Abstract
The 2013 outbreak of avian-origin H7N9 influenza in eastern China has raised concerns about its ability to transmit in the human population. The hemagglutinin glycoprotein of most human H7N9 viruses carries Leu226, a residue linked to adaptation of H2N2 and H3N2 pandemic viruses to human receptors. However, glycan array analysis of the H7 hemagglutinin reveals negligible binding to human-like α2-6-linked receptors and strong preference for a subset of avian-like α2-3-linked glycans recognized by all avian H7 viruses. Crystal structures of H7N9 hemagglutinin and six hemagglutinin-glycan complexes have elucidated the structural basis for preferential recognition of avian-like receptors. These findings suggest that the current human H7N9 viruses are poorly adapted for efficient human-to-human transmission.
In the spring of 2013, an outbreak of human infections caused by avian-origin H7N9 subtype influenza A virus occurred in the eastern provinces of China (1). By the end of May 2013, 132 cases of laboratory-confirmed H7N9 influenza were reported, resulting in 37 deaths (2). These patients generally presented influenza-like illnesses that frequently progressed to acute respiratory distress syndrome and severe pneumonia (3, 4). However, natural infection by H7N9 viruses in avian hosts are asymptomatic, which allows the virus to spread among birds and not be readily detected by surveillance (2).
The H7N9 outbreak has raised concerns about its potential for causing human pandemics or epidemics (5, 6). Compared to H5N1 viruses, H7N9 appears to transmit from birds to humans more readily, with reports of a relatively large number of recent human infections in a short period of time. Fortunately, however, avian influenza viruses such as H7N9 must overcome a species barrier that prevents transmission from human-to-human and thereby attain wide circulation in an antigenically naïve human population. Early reports have suggested that exposure to poultry was responsible for over 75% of the documented human cases of H7N9 influenza, but limited human-to-human transmission cannot be ruled out, especially in a few small clusters of human infections (4). Sequence analysis of the H7N9 viral proteins revealed that the virus has acquired several amino-acid changes associated with adaptation to human receptor binding and transmission in prior human pandemics (7-9). The PB2 protein of the H7N9 virus contains an E627K mutation that is important in other viruses for respiratory droplet transmission among humans (10). Furthermore, the HAs from most H7N9 human isolates have Leu at position 226 (H3 numbering) instead of Gln, which is conserved in avian H7 HAs (7, 8), as well as in other avian subtypes (11, 12). The Gln to Leu mutation is one of the hallmarks of the switch to human receptor binding specificity that occurred in the 1957 H2N2 and 1968 H3N2 human influenza virus pandemics, representing an adaptation believed to be required for efficient human-to-human transmission (11-13). Recent studies suggested that the H7N9 virus could efficiently transmit among experimental ferrets via direct contact (14-16), but respiratory droplet transmission, the mode of transmission relevant to human pandemics, is less efficient as demonstrated by results from five independent studies (9, 14-18). Thus, it is of major interest for public health to understand the extent to which the current H7N9 viruses have evolved to acquire capabilities for human-to-human transmission.
Most avian H7 viruses, including those associated with previous human outbreaks, contain highly conserved avian-specific residues, including Gln226, in their receptor-binding site that enable them to specifically recognize terminal sialic acids in an α2-3 linkage to galactose (19, 20). In contrast, while the first human H7N9 virus isolated contained Gln226 (A/Shanghai/1/2013, Sh1), most other human H7N9 viruses analyzed so far carry Leu226 (e.g., A/Shanghai/2/2013, Sh2), with a few isolates containing Ile226 (e.g., A/Hangzhou/1/2013, Hz1) (7). The Gln to Leu mutation is associated with improved affinity for human receptors where sialic acid is α2-6 linked to galactose (7, 8). In avian H2 and H3 HA, the Q226L mutation by itself greatly decreases HA affinity for α2-3-linked glycans while substantially improving binding to α2-6-linked glycans (21, 22). Recent studies have showed that H7N9 viruses with Leu226 can bind to receptors on the human tracheal epithelium (23) and are able to replicate in the upper respiratory tract of ferrets (14, 15). Unlike previous H7 viruses isolated in humans (H7N7), which exhibit contact transmission between ferrets but no respiratory droplet transmission (24), the human H7N9 viruses exhibit limited transmission by respiratory droplets (9, 14-16, 18), heightening concern that receptor binding changes might support more efficient transmission in humans (24).
Crystal structure of Sh2 H7 HA
We determined the crystal structure of Sh2 H7 HA at 2.7 Å resolution (Fig. 1A, table S1) and found that it is structurally similar to the HA from a highly pathogenic avian H7N7 virus that infected humans (A/Netherlands/219/2003, Neth219) [Protein Data Bank (PDB) entry 4DJ6] (20) (Cα root mean square deviation (RMSD) of 1.2 Å and only 0.4 Å for the receptor-binding domain). The main differences around the receptor-binding site arise from the absence of an N-linked glycosylation site at Asn133 in Sh2 due to a T135A substitution and Leu226 instead of Gln226 (Fig. 1B).
Fig. 1.
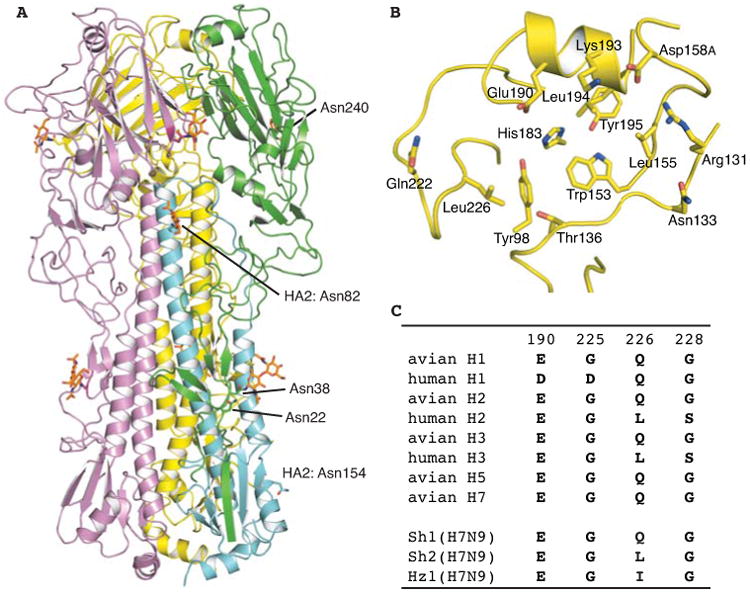
Crystal structure of the HA from a human H7N9 virus (A/Shanghai/2/2013, Sh2). (A) HA trimer is shown in cartoon representation. For one of the protomers, HA1 is colored in green and HA2 in cyan. The other two protomers (B and C) of the trimer are in yellow and magenta respectively. N-glycosylation sites and N-linked glycans are highlighted in sticks and numbered at the Asn attachment site. (B) Human H7 HA has Leu226 in its receptor-binding site. (C) Variation at the four HA positions that are known to mediate the switch in receptor binding specificity for human H1, H2 and H3 pandemic viruses and the corresponding sequences in H1, H2, H3 and H5 subtypes in avian viruses in comparison with human H7N9.
Glycan binding of H7N9 HA
Although studies evaluating receptor binding using whole viruses (14-16, 25, 26) have reported that the Leu226 mutation increases binding of H7N9 viruses to human receptors, binding to avian- and human-type receptors is influenced by the neuraminidase, which preferentially cleaves avian-type receptors (15, 25). To directly examine the intrinsic receptor specificity of the H7N9 HA, we analyzed HA binding to sialosides in plate-based assays and glycan arrays using soluble recombinant trimeric HAs expressed in mammalian cells (Fig. 2). In the plate assay, Sh2 HA strongly prefers α2-3-sialylated di-N-acetyllactosamine (SLNLN), although weak binding to α2-6 SLNLN at high protein concentrations is also observed (Fig. 2A) (27). Substitution of Leu226 to Gln, as in Sh1, or Ile, as in Hz1 strains, slightly increases avidity for α2-3 SLNLN, but completely abrogates binding to α2-6 SLNLN in this assay (Fig. 2A). The preferential binding of recombinant Sh2 HA to α2-3 sialosides in analogous plate assays or in biosensor-based assays has also been recently reported by other groups (28-30).
Fig. 2.
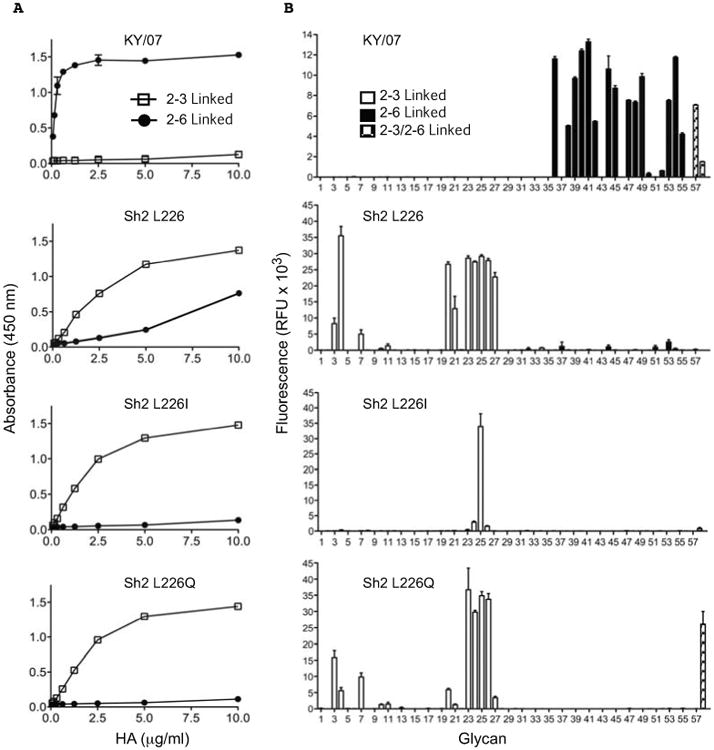
Receptor binding specificity of H7N9 HAs determined by plate-based glycan binding and glycan microarray analyses. (A) ELISA plates coated with either α2-3 or α2-6 linked SLNLN-PAA were probed with recombinant HAs produced in human kidney (HEK293S GnTI-) cells. The HAs were human H1N1 seasonal strain KY/07, or H7N9 Sh2 wild type containing Leu (L) at position 226 and mutants at this position harboring Ile (I) or Gln (Q). Shown are data representative of three independent experiments, each done in triplicate, with binding to α2-6 glycans in solid circles and α2-3 glycans in open squares. (B) The same recombinant HA proteins were used for assessment of receptor binding specificity on a glycan array. The mean signal and standard error were calculated from six independent replicates on the array. α2-3 sialosides are shown in white bars (glycans #3-35), α2-6 sialosides in black (glycans #36-56), and stripped bars denoted mixed bi-antennary glycans containing both α2-3 and α2-6 linked sialylated glycans (glycans #57, 58). Glycans imprinted on the array are listed in table S3.
The H7N9 HA receptor binding properties were further investigated using a custom influenza receptor glycan microarray comprised of diverse α2-3 sialosides (#3-35) and α2-6 sialosides (#36-56) that correspond to N- and O-linked glycans and linear fragments of glycans on mammalian glycoproteins and glycolipids (see list in table S3). Sh2 with Leu226 shows highly restricted binding to a small number of α2-3 sialylated glycans, and no detectable binding to α2-6 sialylated glycans, consistent with the more stringent requirements for binding in this assay (31). This subset of α2-3 sialosides includes sulfated linear glycans (#3, 4 and 7), linear and branched O-linked glycans (#20, 21, 23, 24) and biantennary N-linked glycans (#25-27). Notably, specificity for sulfated glycans #3 (NeuAcα2-3Galβ1-4[6S]GlcNAc) and #4 (NeuAcα2-3Galβ1-4(Fucα1-3) [6S]GlcNAc) are characteristic of H7 avian viruses, including those from poultry and aquatic birds (19, 24, 32). The L226I and L226Q substitutions change the distribution of glycan receptors that Sh2 HA binds, but specificity for α2-3-sialylated glycans is maintained (Fig. 2B) (33). Similar strict specificity for α2-3-sialylated glycans was observed for recombinant H7 HAs expressed in a baculovirus expression system (figs. S1 and S2) (34). In summary, Sh2 HA with Leu226 still maintains strong avian-type receptor specificity, with only very weak binding to human type α2-6 receptors.
H7 HA structures with LSTa and LSTc
Previous structural analyses of HA-receptor recognition and specificity have been limited to a small group of linear glycan receptor analogs including short synthetic fragments of glycans on glycoproteins and glycolipids (3′-SLN, NeuAcα2-3Galβ1-4GlcNAc and 6′-SLN, NeuAcα2-6Galβ1-4GlcNAc) and related oligosaccharides from human milk (LSTa, NeuAcα2-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc, LSTc, NeuAcα2-6Galβ1-4GlcNAcβ1-3Galβ1-4Glc and LSTb, Galβ1-4(NeuAcα2-6)GlcNAcβ1-3Galβ1-4Glc) (Fig. 3A) (30, 35). Notwithstanding, these ligands have provided valuable information on how avian and human HAs differentially recognize α2-3 and α2-6-linked sialosides.
Fig. 3.
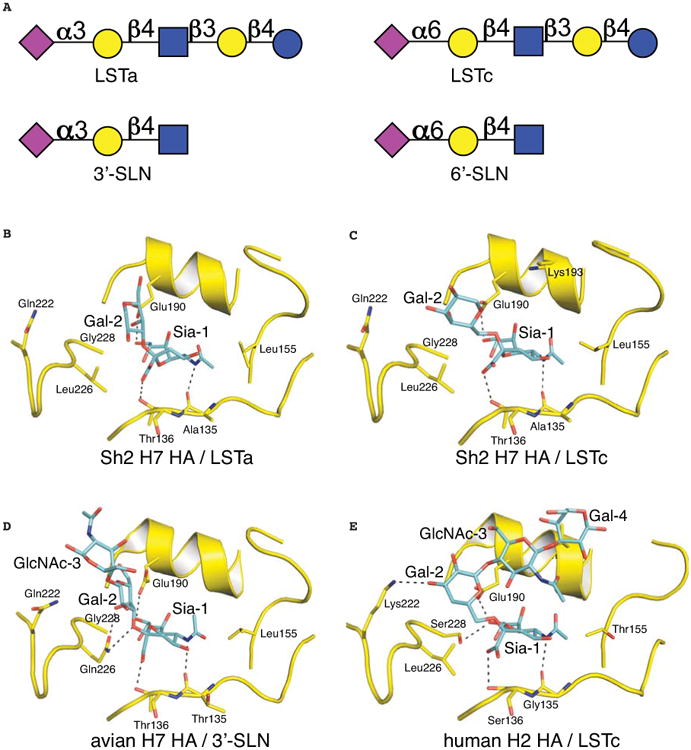
Crystal structures of human Sh2 H7N9 HA in complex with receptor analogs. (A) Glycan structures of four receptor analogs commonly used for structural study. The purple diamond represents sialic acid (Sia), yellow circle represents galactose (Gal), blue squares represent N-acetyl glucosamine (GlcNAc), and blue circle represents glucose (Glc). (B) Avian receptor analog LSTa bound to human H7 HA. (C) Human receptor analog LSTc bound to human H7 HA. (D) Avian receptor analog 3′-SLN bound to avian H7 HA (PDB entry 4DJ7). (E) Human receptor analog LSTc bound to human H2 HA (PDB entry 2WR7).
We determined crystal structures of Sh2 H7 HA with avian receptor analog LSTa at 2.6-2.7 Å resolution and found that only Sia-1 and Gal-2 of LSTa were ordered (Fig. 3B and fig. S3). Comparison of human and avian H7 HA structures reveals several differences in receptor recognition. In avian H7 from Neth219 virus (PDB entry 4DJ7) (20), preferential binding to avian receptors is mediated by Gln226 that hydrogen bonds with the Gal-2 O3 atom at the Sia-Gal linkage and the 4-hydroxyl group of Gal-2 (Fig. 3D). In addition, the Sia-1 carboxyl group is located within 3.5 Å of Gln226. This mode of recognition is highly conserved in other avian HA subtypes (35). In Sh2, the hydrophobic Leu226 does not support such a hydrogen-bonding network and Gal-2 moves away from Leu226, which results in LSTa taking a slightly different trajectory exiting the binding pocket. No significant interactions were visualized with the glycan ligand beyond Sia-1 suggesting a weak interaction that was not improved by increased soaking time and concentration of LSTa (fig. S3, table S1).
Similarly, in the crystal structure of Sh2 with human receptor analog LSTc at 2.9 Å resolution, only Sia -1 and Gal-2 could be modeled (Fig. 3C). Leu226 mediates hydrophobic interactions with the Gal-2 hydrophobic face through its C6 and C4 atoms. Although the specific interactions involving Gal-2 are similar in the human H2 HA complex with LSTc (PDB entry 2WR7) (Fig. 3E) (36), other structural features in H7N9 HA may prevent further stabilization of the glycan receptor analog beyond GlcNAc-3. Similar to avian H7 HA (20), the 150 loop of Sh2 H7 HA is closer to the receptor-binding site than it is in H2 HA (Figs. 3C, 3E).
These Sh2 HA structures with receptor analogs (LSTa and LSTc) then suggest that Leu226 modifies HA recognition of glycans with an α2-3 linkage, but does not adapt the HA receptor binding site for binding of α2-6-linked glycans (Fig. 3, B and C). These structural findings are in general consistent with other recent structural studies on human H7 (25, 29). The most notable difference is the cis configuration of α2-3 linkage in these structures, which may result from use of different receptor analogs (37).
Preferential recognition of avian-like receptors
To further explore the structural basis for retention of avian-type receptor specificity, we determined crystal structures of Sh2 H7 HA at 2.5-2.85 Å resolution with glycans #3, 21 and 23, which were identified as specific binders on the microarray (Fig. 4, table S2). In each case, additional HA-glycan contacts are created distal to the Sia-Gal linkage so that efficient binding to α2-3-linked glycans is achieved despite Leu226 (Fig. 5). Most relevant is glycan #3 (NeuAcα2-3Galβ1-4[6S]GlcNAc), which has been demonstrated to be a receptor analog recognized by all avian H7 viruses (19, 32). Previous efforts to model sulfated glycans in the avian H7 structure positioned the sulfo group in the vicinity of Lys193 for favorable charge interactions (19, 32) and was supported by the crystal structure of H5 HA with a sulfated sialoside (38). Perhaps surprisingly, we found that the sulfo group is located near the 220 loop between Leu226 and Val186 (Figs. 4D, 5C) and hydrogen bonds to the Ser227 main-chain amide and Glu190. The Sia-Gal linkage adopts a trans configuration, similar to linear avian receptor analogs in avian H7 and Sh2 H7 structures (Fig. 5A). The glycan exits the receptor-binding site above the 220 loop and follows a trajectory similar to avian receptors in H7 and other avian HAs. The sulfated GlcNAc-3, however, rotates its linkage with Gal-2 by about 180° to bring the sulfated 6-hydroxyl group over to the side of the 190 helix rather than proximal to the 130 loop. Electron density for the sulfo group is substantially better defined than GlcNAc-3, suggesting that glycan #3 binding to HA is mainly anchored by Sia-1 and the sulfo group, with flexibility in the rest of the receptor.
Fig. 4.
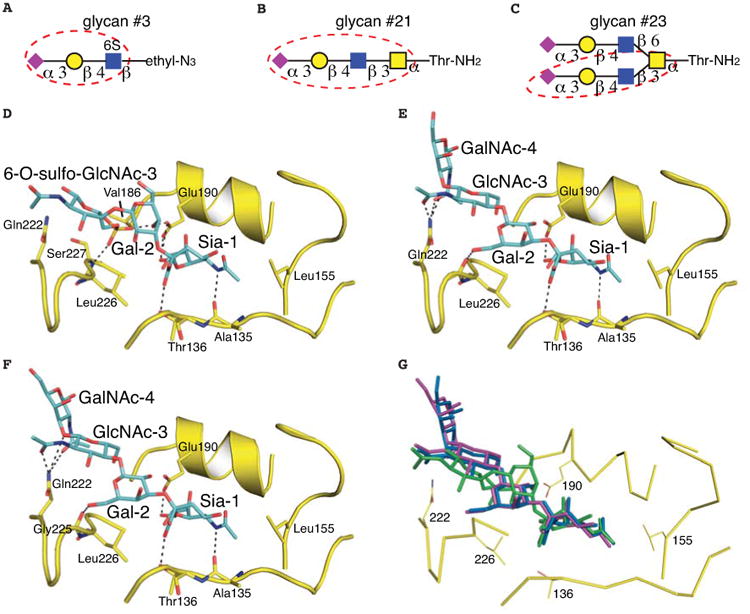
Crystal structures of human Sh2 H7N9 HA in complex with three avian-type receptor glycans recognized in the glycan array. (A-C) Glycan compounds used for structural determination. The glycan portion that could be built in the final model is highlighted by a red dashed circle. The purple diamond represents sialic acid (Sia), yellow circle represents galactose (Gal), and blue and yellow squares represent N-acetyl glucosamine (GlcNAc) and N-acetyl galactosamine (GalNAc), respectively. (D) Sulfated glycan #3 bound to human H7N9 HA. (E) Glycan #21 bound to human H7N9 HA. (F) Biantennary glycan #23 bound to H7N9 HA using one of its two arms. (G) Superposition of glycan #23 (colored in purple), glycan #21 (in blue) and glycan #3 (in green) in the HA receptor binding site. Glycans #21 and #23 share a similar mode of ligand recognition.
Fig. 5.
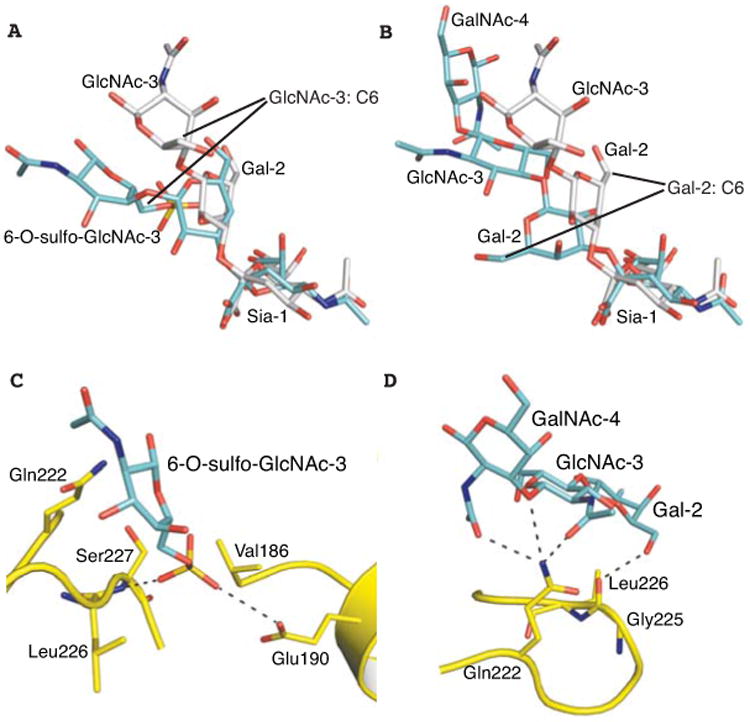
Preferential binding of human Sh2 H7N9 HA to avian-type α2-3-linked glycans is achieved by flexible twisting of the glycan rings that create additional HA-glycan contacts away from Leu226. (A) Superposition of glycan #3 (cyan) and 3′-SLN (grey, from PDB entry 4DJ7) in the HA receptor binding site. (B) Superposition of glycan #21 (cyan) and 3′-SLN (grey) in the HA receptor binding site. (C) The sulfo group of glycan #3 is located near the amide nitrogen of Ser227 and within hydrogen bonding distance of Glu190. (D) Preferential binding to glycan #21 is mediated by hydrophilic interactions between Gln222 and the glycosidic linkage between GlcNAc-3 and GalNAc-4.
The O-linked glycans (#21 and #23) share a common terminal sequence analogous to LSTa. For glycan #21 (NeuAcα2-3Galβ1-4GlcNAcβ1-3GalNAcα-Thr) in complex with Sh2 HA, the Siaα2-3Gal linkage adopts a cis configuration (Fig. 4E). Gal-2 rotates about 180° (Fig. 5B), bringing its 6-hydroxyl group into hydrogen bonding distance with the Gly225 carbonyl oxygen. The receptor analog exits over the 220 loop and turns away from the receptor-binding site between GlcNAc-3 and GalNAc-4, where Gln222 mediates hydrogen bonds with O3 in the linkage as well as the acetamido carbonyls from GlcNAc-3 and GalNAc-4 (Figs. 4E, 5D). Glycan #23 is a biantennary structure with one arm identical to glycan #21 (Fig. 4 B and C). Only this arm is visualized in the electron density map and binds HA similarly to glycan #21 (Fig. 4F-G). Sh2 HA also recognizes other α2-3-linked biantennary glycans (N-linked glycans #24-27) and their longer arms may allow the glycan to span HA trimers to improve avidity.
The HA complexes with these three avian-type receptor glycans reveal different binding modes that highlight the enormous diversity in HA-glycan recognition. Preferential binding for these glycans is mediated by HA-receptor interactions that were not predictable without these crystal structures. These interactions can compensate for unfavorable contacts between Leu226 and the α2-3-linkage, allowing human H7N9 HA to effectively maintain avian-type receptor specificity.
Implication for pandemic risk
The combination of glycan microarrays and x-ray structural determination has enabled us to demonstrate that the HA from current human H7N9 viruses retains specificity for sulfated α2-3-linked sialylated glycans that are recognized by avian H7 viruses (19, 32), and is not optimally evolved for recognition of human-like receptors. Because acquisition of human-type receptor specificity is considered a risk factor for human-to-human transmission, reports of binding to human type (α2-6) receptors (14-16, 23, 25, 26) have raised concern for the potential for H7N9 to become a pandemic virus. However, we and others have found that the human H7 HA is predominantly specific for avian-type (α2-3) receptors with only weak binding to human receptors (28-30). Thus, additional mutations will be needed for the HA to achieve specificity for human-type receptors as observed in human pandemic viruses. We suggest that the intrinsic weak avidity to human receptors in receptor assays is exaggerated in studies with whole virus by preferential cleavage of the avian-type receptors by the neuraminidase (15, 25) and the high valency of HA on the virus that can amplify binding to receptors of weaker affinity. This explanation is consistent with previous observations from analysis of a human H7N7 virus (A/NY/107/03), where the recombinant H7 HA exhibits strong preference for avian-type receptors (39), but whole virus showed significant binding to human-type receptors and manifests contact transmission, but not respiratory droplet transmission, in ferrets (24, 39). The situation for human H7N9 viruses is also quite analogous to H5N1 viruses, where other mutations, in addition to Gln226Leu, are required to achieve human-type receptor specificity that enables respiratory droplet transmission in ferrets (31). Thus, the weak avidity for human-type receptors may contribute to poor transmission by respiratory droplet in the ferret model (14-16, 18), and the lack of sustained transmission in humans (7, 8).
Supplementary Material
Materials and Methods
Figures S1-S5
Tables S1-S3
References (40-47)
Acknowledgments
This work was supported in part by NIH R56 AI099275 (I.A.W.) and the Skaggs Institute for Chemical Biology, the Scripps Microarray Core Facility, and a contract from the Centers for Disease Control (J.C.P.). R.P.d.V. is a recipient of a Rubicon grant from the Netherlands Organization for Scientific Research (NWO). Several glycans used for HA binding assays were provided by the Consortium for Functional Glycomics (http://www.functionalglycomics.org/) funded by NIGMS grant GM62116 (J.C.P.). X-ray diffraction data were collected at the Advanced Photon Source beamline 23ID-B (GM/CA CAT) and the Stanford Synchrotron Radiation Lightsource beamline 12-2. GM/CA CAT is funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. The Stanford Synchrotron Radiation Lightsource is a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH. This is publication 24079 from The Scripps Research Institute. Coordinates and structure factors are deposited in the PDB (4N5J, 4N5K, 4N60, 4N61, 4N62, 4N63, 4N64).
References and Notes
- 1.Emergence of avian influenza A(H7N9) virus causing severe human illness -China, February-April 2013. MMWR Morb Mortal Wkly Rep. 2013;62:366. [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Overview of the emergence and characteristics of the avian influenza A (H7N9) virus, 2013. http://www.who.int/influenza/human_animal_interface/influenza_h7n9/WHO_H7N9_review_31May13.pdf.
- 3.Gao R, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, et al. Preliminary report: epidemiology of the avian influenza A (H7N9) outbreak in China. N Engl J Med. 2013 Epub ahead of print. [Google Scholar]
- 5.Morens DM, Taubenberger JK, Fauci AS. Pandemic influenza viruses--hoping for the road not taken. N Engl J Med. 2013;368:2345. doi: 10.1056/NEJMp1307009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahn RE, Richt JA. The novel H7N9 influenza A virus: its present impact and indeterminate future. Vector Borne Zoonotic Dis. 2013;13:347. doi: 10.1089/vbz.2013.999.ceezad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 8.Kageyama T, et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 9.Steinhauer DA. Influenza: Pathways to human adaptation. Nature. 2013;499:412. doi: 10.1038/nature12455. [DOI] [PubMed] [Google Scholar]
- 10.Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2:160. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matrosovich M, Stech J, Klenk HD. Influenza receptors, polymerase and host range. Rev Sci Tech. 2009;28:203. doi: 10.20506/rst.28.1.1870. [DOI] [PubMed] [Google Scholar]
- 13.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, et al. Infectivity, transmission, and pathology of human H7N9 influenza in ferrets and pigs. Science. 2013;341:183. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe T, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belser JA, et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 2013;501:556. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 18.Richard M, et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 2013;501:560. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gambaryan AS, et al. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012;86:4370. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Carney PJ, Donis RO, Stevens J. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol. 2012;86:8645. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, McBride R, Paulson JC, Basler CF, Wilson IA. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J Virol. 2010;84:1715. doi: 10.1128/JVI.02162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers GN, et al. Host-mediated selection of influenza virus receptor variants. Sialic acid-α2,6Gal-specific clones of A/duck/Ukraine/1/63 revert to sialic acid-α2,3Gal-specific wild type in ovo. J Biol Chem. 1985;260:7362. [PubMed] [Google Scholar]
- 23.Tharakaraman K, et al. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belser JA, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci USA. 2008;105:7558. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong X, et al. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499:496. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J, et al. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499:500. doi: 10.1038/nature12379. [DOI] [PubMed] [Google Scholar]
- 27.As a control, the HA from a representative human seasonal H1N1 strain (A/Kentucky/UR06-0258/2007, KY/07) displays characteristic strong binding to α2-6 SLNLN and negligible binding to α2-3 SLNLN (Fig. 2A).
- 28.Ramos I, et al. H7N9 influenza viruses interact preferentially with α2,3-linked sialic acids and bind weakly to α2,6-linked sialic acids. J Gen Virol. 2013;94:2417. doi: 10.1099/vir.0.056184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Y, et al. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science. 2013;342:243. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 30.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol. 2013 doi: 10.1128/JVI.01854-13. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulson JC, de Vries RP. H5N1 receptor specificity as a factor in pandemic risk. Virus Res. 2013 doi: 10.1016/j.virusres.2013.02.015. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gambaryan AS, et al. 6-sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J. 2008;5:85. doi: 10.1186/1743-422X-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.As a control, human HA KY/07 binds broadly to α2-6-linked glycans on the array (Fig. 2B) and avian H7 HA recognizes a large number and variety of α2-3-sialylated glycans (fig. S2).
- 34.The L226Q mutant produced in insect cells showed robust binding to the array, revealing stronger avidity to most α2-3 glycans, but still no binding to α2-6 sialosides.
- 35.Xu R, Wilson IA. In: Structural Glycomics. Yuriev E, Ramsland PA, editors. CRC Press; 2012. pp. 235–57. [Google Scholar]
- 36.Liu J, et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci USA. 2009;106:17175. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong et al used 3′-SLN as an avian-like receptor analog. Shi et al used α2-3 SLNLN. In our study, we used LSTa.
- 38.Xiong X, et al. Recognition of sulphated and fucosylated receptor sialosides by A/Vietnam/1194/2004 (H5N1) influenza virus. Virus Res. 2013 doi: 10.1016/j.virusres.2013.08.007. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 39.Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. Quantitative description of glycan-receptor binding of influenza a virus H7 hemagglutinin. PLoS one. 2013;8:e49597. doi: 10.1371/journal.pone.0049597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figures S1-S5
Tables S1-S3
References (40-47)


