic and therapeutic exosome applications because it provides a means to minimize clumping of exosome subtypes while simultaneously minimizing exosome sample loss during purification. Because TPM is biocompatible, it can be used to isolate, store, load exosomes and transfuse animal models for in vivo experimentation without having to switch between multiple media resulting in additional exosome isolations, increased aggregation events and potential sample loss.
Our long term goal is to develop exosomal semi-synthetics for theranostic applications. We therefore chose superparamagnetic iron oxide nanoparticles (SPIONs) as a test cargo. In contrast to alternative MRI contrast agents, gadolinium etc., SPIONs are relatively non-toxic as well as being biocompatible and naturally eliminated by iron metabolism pathways in humans [13]. Iron oxide nanoparticles < 20 nm have single domain magnetic properties [34]. This distinction in magnetism separates < 20 nm “superparamagnetic” from > 20 nm “ferromagnetic” iron oxide nanoparticles. Effectively, superparamagnetic SPIONs are not magnetic in the absence of an applied magnetic field such as an MRI scanner. In contrast, multi-domain ferromagnetic particles retain magnetism in the absence of a magnetic field. Thus, 5 nm SPIONs will not magnetically aggregate with one another in vivo. Additionally, the surfaces of SPIONs can be easily conjugated with drugs or other ligands to target tumor or other pathogenic microenvironments for therapy. Combined, the ability of SPIONs to be tracked by MRI but not aggregate in the absence of a magnetic field facilitates SPION mediated drug delivery and imaging of target microenvironments as well as prevents uptake and removal of SPIONs by macrophages allowing for longer circulation times.
SPIONs have been used clinically to detect tumors and lymph node metastasis [13]. It is therefore conceivable that tumor exosome mediated detection and or delivery of therapeutics to tumor microenvironments could be made more effective and specific by carrying SPION cargo. SPIONs are capable of mediating magnetic hyperthermia. Magnetic hyperthermia is the process whereby repeated alterations in the strength and direction of a magnetic field will induce massive heat production by SPIONs. Because tissues are thermally labile, such increases in the temperature of a tumor microenvironment results in tumor cell death or increased susceptibility to subsequent radiation or chemotherapy. Iron oxide mediated magnetic hyperthermia has been used successfully to treat B16 melanomas in mice [35]. Given that the presented data demonstrate a means to load exosomes with SPION5 cargo, our future experimental aim will be to determine whether we can track SPION5 loaded exosomes in vitro and in vivo with MRI. If successful, we can proceed toward exosome mediated therapeutic applications.
In conclusion, use of TPM maximizes exosome colloidal stability while allowing for cargo loading with SPION5. The resultant exosomal density modification with SPION5 provides a simple, label free means to enrich exogenously modified exosomes and facilitates construction of exosomal semi-synthetics for diagnostic or therapeutic applications [1].
Acknowledgments
J.L.H. originated the experimental concepts, hypotheses, and methods, performed experiments, analyzed data, and wrote and edited the manuscript. M.J.S. executed experimental procedures and reviewed data. S.A.W. participated in the conceptual design of the experiments, reviewed results, and wrote and edited the manuscript. All authors read and approved the final version of the manuscript. We would also like to thank Marilyn A. Levy for providing superb technical assistance in obtaining high quality electron microscopy images of exosomes and Grace Hu for performing ICP-OES measurements. Finally, we would like to thank the Elsa U. Pardee Foundation, NIH grant HL073646 and the Saint Louis Institute of Nanomedicine’s Pilot Grant Program for their financial support and encouragement for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Joshua L. Hood, Email: jhood@dom.wustl.edu.
Michael J. Scott, Email: mike@dom.wustl.edu.
Samuel A. Wickline, Email: Wicklines@aol.com.
References
- 1.Hood JL, Wickline SA. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4:458–67. doi: 10.1002/wnan.1174. [DOI] [PubMed] [Google Scholar]
- 2.Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18:199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, Rivoltini L. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008;15:80–8. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 4.Taylor DD, Gercel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. British Journal of Cancer. 2005;92:305–311. doi: 10.1038/sj.bjc.6602316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oshima K, Aoki N, Kato T, Kitajima K, Matsuda T. Secretion of a peripheral membrane protein, MFG-E8, as a complex with membrane vesicles. Eur J Biochem. 2002;269:1209–18. doi: 10.1046/j.1432-1033.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- 6.Hood JL, Pan H, Lanza GM, Wickline SA. Paracrine induction of endothelium by tumor exosomes. Lab Invest. 2009;89:1317–28. doi: 10.1038/labinvest.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE, Zhang HG. Murine Mammary Carcinoma Exosomes Promote Tumor Growth by Suppression of NK cell Function. The Journal of Immunology. 2006;176:1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 10.Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weaver JC. Electroporation: a general phenomenon for manipulating cells and tissues. J Cell Biochem. 1993;51:426–35. doi: 10.1002/jcb.2400510407. [DOI] [PubMed] [Google Scholar]
- 14.Sugar IP, Forster W, Neumann E. Model of cell electrofusion. Membrane electroporation, pore coalescence and percolation. Biophys Chem. 1987;26:321–35. doi: 10.1016/0301-4622(87)80033-9. [DOI] [PubMed] [Google Scholar]
- 15.Stoicheva NG, Hui SW. Electrofusion of cell-size liposomes. Biochim Biophys Acta. 1994;1195:31–8. doi: 10.1016/0005-2736(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 16.Ramos C, Teissie J. Electrofusion: a biophysical modification of cell membrane and a mechanism in exocytosis. Biochimie. 2000;82:511–8. doi: 10.1016/s0300-9084(00)00200-5. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Han D, Cai C, Tang X. An overview of liposome lyophilization and its future potential. J Control Release. 2010;142:299–311. doi: 10.1016/j.jconrel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Mussauer H, Sukhorukov VL, Zimmermann U. Trehalose improves survival of electrotransfected mammalian cells. Cytometry. 2001;45:161–9. doi: 10.1002/1097-0320(20011101)45:3<161::aid-cyto1159>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Thery C, Clayton A, Amigorena S, Raposo G. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Current Protocols in Cell Biology. 2006:3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 20.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–9. [PubMed] [Google Scholar]
- 21.Weiner BB. Measuring the Size & Surface Charge of Exosomes, Microvesicles and Liposomes, Application Note, Brookhaven Instruments, a Nova Instruments company. 2013. [Google Scholar]
- 22.Porterfield JZ, Zlotnick A. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology. 2010;407:281–8. doi: 10.1016/j.virol.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser D, Zarka D, Hedman C, Kallas T. Plasmid and chromosomal DNA recovery by electroextraction of cyanobacteria. FEMS Microbiol Lett. 1995;128:307–13. doi: 10.1111/j.1574-6968.1995.tb07541.x. [DOI] [PubMed] [Google Scholar]
- 25.Simpson RJ. Stabilization of proteins for storage. Cold Spring Harb Protoc. 2010;2010:pdb top79. doi: 10.1101/pdb.top79. [DOI] [PubMed] [Google Scholar]
- 26.Zeta Potential An Introduction in 30 Minutes, Zetasizer Nano series technical note MRK654–01.
- 27.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–18. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 28.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 29.Riteau B, Faure F, Menier C, Viel S, Carosella ED, Amigorena S, Rouas-Freiss N. Exosomes bearing HLA-G are released by melanoma cells. Hum Immunol. 2003;64:1064–72. doi: 10.1016/j.humimm.2003.08.344. [DOI] [PubMed] [Google Scholar]
- 30.Fais S, Logozzi M, Lugini L, Federici C, Azzarito T, Zarovni N, Chiesi A. Exosomes: the ideal nanovectors for biodelivery. Biol Chem. 2013;394:1–15. doi: 10.1515/hsz-2012-0236. [DOI] [PubMed] [Google Scholar]
- 31.Safi M, Courtois J, Seigneuret M, Conjeaud H, Berret JF. The effects of aggregation and protein corona on the cellular internalization of iron oxide nanoparticles. Biomaterials. 2011;32:9353–63. doi: 10.1016/j.biomaterials.2011.08.048. [DOI] [PubMed] [Google Scholar]
- 32.Richards AB, Krakowka S, Dexter LB, Schmid H, Wolterbeek AP, Waalkens-Berendsen DH, Shigoyuki A, Kurimoto M. Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem Toxicol. 2002;40:871–98. doi: 10.1016/s0278-6915(02)00011-x. [DOI] [PubMed] [Google Scholar]
- 33.Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci. 2009;18:24–36. doi: 10.1002/pro.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santhosh PB, Ulrih NP. Multifunctional superparamagnetic iron oxide nanoparticles: promising tools in cancer theranostics. Cancer Lett. 2013;336:8–17. doi: 10.1016/j.canlet.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 35.Balivada S, Rachakatla RS, Wang H, Samarakoon TN, Dani RK, Pyle M, Kroh FO, Walker B, Leaym X, Koper OB, Tamura M, Chikan V, Bossmann SH, Troyer DL. A/C magnetic hyperthermia of melanoma mediated by iron(0)/iron oxide core/shell magnetic nanoparticles: a mouse study. BMC Cancer. 2010;10:119. doi: 10.1186/1471-2407-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]