Abstract
Hepatitis C virus (HCV), a Hepacivirus, is a major cause of viral hepatitis, liver cirrhosis and hepatocellular carcinoma. HCV envelope glycoproteins E1 and E2 mediate fusion and entry into host cells and are the primary targets of the humoral immune response. The crystal structure of the E2 core bound to broadly neutralizing antibody AR3C at 2.65 Å reveals a compact architecture composed of a central Ig-fold β-sandwich flanked by two additional protein layers. The CD81 receptor-binding site was identified by EM and by site-directed mutagenesis and overlaps with the AR3C epitope. The x-ray and EM E2 structures differ markedly from predictions of an extended, three-domain, class II fusion protein fold and therefore provide invaluable information for HCV drug and vaccine design.
Hepatitis C virus (HCV) was discovered in 1989 as the causative agent of non-A, non-B hepatitis (1). It is estimated that 2–3% of the world population is infected with HCV (2) and, in the United States, it has overtaken human immunodeficiency virus 1 (HIV-1) as a cause of death (3). A prophylactic vaccine could help control the HCV pandemic but its development has been technically challenging due to high sequence variability of the viral genome (4) as well as limitations in animal models (5, 6). The envelope glycoprotein E2 is the main target for neutralizing antibody (NAb) responses, but is also the most variable antigen in HCV (7). E2 forms a heterodimer with the other HCV envelope glycoprotein, E1, to mediate cell entry and fusion. Several broadly neutralizing antibodies (bNAbs) have been isolated against E2, raising hopes for the rational design of a broadly effective vaccine (8–10).
Structural characterization of HCV envelope glycoproteins would inform vaccine and drug design but has been challenging because of the difficulty in obtaining homogenous protein preparations. In HCV envelope glycoproteins, N-linked glycans constitute nearly 50% of the E1E2 protein ectodomain molecular weight (11 and 4 N-linked glycosylation sites in E2 and E1, respectively), and a high proportion of the expressed glycoproteins form aberrantly disulfide cross-linked aggregates (11–13). Based on computational models E2 was predicted to be a class II fusion protein characterized by a highly extended conformation (~110–130 Å) of three predominantly β-sheet domains (12, 14).
To gain insight into the HCV E2 structure, which corresponds to residues 384–746 of the viral polyprotein (n.b. E1 is residues 192–383), we designed and expressed 41 different soluble E2 constructs and screened 7 of the well-behaved constructs (15) with various E2-specific Fabs in crystallization trials. Crystals diffracting to 2.65 Å (table S1, fig. S2) were grown from an E2 core [(E2c, spanning residues 412–645, see supplementary online material (15)] in mammalian cells (HEK 293F) with the HCV prototypic strain H77 sequence in complex with bNAb AR3C Fab (4, 8). The engineered E2c had truncations at the N- and C- termini, substitution of the potentially flexible variable region 2 (residues 460–485) with a Gly-Ser-Ser-Gly linker, and removal of N448 and N576 glycosylation sites of E2 (fig. S3) (15). E2c maintains a native E2 fold as verified by binding to a panel of monoclonal antibodies (mAbs) specific to conformational epitopes in E2 and to the CD81 receptor, and by functional inhibition of HCV infection presumably through receptor competition (fig. S1). Two equivalent E2 complexes (A and B) compose the crystal asymmetric unit, making extensive crystal contacts with each other between two sides of E2 and the heavy-chain constant domain of AR3C (fig. S2) (16).
Overall, the E2c structure is globular but contains many regions with no regular secondary structure despite the presence of 8 disulfide bonds. In fact, nearly 62% of all E2c residues are either in loops or disordered (Fig. 1 and fig. S3). The disordered regions are at the E2c N-terminus (residues 412–420), a short segment (spanning positions 454–491) surrounding the severely truncated E2 VR2, and a loop (586–596). Flexibility is also observed for 6 N-linked glycans in E2c: N417 and N423 are completely disordered, N532, N540 and N623 have only one ordered GlcNAc, and N556 has two ordered GlcNAcs. Glycan N430 in the crystal interface can be modeled as Man6GlcNAc2. Despite flexibility in parts of the structure, E2c has an overall well-defined architecture consisting of a central β-sandwich flanked by front and back layers consisting of loops, short helices and β-sheets.
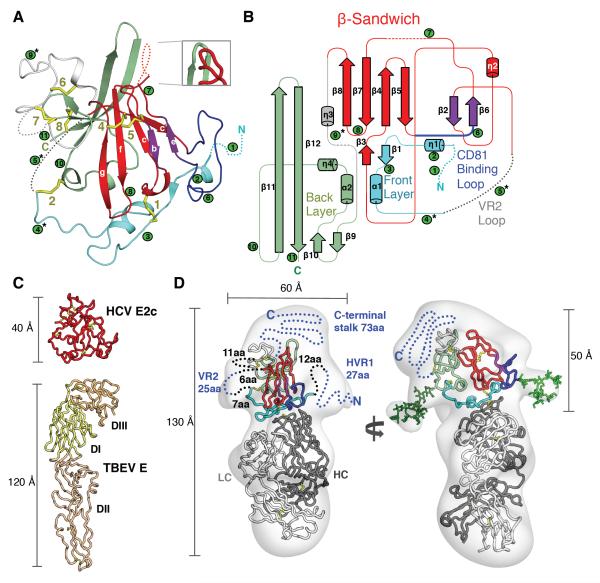
Fig. 1. Structures of HCV E2 and comparison with the class II fusion fold.
(A) The crystal structure of HCV E2c is displayed as a cartoon representation and colored by structural components: the front layer is formed by the N-terminal region residues 421–453 (cyan); the outer (purple) and inner (red) sheet form the Ig β-sandwich; the CD81 receptor binding loop is a bilobed structure (blue); a flexible region (white) encompasses variable region 2 (VR2); and the back layer (597–645) is formed by two short helices, loops and a 4-stranded β-sheet (light green). Labeling of β-sandwich strands follows Ig-fold conventions. Disulfide bonds are shown as yellow sticks and numbered from the N-terminus. N-linked glycans are indicated by green circles and are also numbered from the N-terminus. Asterisks indicate N-linked glycans deleted in the construct. Disordered regions in the structure are shown by dotted lines. An inset shows the structure of a loop in the E2c structure from complex B that is disordered in complex A. (B) Topology diagram of E2c following the same coloring scheme as in the cartoon representation. (C) Scaled comparison of HCV E2c (top) and TBEV E protein, the canonical class II fusion protein (PDBID: 1SVB). DI–III indicates domains I–III. (D) The 16 Å EM density map of HCV E2ΔTM bound to Fab AR3C (transparent gray surface) is shown from two perspectives with the crystal structure of HCV E2c bound to Fab AR3C fitted into the EM density. The crystal structure is displayed as a ribbon, with the E2c colored as in Figure 1. Dotted lines indicate the main portions of E2ΔTM that are absent in the E2c crystal structure. The numbers of amino acids (aa) in the missing regions are shown. Blue dotted lines regions in E2ΔTM are not in the E2c construct and black dotted lines indicate regions that are in the E2c construct but are disordered in the crystal structure. Two protrusions in the EM density are in the vicinity of N-linked glycan sites and high mannose glycans (green ball-and-sticks) are modeled at those positions. Measurements of dimensions are rounded to the nearest ten.
The E2 β-sandwich (residues 492–566) contains four strands designated as the inner sheet (g, f, c, c') and two strands exposed to solvent designated as the outer sheet (e, b) (Fig. 1 and fig. S4). The overall strand connectivity places it within the C2-set of Immunoglobulin (Ig)-folds, which are characterized by the presence of the c' strand in the inner sheet, as in CD4 domain 2, instead of the d strand in the outer sheet as, for example, in the C1-set (17, 18) (fig. S4). The loop connecting strands c' to e contains 17 amino acids and exhibits a bilobed structure that extends from a narrow stalk formed by the N- and C- termini. The C-terminal lobe contains receptor-binding residues (Tyr527, Trp529, Gly530 and Asp535) and is adjacent to the front layer that also contains key receptor-binding residues (Fig. 1) (19) (see supplementary text (15) for detailed structural description of the front and back layers).
The Ig-fold β-sandwich is the only structural element in E2c that is shared with presumed structural homologues that include class II viral fusion proteins, which have a three-domain architecture (domains I–III) (figs. S4). Ig-folds are found in domain III of fusion envelope proteins from flaviviruses (e.g. Tick-Borne Encephalitis Virus (TBEV) and West Nile Virus) (20, 21), domain III of E1 fusion proteins and all domains of E2 proteins from rubivirus (Rubella) (22) and alphaviruses (e.g. Chikungunya and Sindbis viruses) (23, 24), and E2 domain B from pestivirus (bovine viral diarrhea virus) (25, 26). However, the compact HCV E2c protein, which maximally spans ~50 Å, does not adopt the extended three-domain class II fusion protein fold (Fig. 1C), which measures 100–120 Å (27).
To confirm that full-length E2 is also compact, and to visualize regions that are absent in the E2c crystal structure, the complete E2 ectodomain (E2ΔTM, residues 384–717) bound to Fab AR3C was analyzed by negative-stain EM. Asymmetry of the complex due to the protruding Fab allowed for unambiguous fitting of the E2c-Fab AR3C crystal structure into the 16 Å resolution EM 3D reconstruction, leaving ~30% of the EM volume unassigned (Fig. 1D), which is consistent with the expected 21% difference in mass between fully glycosylated E2c-Fab AR3C and E2ΔTM-Fab AR3C (28). The EM reconstruction accounts for most of the E2ΔTM protein and likely some of its N-linked glycans (Fig. 1D). Overall, the EM reconstruction shows that the E2ΔTM construct also displays a compact globular shape, confirming that the complete E2 ectodomain also does not adopt the highly extended, three-domain class II fusion fold (Fig. 1C).
The EM reconstruction enables us to approximately define regions of E2 that are absent in the E2c structure (Fig. 1D). The disordered and truncated N-terminal region (residues 384–421), which includes hypervariable region 1 (HVR1), likely fits into a bulb of density next to the β-sandwich, consistent with epitope mapping of Fab AR1A (8), which identified Thr416, as well as Val538 and Asn540 on the top sheet of the β-sandwich, as key interacting residues. The region (residues 454–491), which is largely truncated in the E2c construct, contains VR2 (460–485) and the N476 glycan and is readily accommodated in the EM density on the opposite face of the β-sandwich. The largest portion of EM density not accounted for by the crystal structure is situated behind the back layer and VR3, where extensive crystal contacts are made between two E2 subunits in the asymmetric unit of the crystal structure. The 73-residue C-terminal stalk region that is also absent in the E2c construct would fit at this location, forming a final layer to the overall protein architecture.
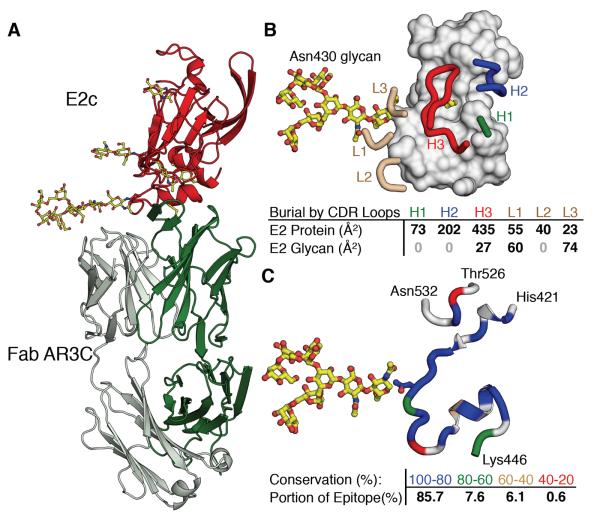
In the crystal structure, E2c is bound to antibody AR3C, which belongs to a group of bNAbs that recognize antigenic region 3 (AR3) of E2 and cross-neutralizes HCV genotypes by blocking CD81 receptor binding (8). The crystal structure defines the common surface that these bNAbs recognize, revealing a prime target for vaccine design. Within the binding interface, Fab AR3C buries 828 Å2 of E2 protein surface and 161 Å2 of E2 glycans (Fig. 2). Overall, residues that are 80–100% conserved across HCV genotypes make up 86% of the buried surface area in the AR3C epitope (Fig. 2C), including critical residues previously identified by alanine scanning mutagenesis (8). The epitope is relatively flat, encompassing most of the front layer, a serpentine stretch of residues 421–446, and a portion of the CD81 receptor-binding loop. The protein-protein interface is composed mainly of the heavy-chain variable domain (86%), which corroborates data indicating AR3C binding activity is not compromised when its light chain is swapped (fig. S1 and Fig. 2). The bNAb interaction is dominated by the CDR H3 loop, which buries a strand of highly conserved residues near the E2 N-terminus in the front layer and Trp529 in the CD81 receptor binding loop, together accounting for 44% of the total buried surface (Fig. 2). Antibody binding to the N-terminal strand is mediated by main-chain interactions (fig. S5), thus tolerating sequence variation in E2. The CDR H3 loop adopts a β-hairpin fold and is stabilized by a disulfide (29), which is encoded in 17% of human IGHD2 germline alleles (30) (fig. S5), suggesting similar antibodies could be raised by vaccination. The CDR H1 and H2 loops of AR3C are encoded by the germline IGHV1-69 gene, which has been utilized in bNAbs against several viruses including influenza (31) and HIV-1 (32). CDR H2 loops encoded by this VH gene have a hydrophobic tip, which tends to interact with hydrophobic clusters on the antigen and has been proposed as a primordial pattern recognition receptor (33). CDR H2 also interacts with hydrophobic residues on the N-terminal side of the front layer and, together with the CDR H1 loop, contacts hydrophobic residues on the α1 C-terminus (see (15) for discussion of α1 recognition and antibody AR3C germline genes).
Fig. 2. HCV E2 interaction with Fab AR3C.
(A) Overall structure of E2c (red) bound to Fab AR3C is displayed as a cartoon representation with the heavy and light chains of Fab colored dark and light green, respectively. N-linked glycans are shown in a ball-and-stick representation with carbon, oxygen and nitrogen atoms colored yellow, red and blue. (B) Interactions between Fab AR3C and E2c. The CDR loops of AR3C are displayed as thick tubes over the gray molecular surface of E2. A relatively unusual disulfide bond in CDR H3 for known human antibody structures is shown in yellow. The N430 glycan that interacts mainly with the light chain of AR3C is shown in yellow and red. Below the diagram is a table listing the surface areas buried on E2 by the different CDR loops. (C) The AR3C epitope on E2 is shown as a cartoon representation from the same perspective as in (B) and colored according to sequence conservation. Residues that are not buried by AR3C are colored gray. Below is a table listing the fractions of the surface on the E2 core protein with respect to sequence conservation (binned and color coded) that are buried by AR3C.
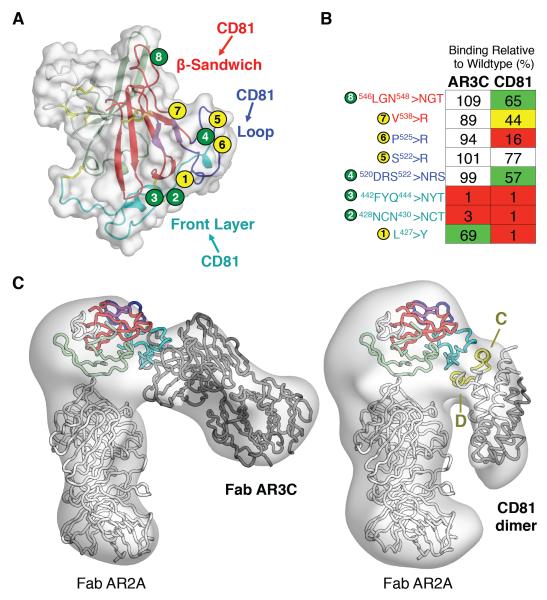
CD81 receptor-binding residues identified in prior mutagenesis studies (fig. S3) (19, 34) mainly mapped to the AR3C epitope; however some were also found in the β-sandwich. To determine the location of the binding site, we performed site-directed mutagenesis and negative-stain EM. First, mutagenesis-driven modeling based only on the published putative receptor interacting residues indicated three possible binding sites: one side of the β-sandwich, an isolated portion of the CD81 receptor-binding loop, and the front layer (Fig. 3A and table S2). Thus, mutations were introduced to the full-length E1E2 heterodimer (Fig. 3B). Mutations that totally abrogated CD81 binding were addition of glycosylation sites at position 442 or 428 or an L427Y mutation in the front layer. A P525R, but not P525A, mutation in the N-terminal lobe of the CD81 receptor-binding loop greatly reduced binding, suggesting it may either bind CD81 or be involved in the correct folding of the C-terminal lobe for binding CD81 via Tyr527, Trp529, Gly530 and/or Asp535 (19). Together with previous mutagenesis data (19), these results suggest that CD81 interacts with the front layer and the CD81 receptor-binding loop (Fig. 3, A and B, and fig. S6).
Fig. 3. CD81 receptor-binding site.
(A) Three potential CD81 binding sites on E2 were indicated from previously published alanine-scanning results (19): a surface on the β-sandwich, the top of the CD81 binding loop, and the front layer. E2 mutations used to evaluate the possible binding sites are shown as circles numbered from the N-terminus. A yellow circle indicates substitution by a bulky amino acid and a green circle indicates the introduction of an N-linked glycosylation site. E2 is depicted as a cartoon within its molecular surface and colored as in Fig. 1A. (B) The substitutions shown in (A) are described and their effects on CD81 and Fab AR3C binding are tabulated. Numbers indicate % binding of CD81 or Fab AR3C to variants relative to wild type E1E2 in an ELISA. Binding is color coded: 0–25% = red, 26–50% = yellow, 51–75% = green, >75% = white. (C) Negative-stain EM reconstructions of deglycosylated E2ΔTM bound to Fabs AR2A and AR3C (left) or bound to Fab AR2A and CD81 dimer (right). The crystal structure of E2c bound to Fab AR3C and a model of Fab AR2A displayed as ribbons are fitted within the electron density. E2c is colored as in Fig. 1. Helices C and D of CD81 are highlighted in yellow because they contain residues important for binding to E2 (34). In the CD81 complex, the density suggests that a CD81 dimer is present as in CD81 crystal structures (35, 36).
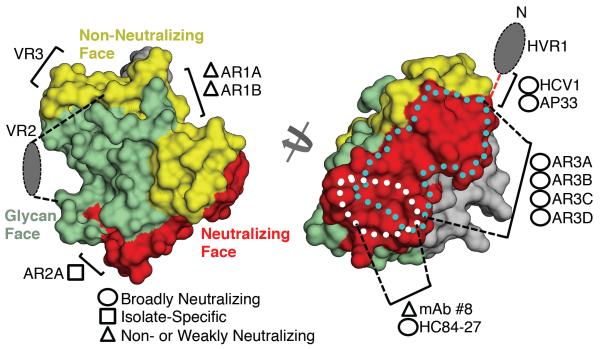
To further delineate the receptor binding site, we performed negative-stain EM on a ternary complex of deglycosylated E2ΔTM, the large external loop (LEL) of CD81, and Fab AR2A (Fig. 3C and fig. S7). Docking of the E2c crystal structure into the 19 Å resolution EM reconstruction was aided by determination of the Fab AR2A interaction in the 20 Å EM reconstruction of E2ΔTM bound to Fabs AR2A and AR3C (Fig. 3C, left). The EM density for CD81 was too extensive for a monomer, but consistent with a CD81 dimer, similar to that in CD81 crystal structures (35, 36). One of the CD81 monomers interacts with the E2c front layer, consistent with the mutagenesis data (Fig. 3C and fig. S8). Only one orientation of the CD81 dimer has a reasonable fit to the EM density (37), and results in helices C and D being adjacent to E2 α1 and CD81 receptor-binding loop. These helices contain critical E2 interacting residues (34). Therefore, CD81 binds to the same surface as AR3C and some other bNAbs (8), suggesting it is a site of vulnerability that could be exploited in immunogen design. This exposed surface is relatively hydrophobic, has relatively low sequence variability, and is free of N-linked glycans (Fig. 4 and fig. S9). This site may also include residues 412–420, which are disordered in the crystal structure, but are bound by bNAbs HCV1 and AP33 (38, 39). However, similar to HIV-1 gp120 and influenza HA, HCV E2 contains multiple highly variable regions (HVR1, VR2, VR3) (40) and N-linked glycans to escape from immune recognition (Fig. 4 and fig. S9). For example, an extensive glycan shield masks an exposed face on the E2 surface from NAbs via 7 of the 11 N-linked glycans on E2 (Fig. 4 and fig. S9. Also, on the opposite side of the β-sandwich, a relatively hydrophobic surface is likely occluded by the N-terminal HVR1 (fig. S9); deletion of HVR1 is not lethal to the virus and greatly increases the sensitivity of the virus to antibody neutralization (41). Finally, between the glycosylated and HVR1-occluded faces of E2, a separate oblong surface, formed by the outer sheet of the β-sandwich and portions of VR3 (Fig. 4 and fig. S9) with relatively high sequence variability, is recognized by several weakly or non-NAbs (8). These non-NAbs can bind soluble recombinant E1E2 at high affinity (8), suggesting their epitopes must be occluded on the virus.
Fig. 4. HCV E2 antigenic surface.
The E2c structure is displayed as a molecular surface colored according to different antigenic properties described in the text. Missing HVR1 and VR2 regions are represented as colored ovals and labeled. The mAb binding sites mapped from alanine scanning studies or from crystal structures bound to peptides, as described in the main text, are indicated. On the neutralizing face (right), three antigenic regions are shown: residues 412–423 (red dashed line connecting HVR1 and the rest of E2), the AR3C epitope in the front layer that overlaps with the CD81 binding site (cyan dotted line), and α1 in the front layer (white dotted line). The gray surface is relatively hydrophobic and conserve and may be an occluded zone covered by HVR1 in the full-length E2 protein, as suggested by the EM reconstruction of E2ΔTM (Fig. 1C).
Until recently, HCV was the sole identified member of the Hepacivirus genus in the Flaviviridae family. Non-primate hepaciviruses have now been identified in dogs, horses and rodents, providing insights into the evolution of this viral genus (42). Despite sharing a common Ig-fold, the compact globular HCV E2 structure clearly distinguishes HCV from the related virus genera Pestivirus and Flavivirus that have extended, multi-domain envelope protein structures, undermining the proposals of E2 as a class II fusion protein (12, 14). To determine whether the Hepacivirus envelope protein uses a novel fusion mechanism will require structural characterization of E1 and the E1E2 complex in the context of the virus. However, the E2 structure and the characterization of broadly neutralizing epitopes and the CD81 binding site now provide new opportunities for HCV vaccine and drug design.
Supplementary Material
Acknowledgements
We thank H. Tien, T. Clayton and M.C. Deller for help in setting up crystallization screens using the CrystalMation and Douglas robots; J. Robbins for help with protein purification; N. Laursen for useful discussions on refinement; L. Jaroszewski for help in analyzing the HCV E2c protein fold; P. Verdino for discussion of Ig-folds; and J.P. Verenini for help in manuscript formatting. This work is supported by NIH grants AI079031 and AI080916 (to M.L.), AI071084 (to D.R.B.), AI084817 and U54 GM094586 (to I.A.W.), and the Skaggs Institute (I.A.W.). L.K. is grateful to the American Foundation for AIDS Research for a Mathilde Krim Fellowship in Basic Biomedical Research and R.U.K. to the Swiss National Science Foundation for a post-doctoral fellowship. The EM data were collected at the US National Resource for Automated Molecular Microscopy (NRAMM) at The Scripps Research Institute, which is supported by the US National Institutes of Health (NIH) through the National Center for Research Resources' P41 program (RR017573) at the National Center for Research Resources. X-ray data sets were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12–2, a Directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research; NIH's National Center for Research Resources, Biomedical Technology Program (P41RR001209); and the National Institute of General Medical Sciences (NIGMS). Coordinates and structure factors for the E2c complex with Fab AR3C have been deposited with the Protein Data Bank under accession code 4MWF. The EM reconstruction densities for the E2ΔTM-Fab AR3C, E2ΔTM-Fab AR3C-Fab AR2A, and E2ΔTM-FabAR2A-CD81 LEL complexes have been deposited with the Electron Microscopy Data Bank under accession codes EMD-5759, EMD-5760 and EMD-5761 respectively. The content is the responsibility of the authors and does not necessarily reflect the official views of the NIGMS, NCI or NIH. This is manuscript #24038 from The Scripps Research Institute.
Footnotes
The authors declare no competing financial interests.
References and Notes
- 1.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D. The global burden of hepatitis C. Liver Internat. 2009;29:74. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 3.Ly KN, et al. The increasing burden of mortality from viral hepatitis in the United States between 1999 and 2007. Ann. Intern. Med. 2012;156:271. doi: 10.7326/0003-4819-156-4-201202210-00004. [DOI] [PubMed] [Google Scholar]
- 4.Kuiken C, Hraber P, Thurmond J, Yusim K. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 2008;36:D512. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton M, Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature. 2005;436:961. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 6.Billerbeck E, de Jong Y, Dorner M, de la Fuente C, Ploss A. Animal models for hepatitis C. Curr. Top. Microbiol. Immunol. 2013;369:49. doi: 10.1007/978-3-642-27340-7_3. [DOI] [PubMed] [Google Scholar]
- 7.Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Methods Mol. Biol. 2009;510:33. doi: 10.1007/978-1-59745-394-3_4. [DOI] [PubMed] [Google Scholar]
- 8.Law M, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 2008;14:25. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 9.Broering TJ, et al. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J. Virol. 2009;83:12473. doi: 10.1128/JVI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keck ZY, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalak JP, et al. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 1997;78:2299. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 12.Krey T, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whidby J, et al. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J. Virol. 2009;83:11078. doi: 10.1128/JVI.00800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagnik AT, et al. A model for the hepatitis C virus envelope glycoprotein E2. Proteins. 2000;40:355. doi: 10.1002/1097-0134(20000815)40:3<355::aid-prot20>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 15.Materials and methods, and supplementary text are available as supplementary material on Science Online.
- 16.Complex A has better defined electron density and lower B-values, likely from additional crystal contacts between the tips of glycan N430 and a neighboring symmetry mate (fig. S2); the same glycan in complex B is mostly disordered. The N430 interaction may explain why enzymatic deglycosylation, which typically aids crystallization, does not produce crystals.
- 17.Halaby DM, Poupon A, Mornon J. The immunoglobulin fold family: sequence analysis and 3D structure comparisons. Protein Eng. 1999;12:563. doi: 10.1093/protein/12.7.563. [DOI] [PubMed] [Google Scholar]
- 18.One difference from the standard C2-set connectivity is the absence of strand a from the outer sheet of the E2 β-sandwich, which could be due to disorder or to truncation of E2 VR2 in the construct. A more important difference is the absence of a canonical disulfide connecting outer and inner sheets at strands b and f that is present in most Ig-folds, although observed together with C2-set strand connectivity in some bacterial, actinoxanthine-like and fibronectin III-like domains comprising the C3, C4 and FN3 – sets. However, the E2 β-sandwich has disulfides linking the loop prior to strand b with strand g (residues 494–564), and strand c with strand f (508–552), which are typically observed in the C2-set but not for the C3, C4 or Fn3 sets. This unique disulfide system found in the E2 β-sandwich may have constrained the rotational angle between inner and outer sheet strands to be nearly parallel, rather than the −30° offset commonly observed in Ig-folds. Together, these features suggest the E2 β-sandwich is C2-set-like but with differences such as lack of strand a, lack of disulfide connection between strands b and f, and lack of −30° offset between front and back sheets that prevent finding significant matches with equivalent strand connectivity to other structures in the PDB using structural homology programs DALI and FATCAT (15).
- 19.Owsianka AM, et al. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 2006;80:8695. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 21.Nybakken GE, et al. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DuBois RM, et al. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature. 2013;493:552. doi: 10.1038/nature11741. [DOI] [PubMed] [Google Scholar]
- 23.Voss JE, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3:30. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Wang J, Kanai R, Modis Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl. Acad. Sci. USA. 2013;110:6805. doi: 10.1073/pnas.1300524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaney MC, Rey FA. Class II enveloped viruses. Cell. Microbiol. 2011;13:1451. doi: 10.1111/j.1462-5822.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- 28.This calculation assumes all N-linked glycans are Man9GlcNAc2 due to the kifunensine treatment during protein production.
- 29.Almagro JC, et al. Characterization of a high-affinity human antibody with a disulfide bridge in the third complementarity-determining region of the heavy chain. J. Mol. Recognit. 2012;25:125. doi: 10.1002/jmr.1168. [DOI] [PubMed] [Google Scholar]
- 30.This calculation was made from searching the IMGT database on March 27, 2013. There were n=8832 IGHD germline genes in the database, of which 1514 contained the disulfide motif.
- 31.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummer HE, Wilson KA, Poumbourios P. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 2002;76:11143. doi: 10.1128/JVI.76.21.11143-11147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitadokoro K, et al. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitadokoro K, et al. Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop. Biol. Chem. 2002;383:1447. doi: 10.1515/BC.2002.164. [DOI] [PubMed] [Google Scholar]
- 37.At this resolution, we have approximated the known CD81 dimer structure into the EM reconstruction but some differences are apparent that will need higher resolution structure information to discern subtle differences in free and bound conformations of CD81.
- 38.Kong L, et al. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J. Virol. 2012;86:13085. doi: 10.1128/JVI.01939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong L, et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc. Natl. Acad. Sci. USA. 2012;109:9499. doi: 10.1073/pnas.1202924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.HVR1 has been defined as the most variable region of the HCV genome. However, relative to the entire genome, VR2 and VR3 are not considered hypervariable. Here, we define these regions as variable with respect to E1 and E2 proteins only, and not with respect to the rest of the HCV genome.
- 41.Bankwitz D, et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 2010;84:5751. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapoor A, et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio. 2013;4:e00216–13. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.