Introduction
Nervous system function is dependent on the intricate network of connections formed between neurons. Axons often adopt a branched morphology in their target area with axonal arbors decorated by synapses. Based on electron microscopy observations that synapses are often present at branch points, Vaughn et al., hypothesized that synapse formation might promote the elaboration of axonal and dendritic branches (Cline and Haas, 2008; Vaughn, 1989). Consistent with this idea, in vivo imaging of developing retinal ganglion cell (RGC) showed that synapse formation and axonal arbor formation occur simultaneously during development. New axonal branches initiate from synapses and branches with synapses are more stable than synapse-free branches (Meyer and Smith, 2006). These observations point to molecular mechanisms that link synapse formation and branching.
Although little is known about such links, the mechanisms of presynapse formation and axonal branch formation have been studied extensively. De novo branches that form from the main axon shaft are termed collateral axonal branches. Formation of collateral branches requires cytoskeletal organization at branch sites. Often, actin assembly initiates filopodia or lamellipodia formation followed by microtubule invasion which marks the maturation of the collateral branch (Gallo, 2011).
The importance of F-actin during synapse formation has been shown by studies where depolymerizing F-actin during a critical developmental time window causes synapse loss (Zhang and Benson, 2001). As actin is ubiquitous, it is not surprising that F-actin plays many roles during synaptogenesis. F-actin can interact with presynaptic active zone proteins and affect the recruitment of active zone components to synapses (Chia et al., 2012; Zhang and Benson, 2001). Conversely, active zone proteins may regulate F-actin organization at synapses. For example, the vertebrate active zone protein Piccolo can bind actin regulator profilin (Waites et al., 2011). Similarly in Drosophila, a perisynaptic SH3 adaptor protein, Nervous Wreck, controls synapse morphology by binding to Wasp, a key regulator of F-actin dynamics (Coyle et al., 2004). Presynaptic F-actin may also be required for clustering synaptic vesicles around the active zone (Doussau and Augustine, 2000; Murthy and De Camilli, 2003). Recent studies have linked various transmembrane cell adhesion molecules that facilitate recognition and interaction of pre- and postsynaptic membranes to intracellular F-actin rearrangements at synapses. These include the fly immunoglobulin superfamily protein Basigin (Besse et al., 2007), UNC-40/DCC (Stavoe et al., 2012), SYG-1/NEPH1 (Chia et al., 2012), Tenuerins (Mosca et al., 2012), and cadherins (Sun and Bamji, 2011).
Like synapse assembly, the formation of collateral axonal branches rely heavily on F-actin dynamics (Gallo, 2011). Treating cortical neurons in vitro with Latrunculin, a drug that inhibits F-actin dynamics, resulted in a loss of axon branching but did not affect the elongation of the core axon shaft (Dent and Kalil, 2001). The actin nucleation factor, Arp2/3 complex, has also been shown to be required for branch formation in embryonic chicken dorsal root ganglia neurons (Spillane et al., 2011). Knocking down Ena/VASP, another F-actin nucleation factor, drastically affected branching of RGC axons in Xenopus (Dwivedy et al., 2007).
Although the phenomenon of synapse-directed arborization has been observed, few studies have explored pathways that mechanistically link axon arbor growth and synaptogenesis. Here we demonstrate that the transmembrane cell adhesion molecule SYG-1/NEPH1 can recruit the WASP-family verprolin-homologous protein (WVE-1/WAVE) regulatory complex (WRC), a well-known activator of the Arp2/3 complex, to nascent synapses. This interaction is mediated by a conserved amino acid sequence, the WRC interacting receptor sequence (WIRS), in the cytoplasmic tail of SYG-1. This SYG-1/WRC interaction controls the assembly of an Arp2/3 mediated F-actin patch that localizes to developing synapses and is required for both downstream axonal arborization and synapse assembly. Hence, our data supports the synaptotropic model by identifying a common downstream modulator shared by both processes and is recruited to nascent synapses by synaptic cell adhesion receptors.
Results
Local assembly of F-actin by SYG-1/SYG-2 interaction is required for presynaptic assembly and branch formation
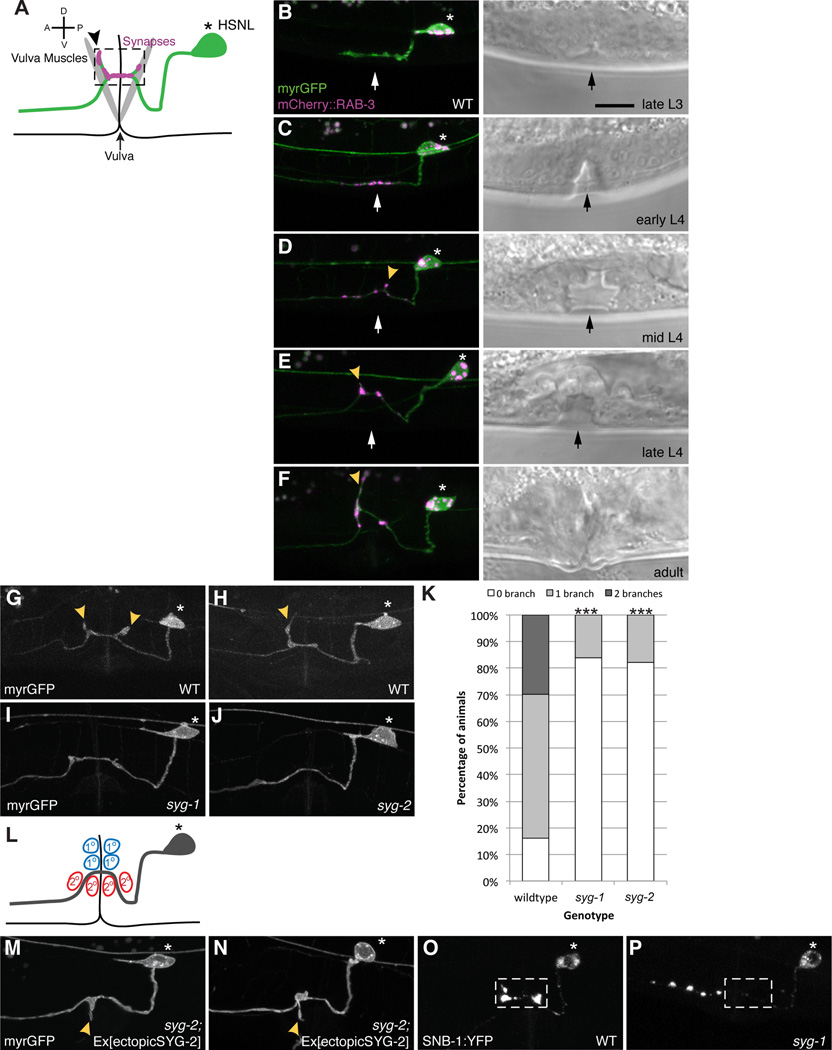
To investigate the processes that coordinate synapse formation and collateral axon branch formation in vivo, we studied the C. elegans egg-laying motorneurons HSN. The cell bodies of HSN are located posterior to the vulva and each extends an axon anteriorly into the nerve ring. As the axon extends past the vulva, HSN forms clusters of en passant synapses onto the vulva muscles (Figure 1A). At the synaptic region, HSN also elaborates one or two stereotyped axonal branches dorsally. To understand the temporal relationship between synaptogenesis and branch formation during development, we expressed both a synaptic vesicle marker, mCherry::RAB-3, and a plasma membrane marker, myristolated GFP, in HSN using cell-specific promoters to track the development of the HSN neuron (Figures 1B–1F). In the late L3 stage, the HSN axon grows across the developing vulval from posterior to anterior, with no detectable RAB-3 clusters and no axonal branches (Figure 1B). In early L4 animals, the axon growth cone continues to extend anteriorly towards the nerve ring, RAB-3 clusters begin to accumulate at the vulva region (Figure 1C). Other synaptic markers such as SNB-1/synaptobrevin (Shen and Bargmann, 2003) (Figure 1O) and active zone markers including SYD-2/liprin-α (data not shown) also accumulate, suggesting that bona fide presynaptic terminals form at this stage. Interestingly, no axonal branches are visible at this stage. During the mid L4 to adult stage, the intensity of the RAB-3 clusters increases. In the mean time, branches form along the synaptic region, which increase in length into the adult stage (Figures 1D–1F). These observations suggest that the onset of synaptogenesis, signified by the clustering of synaptic vesicles and active zones proteins in the synaptic region, precedes axonal collateral branch formation.
Figure 1. Interaction between SYG-1/SYG-2 is required for presynaptic assembly and branch formation.
(A) Schematic of HSN. * Denotes the cell body and synapses (pink) form in the synaptic region (dashed box) onto the vulva muscles. Black arrowhead points to axonal collateral branch. (B–F) Representative images depicting the development of HSN neuron. Myristolated GFP highlights the morphology of HSN. Yellow arrowheads point to axonal branches with synapses labeled by mCherry::RAB-3 (pink). Black and white arrows denote the vulva. During the late L3 stage, the main axon is growing across the developing vulva with no visible accumulation of synaptic material (pink). At the early L4 stage, synaptic vesicles begin accumulating at the synaptic region around the vulva. In the mid L4 stage, in some animals, one or two collateral axonal branches extend and become quite pronounced by the late L4 stage. These branches continue to lengthen into the adult stage and accumulate synaptic material, (G, H) Myristolated GFP labels the morphology of HSN. HSN elaborates one or two axonal branches (yellow arrows) that always develop from the synaptic region. (I, J) Branches fail to form in syg-1 or syg-2 mutants. (K) Graph quantifies the percentage of animals in each genotype that elaborates zero, one or two branches. Statistics for each mutant was from comparison with the wildtype values (***p<0.001 with n>100, Fisher’s exact test). (L) Schematic showing the location of primary (1°, red) and secondary (2°, blue) vulva epithelial cells. In wildtype, the 1° vulva epithelial cells express SYG-2. (M, N) Ectopic expression of SYG-2 in 2° vulva cells in syg-2 mutants causes ectopic branches that elaborate ventrally. (O) A wildtype HSN neuron with synapses labeled by synaptobrevin::YFP. (P) syg-1 mutants show ectopic accumulations of synaptobrevin::YFP along the axon anterior to the normal synaptic region around the vulva. Scale bars represent 10 µm.
Our previous work showed a pair of immunoglobulin synaptic adhesion molecules, SYG-1 and SYG-2, specify the location of HSN synapses. SYG-2 is expressed in the primary epithelial cells located immediately dorsal to the HSN axon which expresses SYG-1. SYG-2 binds and localizes SYG-1 to specify the HSN synaptic region (Shen and Bargmann, 2003; Shen et al., 2004). To understand the molecular mechanisms underlying branch formation, we examined syg-1 and syg-2 loss-of-function mutants and found that about 80% of the mutants have no branch whilst only 16% of wildtype animals lack branches (Figures 1G–1K). This result suggests that SYG-1 and SYG-2 are not only required for assembling synapses, but also critical for branch formation. To further address if the SYG-1/SYG-2 interaction is sufficient to trigger branch formation, we ectopically expressed SYG-2 in the secondary vulva epithelial cells, which localizes to the ventral side of HSN axon (Figure 1L), in a syg-2 mutant. This has previously been shown to induce ectopic synapse formation due to specific recruitment of SYG-1 to ectopic SYG-2 expressing sites (Shen et al., 2004). We found that this manipulation is sufficient to induce ventrally directed branches that are not found in wildtype animals (Figures 1M and 1N). Taken together, these data argue that the SYG-1/SYG-2 interaction instructs both synapse formation and axon branch formation.
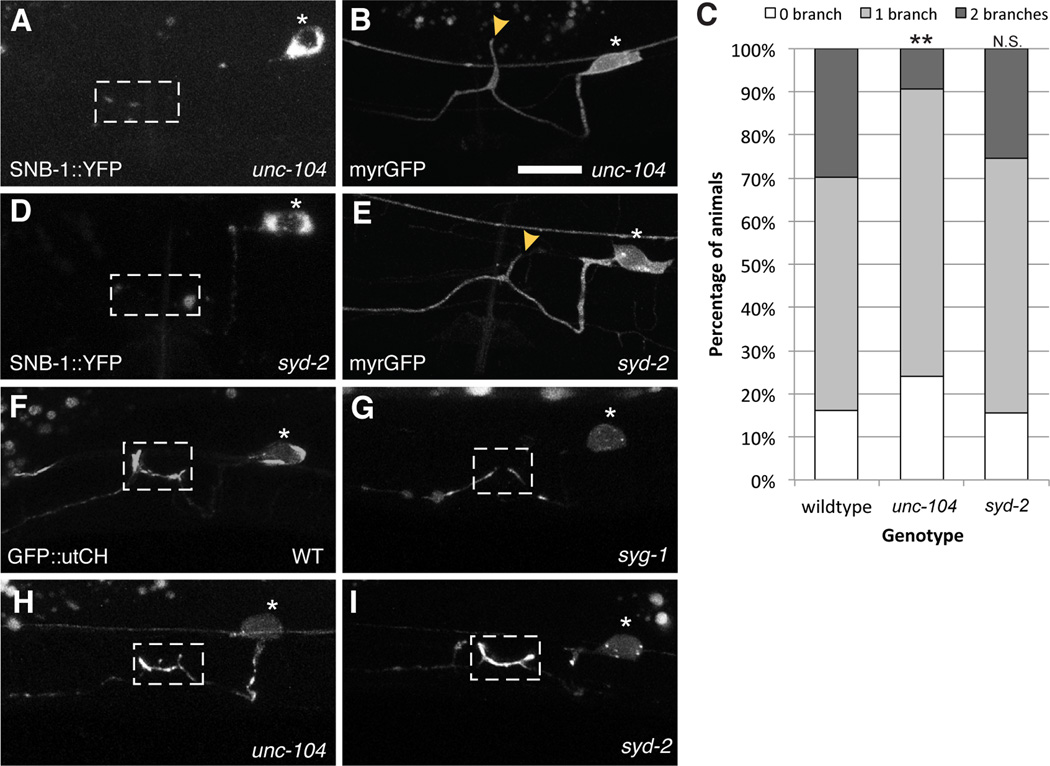
There are two possible mechanisms that can produce tight spatial and temporal correlation between synaptogenesis and branch formation. Firstly, local accumulation of presynaptic material by the SYG-1/SYG-2 interaction might directly induce branch formation. This hypothesis is also supported by the loss of synapses from the HSN synaptic region in syg-1 mutants (Figure 1O and 1P). Alternatively, synaptogenesis and branch formation might be parallel events that are initiated by the SYG-1/SYG-2 interaction. To distinguish between these possibilities, we examined branch formation in unc-104 and syd-2 mutants to understand if loss of synaptic material might affect branching. The motor protein UNC-104/kinesin-3 is required to transport synaptic vesicles but not active zone proteins to presynaptic terminals in HSN (Patel et al., 2006). Loss of UNC-104 results in synaptic vesicles becoming completely trapped in the HSN cell body (Figure 2A). However, unc-104 mutants displayed only a subtle branching phenotype (Figure 2B and 2C). Similarly, loss of syd-2 prevents recruitment of synaptic vesicles and most active zone proteins to synapses (Figure 2D) (Patel et al., 2006). Despite these defects, syd-2 mutants still show normal branching (Figure 2E). These data suggest that neither synaptic vesicles nor active zone proteins are essential for branch formation. Therefore, the data suggests that synapse formation and axonal branching may be initiated by the SYG-1/SYG-2 interaction in parallel.
Figure 2. Synaptic vesicles and active zone proteins are not required for collateral branch formation.
(A) In kinesin motor unc-104 mutants, synaptic vesicles labeled by synaptobrevin::YFP fail to get transported to the synaptic region. (B) Loss of unc-104 results in a partial reduction in branch formation. (C) Graph quantifies the percentage of animals in each genotype that elaborate zero, one or two branches. Statistics for each mutant was from comparison with the wildtype values (**p<0.01 with n>100, Fisher’s exact test). (D) syd-2 mutants fail to accumulate synaptic vesicles and active zone molecules (E) but branches are unaffected in syd-2 mutants. (F) GFP::utrophinCH labels synaptic F-actin that is enriched at presynaptic specializations in the L4 stage. (G) This F-actin localization is loss in syg-1 mutants (H, I) but is unaffected in unc-104 and syd-2 mutants. Yellow arrowheads point to collateral branches. Scale bars represent 10 µm. See also Figure S1.
We next asked how the SYG-1/SYG-2 interaction might initiate the two processes. We previously showed that SYG-1 is not only required to initiate synapse assembly, but it is also required to pattern an F-actin network at the HSN synaptic region during development (Chia et al., 2012). This local F-actin is crucial for presynaptic assembly. In syg-1 mutants, F-actin, which is labeled by GFP fused to the calponin homology domain of F-actin binding protein utrophin (GFP::utCH), is no longer enriched at the synaptic region (Figure 2F and 2G). On the contrary, the F-actin patch is unperturbed in either unc-104 or syd-2 mutants (Figures 2H and I) (Chia et al., 2012), suggesting that F-actin assembly is an upstream event of the local accumulation of synaptic vesicles and active zone proteins. Thus, the F-actin network might be important for initiating both synapse assembly and axon branching.
To test if the F-actin network is required for HSN branching, we injected Latrunculin A, a drug that disrupts F-actin organization, locally into the vulva region. We had previously shown that this treatment disrupts F-actin at synapses in HSN (Chia et al., 2012). We observed a slight but significant decrease in branching in animals injected with Latrunculin A as compared to untreated animals and animals injected with DMSO as a control (Supplementary figure 1).
Taken together, loss of the local F-actin but not synaptic material affects axonal branching, Furthermore, these results hint that synapse formation and axon branching are parallel events but are spatially linked by the F-actin structure downstream of SYG-1. In the vertebrate system, F-actin has been shown to be important for both synapse assembly as well as axon arborization (Gallo, 2011).
WRC is required for assembling an Arp2/3 mediated actin network at synapses
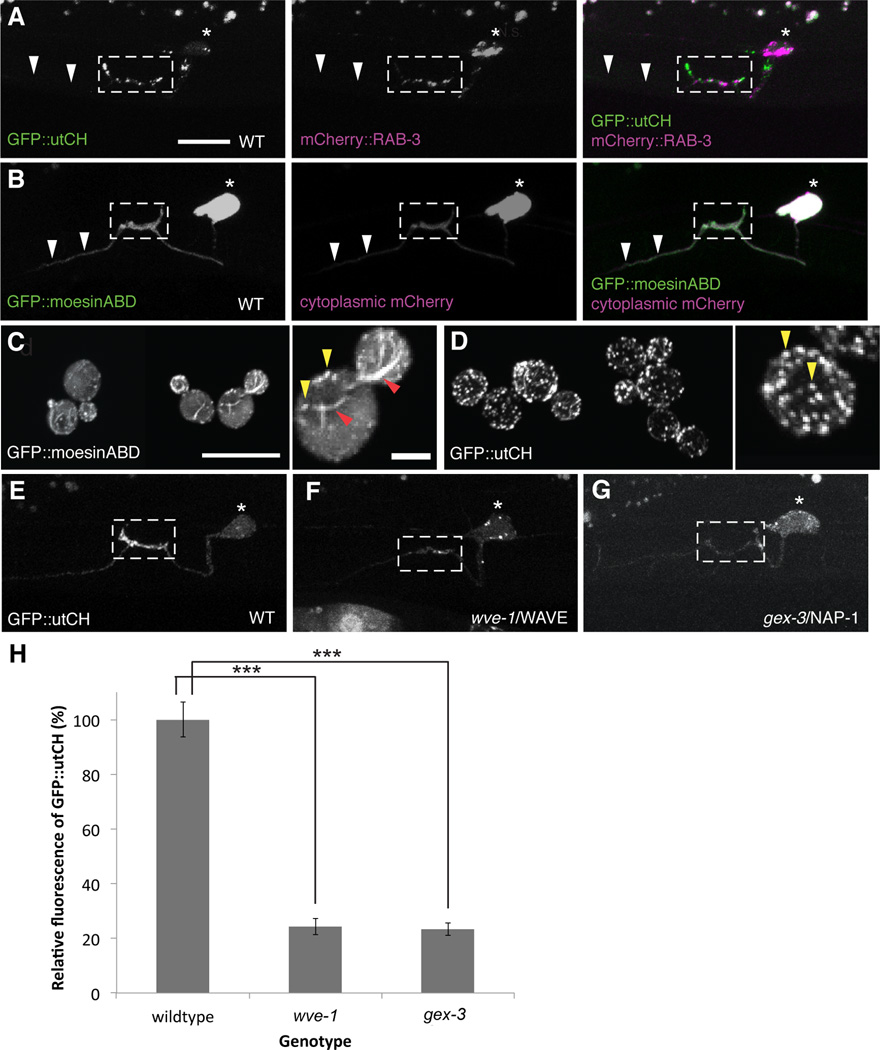
To further dissect the molecular pathway involved in establishing this F-actin network, we first sought to understand the nature of the F-actin at HSN synapses. Cells can generate a diverse array of F-actin networks that differ in geometry, mechanics and dynamics for various cellular functions. These F-actin structures bind and interact with different subsets of proteins that can also be used to label F-actin in vivo. When expressed in HSN, GFP::utCH distinctively labels F-actin enriched at the synaptic region labeled by mCherry::RAB-3 (Figure 3A). This localization is unlike another well-established in vivo F-actin probe, the actin-binding domain of moesin (GFP::moesinABD), which labels the entire HSN axon with no obvious enrichment at the synaptic region, similar to a cell morphology marker, cytoplasmic mCherry (Figure 3B). This suggests that GFP::utCH may bind to a specific subpopulation of F-actin that is found locally at synapses. To identify the specific F-actin structure that utCH binds to, we expressed both GFP::moesinABD and GFP::utCH in Saccharomyces cerevisiae. Three distinct actin networks are known to assemble in yeast: Arp2/3-dependent branched F-actin involved in clathrin-mediated endocytosis; formin-mediated actin cables required for vesicle trafficking; and a formin mediated actin contractile ring necessary for cytokinesis (Michelot and Drubin, 2011). GFP::moesinABD, when expressed in yeast cells labeled all three structures (Figure 3C). Interestingly, GFP::utCH only labeled the Arp2/3-dependent endocytic patches in yeast cells (Figure 3D). Together, these results suggest that the synaptic F-actin network might comprise of Arp2/3-dependent, branched F-actin.
Figure 3. WRC is required for assembling an Arp2/3 mediated actin network at synapses.
(A) GFP::utrophinCH labels the F-actin network that is enriched at synapses labeled by mCherry::RAB-3. White arrows point to the anterior axon that has very little GFP::utCH staining. (B) GFP::moesinABD labels the entire HSN neuron (white arrows show bright labeling along the entire axon) with no significant enrichment at presynaptic sites as compared to cytoplasmic mCherry. Scale bars represent 10 µm. (C, D) This difference in actin binding is observed in yeast where GFP::moesinABD binds F-actin cables (red arrowheads) and endocytic F-actin patches (yellow arrowheads) whilst GFP::utCH binds only endocytic F-actin patches Scale bars represent 10 µm and higher magnification image is 2 µm. (E–G) Localization of GFP::utCH at the synaptic region is loss in wve-1 and gex-3 mutants compared to WT. (H) Graph quantifies the average fluorescence intensity for GFP::utCH. wve-1 and gex-3 showed a 66 ± 3% and 67 ± 2% reduction in utCH fluorescence respectively. Each bar represents the average fluorescence value and error bars are ± S.E.M. (***p<0.001 with n>20, Two-tailed Student’s t-test). See also Figure S2 and S3.
The actin nucleating activity of the Arp2/3 complex is tightly regulated by various cytosolic proteins, including the WASP and WAVE protein complexes and their upstream regulators, the Rho family of small GTPases (Derivery and Gautreau, 2010; Padrick and Rosen, 2010; Takenawa and Suetsugu, 2007). To understand the molecular mechanisms that establish the synaptic F-actin network, we performed a candidate screen for factors that affect the localization of synaptic F-actin. We found that the localization of GFP::utCH at the synaptic region is drastically reduced in wve-1/WAVE mutants (24 ± 3% of wildtype) (Figures 3E, 3F and 3H), suggesting that WAVE is required to assemble the synaptic F-actin network. In cells, the WAVE protein is constitutively incorporated into a five component complex, the WAVE Regulatory Complex (WRC) that is required for both its regulation and function (Chen et al., 2010b; Eden et al., 2002; Kurisu and Takenawa, 2009). We observed similar reductions in GFP::utCH labeling at synapses in gex-3/NAP1 mutants (23 ± 2% of wildtype), another component of the WRC (Figures 3G and 3H). Furthermore, the fluorescence level in the cell body is comparable between wildtype and wve-1 or gex-3 mutants, suggesting that the WRC does not regulate overall levels of F-actin, but is specifically required for enrichment of F-actin at the presynaptic region. WRC mutations do not alter the distribution of GFP::moesinABD in HSN (Supplementary figure 2), further indicating that the WRC specifically patterns synaptic F-actin. We also examined a partial loss-of-function mutant of the wsp-1/WASP gene and observed no defect in GFP::utCH enrichment at synapses (data not shown). However, since this mutant allele does not completely eliminate the function of WSP-1 and the null mutant is embryonic lethal, we are unable to comment on the role of wsp-1.
The WRC is inactive in its basal state and activated by the small GTPase Rac (Chen et al., 2010b; Eden et al., 2002; Ismail et al., 2009; Lebensohn and Kirschner, 2009). Three Racs exist in the C. elegans genome: ced-10, mig-2 and rac-2, which function in a partially redundant manner to regulate axon guidance (Shakir et al., 2008). We examined synaptic F-actin recruitment in single or compound mutants of these genes. While the mig-2 single mutant showed slightly reduced F-actin staining, the ced-10 and rac-2 single mutants showed no statistically significant reduction (Supplementary figure 3). However, in both mig-2;rac-2 and ced-10;rac-2 double mutants, F-actin is dramatically reduced, suggesting that these small GTPases function redundantly to regulate synaptic F-actin (Supplemental Figure 3). Taken together, the data above shows that the synaptic F-actin network is dependent on a signaling pathway that involves SYG-1, the Rac GTPases and the WRC.
The WRC is required for both presynaptic assembly and axonal branch formation
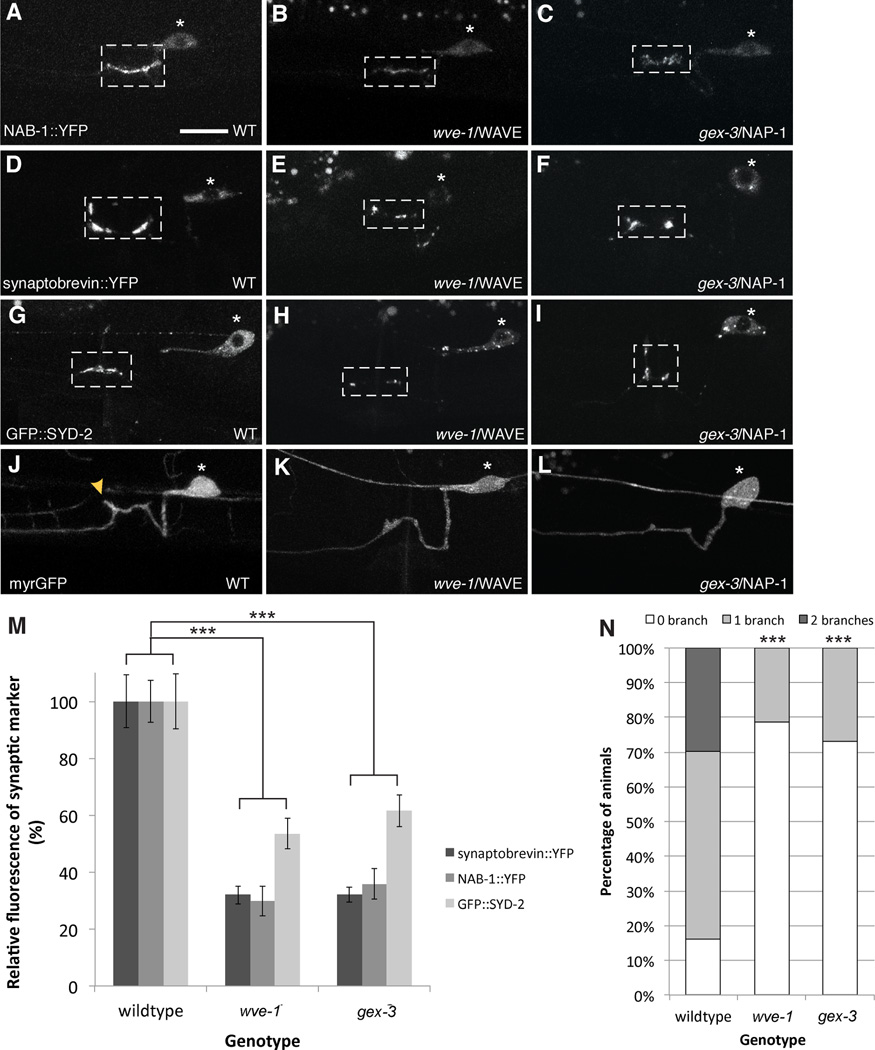
Since the WRC is involved in local F-actin assembly it suggests that the WRC may be required for both synapse formation and axonal branching. We found previously that the presynaptic actin recruits scaffolding molecule NAB-1, which in turn sequesters active zone proteins and synaptic vesicles. We found that NAB-1 recruitment to synapses is significantly reduced in wve-1 and gex-3 mutants, suggesting that the reduced F-actin impact the amount of the NAB-1 at synapses (Figures 4A–4C and 4M). Both wve-1 and gex-3 mutants also showed reduction in synaptic vesicle marker, SNB-1 as well as active zone molecule, SYD-2, suggesting that the WRC is required for synapse assembly (Figures 4D–4I and 4M). In contrast to these presynaptic markers, we found that the localization of SYG-1 remained unaffected in wve-1 or gex-3 mutants, suggesting that the WRC functions downstream of SYG-1 (Supplementary figure 4).
Figure 4. WRC is required for both presynapse assembly and axonal branch formation.
(A) F-actin dependent active zone protein NAB-1::YFP localizes to synapses in WT animals. Loss of (B) wve-1 or (C) gex-3 results in failed recruitment of NAB-1 to presynaptic sites. (D) A wildtype neuron with synapses labeled by synaptobrevin::YFP. (E, F) wve-1 and gex-3 mutants show partial loss of synaptobrevin::YFP. (G–I) Similarly for active zone molecule SYD-2, wve-1 and gex-3 mutants display a partial reduction in the recruitment of GFP::SYD-2 to synapses. (J) Myristolated GFP highlights the morphology of HSN. (K, L) Most wve-1 and gex-3 mutants fail to extend collateral axonal branches. Yellow arrowheads point to branches. Scale bars represent 10 µm. (M) Graph quantifies the relative average fluorescence of synaptobrevin::YFP, NAB-1::YFP and GFP::SYD-2 in wildtype, wve-1 and gex-3 mutants. Each bar represents the average fluorescence value and error bars are ± S.E.M. (***p<0.001 with n>20, Two-tailed Student’s t-test). See also Figure S4. (N) Graph quantifies the percentage of animals that elaborate zero, one or two branches. wve-1 and gex-3 mutants have significantly less branches as compared to wildtype. Statistics for each mutant was compared against wildtype (***p<0.001 with n>100, Fisher’s exact test). See also Figure S4 and S5.
We next examined if the WRC is also required for axonal branch formation. We observed that about 70% of the wve-1 and gex-3 mutant animals fail to from collateral branches (Figures 4J–4L and 4N). This loss of branching is not due to general defects in axon outgrowth as the main HSN axon shaft is able to extend to its normal length along the ventral cord. Furthermore, we observed that enrichment of GFP::utCH at the growing tip of the HSN axon is unaffected in wve-1 mutants (Supplementary figure 4).
Taken together, these data suggest that the WRC is required to assemble a local F-actin network directed by cell adhesion protein SYG-1, which is important for both synapse assembly and collateral axon branching in HSN. This requirement for the WRC was also observed in the VC4/5 neurons, two ventral cord neurons that synapse onto the vulva muscles (Supplementary figure 5). SYG-1 is not required for the branching and synapse formation of the VC4/5 neurons, suggesting that the WRC has more general functions (A.H. and K.S. unpublished data), consistent with prior work showing that collateral branches often initiate from actin patches (Ketschek and Gallo, 2010).
The cytoplasmic tail of SYG-1 can bind the WRC
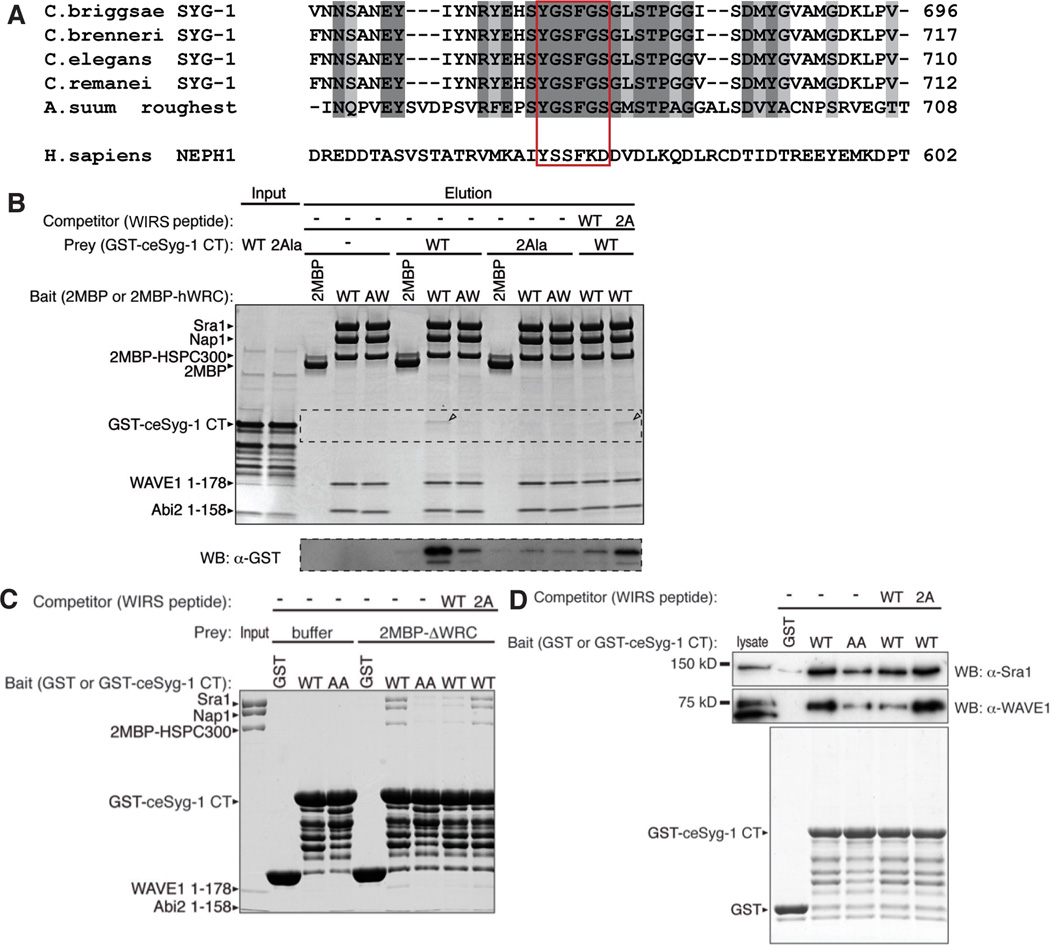
The data above argue strongly that the WRC functions downstream of SYG-1 to build a synaptic actin network that is required for both synapse assembly and collateral axon branching. To further understand how SYG-1 specifies the assembly of a synaptic F-actin network, we asked if SYG-1 might directly bind to the WRC. Recent work by Chen et. al., had identified a consensus peptide motif termed the WRC interacting receptor sequence (WIRS), which binds to a conserved site on the surface of the WRC (Chen et al., 2013). This motif is found in the intracellular tails of a large number of diverse neuronal receptors, including protocadherins, ROBO, netrin receptors, neuroligins and various channels. Structural and biochemical studies established that WIRS binds to a composite surface pocket formed by the Abi and Sra subunits of the WRC. This pocket is nearly 100% conserved in metazoans, including Drosophila and C. elegans and the interaction recruits the WRC to the cell membrane. Based on the definition of the WIRS consensus motif, Φ-x-T/S-F-x-x (Φ=preference for bulky hydrophobic residues; x=any residue), we found that the SYG-1 cytoplasmic tail contains a potential WIRS (peptide sequence YGSFGS) that is conserved throughout the nematode phyla (Figure 5A). Even though human NEPH1 and C. elegans SYG-1 share very little sequence homology in their cytoplasmic tails, human NEPH1 also had a similar WIRS (peptide sequence YSSFKD).
Figure 5. SYG-1 cytoplasmic tail contains a WRC interacting receptor sequence (WIRS) that specifically bind the WRC.
(A) Amino acid sequence alignment of SYG-1/Roughest/NEPH1 homologs. Dark shaded sequences are identical and light shaded sequences are conserved. The WIRS sequence is highlighted in the red box. (B) Pull-down using immobilized di-MBP-tagged human WRC complex as bait (wild type 2MBP-hWRC, or containing R106A/G110W mutations in the Abi2 subunit (AW), which impair binding to WIPS motifs). The top gel is an SDS-PAGE gel stained by coomassie blue and the bottom gel was blotted using mouse anti-GST conjugated to HRP. GST-tagged C. elegans SYG-1 cytoplasmic tail (GST-ceSYG-1 CT) is pulled down by MBP-hWRC (band in the coomasie blue gel is highlighted by black arrow). Making the AW mutation in hWRC interface that interferes with WIRS binding decreases this binding. Mutating the WIRS sequence in GST-ceSYG-1 CT (2Ala) also decreases the binding efficiency. Competitors were chemically synthesized 15 amino acid WIRS peptides (WT for wildtype and 2A for the mutant peptide) and only the wildtype peptide was able to compete for binding. (C) Coommasie blue stained gel from pull-down using immobilized GST or GST-ce-SYG-1 CT as bait and 2MBP-hWRC as prey (wild type and mutants as in panel B), with or without WIRS peptide competitor. The hWRC complex is pulled down by GST-ceSYG-1 CT (D) Pull-down using immobilized GST or GST-ce-SYG-1 CT as bait (wild type and mutants as in panels B and C) and mouse brain lysate as prey, with or without WIRS peptide competitor. Top two gels are western blots with anti-rabbit Sra1 and anti-mouse WAVE1 antibodies respectively; bottom gel is coomassie blue stained to show preys. See also Figure S6.
To verify if the SYG-1 tail specifically binds to the WRC, we performed pull-down experiments by immobilizing purified recombinant human WRC fused to a tandem maltose binding protein repeat (2MBP-hWRC) and ask if it can retain purified GST-tagged C. elegans SYG-1 cytoplasmic tail (GST-ceSYG-1-CT). After washing, the 2MBP-hWRC was able to weakly retain the SYG-1 cytoplasmic tail (Figure 5B). Similarly, immobilized GST-ceSYG-1-CT was able to retain 2MBP-hWRC (Figure 5C).
We next performed a series of experiments to learn whether binding is mediated by interactions of the SYG-1 WIRS motif and the WRC WIRS-binding surface. First, we tested the binding using a WRC with two point mutations on the WIRS binding surface, which specifically eliminate the interaction between the WRC and WIRS (2MBP-hWRCAW, contains R106A/G110W in the Abi2 subunit). We found that the AW mutant under the same conditions exhibited much less binding to the SYG-1 cytoplasmic tail, suggesting that SYG-1 binds to the conserved WIRS binding interface in the WRC. Reciprocally, we found that mutating two conserved residues in the SYG-1 WIRS sequence (ce-SYG-1-CT-2Ala, peptide sequence YGAAGS) also impair binding to the wild type WRC (Figure 5B). In addition, we performed a competition assay with a synthetic WIRS-containing peptide from protocadherin 10 (PCDH10, the first identified WIRS-containing protein) (Chen et al., 2013) and found that wildtype PCDH10 WIRS peptide can efficiently compete for binding with GST-ceSYG-1-CT in the pull down, whilst a peptide with a disrupted PCDH10 motif is unable to do so (Figure 5B).
Finally, to show that the SYG-1 WIRS sequence can interact with endogenous proteins, we performed the pull down using mouse brain lysates with immobilized GST-ce-SYG-1-CT. Using antibodies against Sra1 and WAVE1, two subunits of the WRC, we observed that SYG-1-CT was able to pull down both components (Figure 5D). Likewise in this experiment, mutating the SYG-1CT WIRS sequence or adding a wildtype competing PCDH10 WIRS peptide reduced the pull down efficiency of the WRC.
Although we did not have access to recombinant C. elegans WRC, we found that the SYG-1 cytosolic domain can specifically bind to reconstituted Drosophila WRC (Supplementary Figure 6). As the WIRS/WRC binding surface is completely conserved and we have observed interaction between human, mouse and fly WRC with the SYG-1 cytoplasmic tail, it is highly likely the C. elegans WRC also binds to SYG-1 WIRS (Chen et. al, 2013, (Ismail et al., 2009).
Together, the biochemical data above strongly indicates that the SYG-1 cytoplasmic tail binds to the conserved WIRS-binding surface of the WRC.
Local F-actin assembly requires interaction between SYG-1 WIRS and the WRC
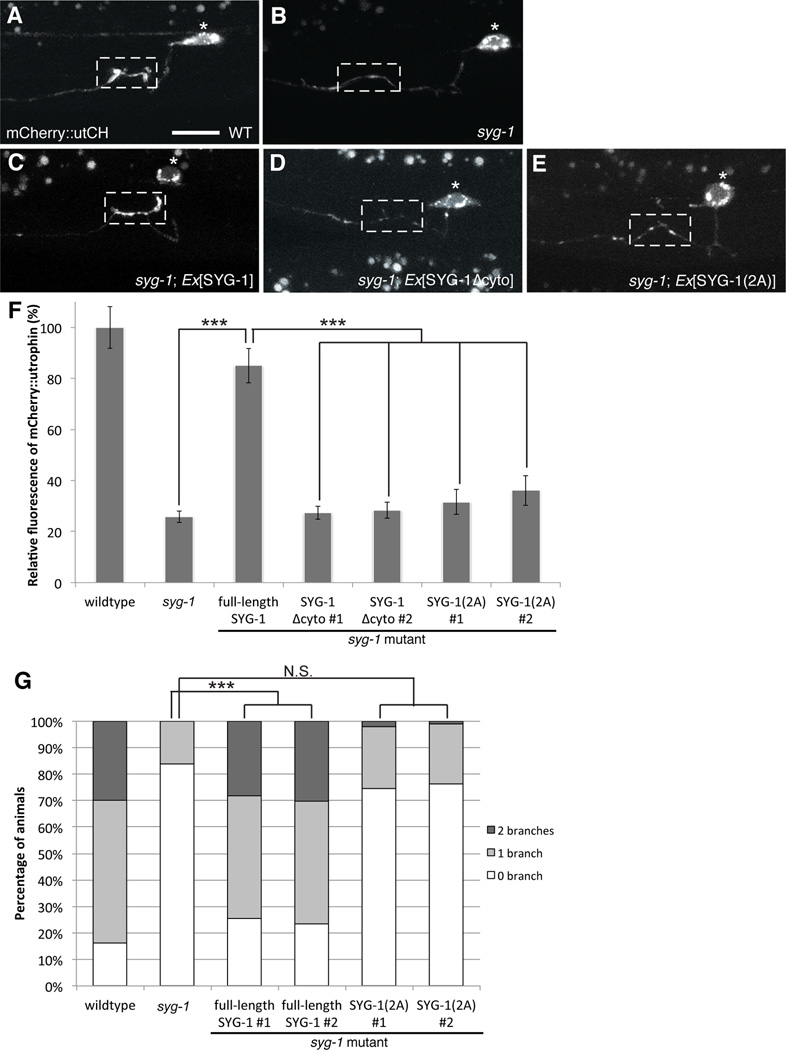
To determine if direct interaction between the WRC and SYG-1 WIRS motif is required in vivo, we performed structure/function experiments on SYG-1 and examined their ability to rescue F-actin assembly. In syg-1 mutants, the recruitment of synaptic F-actin is defective compared to wildtype animals as detected by the loss of mCherry::utCH labeling (75 ± 2% reduction compared to wildtype) (Figures 6A–B). Expressing a transgene containing full-length SYG-1 rescues this defect and restores F-actin assembly at the synaptic region (Figure 6C and 6F). SYG-1 lacking the entire cytoplasmic tail cannot rescue F-actin assembly (Figures 6D and 6F). More specifically, a SYG-1 protein containing di-Ala mutations in the WIRS motif, which did not bind WRC in vitro (Figure 5), also failed to rescue the defects in F-actin assembly in syg-1 mutants (Figures 6E and 6F). Deleting the cytoplasmic tail or mutating the WIRS sequence of SYG-1 does not affect its ability to cluster at the HSN synaptic region, a function that is solely dependent on its extracellular domain (Supplementary Figure 6). Thus, the SYG-1 WIRS interaction with the WRC is important for local F-actin assembly at the synaptic region. Finally, to understand if this interaction is also required for axon collateral branching, we expressed the various SYG-1 transgenes in syg-1 mutants and examined HSN branching with a neuronal morphology marker. Consistently, mutating the WIRS sequence in the SYG-1 cytoplasmic tail failed to rescue the loss of branching in syg-1 mutants (Figures 6G). Together, the data suggest that the SYG-1 WIRS motif interacts with the WRC to locally generate a synaptic F-actin network that is required for both presynaptic assembly and collateral axonal branching.
Figure 6. Local F-actin assembly requires interaction between SYG-1 WIRS and the WRC.
Structure-function analysis of SYG-1 (A) mCherry::utCH labels synaptic F-actin in wildtype worms. (B) This enrichment is lost in syg-1 mutants. (C) This defect is rescued by HSN-specific expression of a transgene carrying full-length SYG-1. (D) Expression of SYG-1 lacking its cytoplasmic tail SYG-1Δcyto, fails to rescue. (E) Similarly, expression of SYG-1 with two alanine mutations SYG-1(2A) in the WIRS sequence fails to restore F-actin localization to synapses. Scale bars represent 10 µm. (F) Graph quantifies the relative average fluorescence intensity of mCherry::utCH. Each bar represents the average fluorescence value and error bars are ± S.E.M. For lines expressing SYG-1Δcyto and SYG-1(2A) transgenes, two independent lines were quantified. (***p<0.001 with n>25, Two-tailed Student’s t-test). (G) Graph quantifies the percentage of animals that elaborate zero, one or two branches. Statistics for each mutant was compared against the wildtype values. (***p<0.001 with n>100, Fisher’s exact test). See also Figure S6
Discussion
WRC regulates F-actin and neuronal development
F-actin is found in various subcellular locations of neurons (Letourneau, 2009; Zhang and Benson, 2002). For example, cortical F-actin can be detected just underneath the plasma membrane in the neuronal cell body. In developing neurons, F-actin is also enriched at growth cones and branching sites (Dent et al., 2011; Gallo, 2011). In mature neurons, F-actin is enriched at presynaptic terminals and dendrite spines (Hotulainen and Hoogenraad, 2010; Sankaranarayanan et al., 2003). Within the pre- and postsynaptic terminals, spatially distinct populations of F-actin have been described, such as active zone associated F-actin, synaptic vesicle associated or endocytic zone associated actin on dendritic spines (Bleckert et al., 2012; Chia et al., 2012; Frost et al., 2010). Microscopically, F-actin can adopt several different forms including branched networks and bundled, unbranched filaments (Bloom et al., 2003; Chia et al., 2012; Sankaranarayanan et al., 2003; Waites et al., 2011). In a recent paper using super-resolution microscopy, actin and spectrin were found to form periodic ring-like structures that decorate the cortical cytoskeleton of axons in cultured hippocampal neurons. This cytoskeletal organization was not observed in dendrites (Xu et al., 2012). Molecularly, distinct molecular programs construct different forms of F-actin (Chhabra and Higgs, 2007; Michelot and Drubin, 2011). For example, studies in fibroblasts showed that branched actin networks are built by an Arp2/3 dependent mechanism, while unbranched actin bundles require molecules including formins (Derivery and Gautreau, 2010; Pruyne et al., 2002). We also found that different in vivo molecular markers of F-actin can distinguish these structures in yeast. UtrophinCH preferentially labels Arp2/3 dependent endocytic patches whilst moesinABD labels all forms of F-actin in yeast (Figures 3C and 3D).
These results raise two interesting questions in terms of the F-actin organization in neurons. First, are there distinct F-actin structures at different subcellular locations? Second, if so, what molecular mechanisms are responsible for assembling these different structures? Our results showed that F-actin found at HSN synapses is specifically labeled by utrophinCH. Consistently, utrophinCH labels Arp2/3-dependent F-actin structures in yeast and establishment of the F-actin network at synapses is dependent on the WRC, a multi-protein complex that activates the Arp2/3 complex to initiate the formation of branched F-actin (Miki et al., 1998; Padrick and Rosen, 2010; Pollard, 2007).
The roles of F-actin regulators in the development and function of presynaptic terminals have been reported in multiple systems. In Drosophila neuromuscular junctions (NMJ), the actin-capping protein adducin affects the stabilization and growth of synapses. In addition, the formin-related protein diaphanous controls the growth of NMJ by regulating both actin and microtubules (Pawson et al., 2008; Pielage et al., 2011). Interestingly, WASP and the Arp2/3 complex also play important roles at the fly NMJ. Mutations in these proteins as well as in the BAR protein, Nervous-Wreck, cause the formation of a highly ramified cluster of synaptic boutons, likely through regulating endocytosis, as endocytosis mutants exhibits similar phenotypes (Coyle et al., 2004; Rodal et al., 2008). The vertebrate active zone molecule Piccolo has also been shown to promote assembly of presynaptic F-actin and regulate presynaptic neurotransmitter release (Waites et al., 2011). The diversity of actin regulators at presynaptic terminals is likely evolved to accommodate different size, shape and functional properties of various synapses.
Besides presynaptic terminals, utrophinCH also labels the axonal growth cone during HSN axon outgrowth, suggesting that branched actin is also enriched there. However, we found little axon guidance defects in wve-1 mutants, possibly due to redundant pathways. In many processes, the WRC often functions redundantly with the other major Arp2/3 activator, WASP (Kurisu and Takenawa, 2009; Tang et al., 2013). Consistent with this notion, WVE-1 has also been shown to act together with WSP-1/WASP during axon guidance of sensory neuron PDE (Shakir et al., 2008). A recent study in C. elegans also showed that partial loss of wsp-1 causes neurons to be defective in acetylcholine transmission suggesting that WSP-1 is involved in synapse function (Zhang and Kubiseski, 2010). Consistent with our observation that F-actin assembly at synapses is unaffected with partial loss of wsp-1, the study also found that synapses labeled by synaptic vesicle marker, synaptogyrin, appeared unaffected in wsp-1 mutants. Fine control of F-actin assembly may also occur at the level of molecules that control the proteins that directly nucleate actin filaments. For example, the Rac GTPases CED-10/Rac and MIG-2/RhoG were shown to function in parallel pathways regulating WVE-1 and WSP-1 respectively in axon guidance (Shakir et al., 2008).
Cell adhesion molecules dictate F-actin organization to promote synapse formation and neuronal arborization
Various studies have shown that synaptogenesis occurs side by side with neuronal arborization. Direct observations of axon arbors in developing RGCs showed that synapses promote axon branch formation and increased branch stability (Meyer and Smith, 2006). These studies support the synaptotropic hypothesis that synapse formation can promote the elaboration of neuronal processes (Cline and Haas, 2008; Vaughn, 1989). Furthermore, molecular studies in RGCs also showed that the Netrin-DCC signaling pathway promotes addition of new synapses whilst also increasing branch dynamics, suggesting that synapse formation and branching events might be linked (Manitt et al., 2009). Although these studies provide observations that synapse formation and axonal branching occur together, it is not clear what molecular mechanisms link these two events or whether each of these processes are dependent on one another.
Our findings provide new mechanistic insights into how synapse-directed arborization can be achieved, as both synaptogenesis and axonal branching require the same local synaptic F-actin network to proceed. A homologous process where cell adhesion molecules locally induce F-actin rearrangements also occurs during myoblast fusion where the SYG-1 homologs, sticks and stones (sns) and hibris (hbs) organize a podosome-like F-actin structure to invade the apposing muscle founder cell (Jin et al., 2011; Sens et al., 2010). Both WAVE and WASP are involved in this function downstream of the adhesion molecules, suggesting that the WRC/WIRS interaction in the SYG-1/SYG-2 family of molecules might be conserved and used in different developmental contexts to couple membrane interactions with diverse F-actin based cellular responses.
During development, multiple sets of adhesion and diffusible cues pattern axonal projection and synaptic connections. Several pieces of evidence argue that the presynaptic F-actin network might be a common component to couple axon arborization and synapse formation. For example, a recent study showed that stabilization of filopodia by neurexin-neuroligin adhesion complexes are required to promote both synaptogenesis and dendrite arborization (Chen et al., 2010a). Furthermore, UNC-40/DCC receptor downstream of netrin signaling is required for formation of axon arbors, presynaptic terminals and neurosecretory terminals. It is also noteworthy that both neuroligins and UNC-40/DCC contain conserved WIRS motifs in their cytoplasmic tails (Chen et. al., 2013). Both Ena/VASP and MIG-10/Lamellipodin, regulators of F-actin polymerization, function downstream of axon arborization (Nelson and Colon-Ramos, 2013; Stavoe et al., 2012). Even poly-D-lysine-coated beads can induce presynaptic differentiation and local assembly of F-actin in vitro, suggesting that cell adhesion alone may be sufficient to induce F-actin assembly (Lucido et al., 2009). Together with our data, these studies highlight the importance of cell adhesion molecules in specifying the subcellular location of F-actin rearrangements to coordinate various processes during nervous system development.
Together, we provide a molecular pathway where synaptic adhesion molecule SYG-1 spatially link downstream synapse formation and axonal collateral branch formation by locally assembling an F-actin network. SYG-1 exerts this function through a direct interaction with the WRC via a WIRS motif in its cytosolic tail. We propose that this interaction may potentially restrict the actin regulation activity of the WRC to desired subcellular domains.
Experimental Procedures
Worm strains
All strains were maintained at 20°C on OP50 E. coli nematode growth medium plates. N2 Bristol stain worms were used as the wildtype reference and the following mutants were used: wve-1(ok3308)I, gex-3(zu196)IV, syg-1(ky652)X and syg-2(ky673)X.
Transgenic lines
wyIs291 [Punc-86::gfp::utCH; Podr- 1::gfp], wyEx4096 [Punc-86::gfp::utCH; Punc-86::mCherry::RAB-3 ; Podr- 1::gfp], wyEx4099 [Punc-86::gfp::utCH; Punc-86::SYG-1::mCherry ; Podr- 1::gfp], wyEx4445 [Punc-86::gfp::utCH; Punc-86::mCherry::NAB-1 ; Podr- 1::dsred], wyEx3840 [Punc-86::gfp::utCH; Punc-86::mCherry::RAB-3 ; Podr- 1::gfp]. SYG-1 lines – wyEx241 [Punc-86::syg-1Δcyto::CFP; Podr-1::gfp], wyEx5316 [Punc-86::syg-1; Podr-1::gfp], wyEx5973 [Punc-86::syg-1(2A); Podr-1::gfp], wyEx5367 [Punc-86::syg-1::mCherry; Podr-1::gfp], wyEx6141 [Punc-86::syg-1Δcyto::mCherry; Podr-1::gfp], wyEx6143 [Punc-86::syg-1(2A)::mCherry; Podr-1::gfp]. wyIs97 [Punc-86::myrgfp; Punc-86::mCherry::rab-3; Podr- 1::gfp], kyIs235 [Punc-86::snb-1::yfp; Punc-4::lin-10::dsred; Podr- 1::dsred], wyIs12 [Punc-86::gfp::syd-2; Podr- 1::gfp], wyEx7 [Punc-86::gfp::syd-2; Podr- 1::gfp] kyEx673 [Pegl-17::syg-2; Podr- 1::gfp].
Molecular Biology
Expression plasmids for transgenic worm lines were made using the pSM vector, a derivative of pPD49.26 (A. Fire). The unc-86 promoter was cloned between SphI/XmaI and genes of interests were cloned between NheI/KpnI. Plasmids were injected into animals at 1 ng/µl together with co-injection markers at 40ng/ul. Yeast expression plasmids were made in pRS413 and genes of interest were cloned between BamHI/XhoI.
Fluorescence quantification and confocal imaging
All fluorescence images of HSN synapses were taken in live worms immobilized with 10uM levamisol with a 63×/1.4NA objective on a Zeiss Axioplan 2 Imaging System or a Plan-Apochromat 63×/1.4 objective on a Zeiss LSM710 confocal microscope using similar imaging parameters for the same marker across different genotypes. Fluorescence quantification was determined using Image J software (NIH) (n>20).
Latrunculin A injections
Early L4 animals were isolated and microinjected with either 1mM latrunculin A (Sigma) in DMSO or DMSO alone, into the pseudocoelom of the worm at a site posterior of the vulva. 16 hours post-injection, animals were scored for branches. Statistical significance was determined using the Fisher’s exact test (n>30).
Protein Purification
Human WRC was purified as described (ref to PMID:21107423; PMID:19363480) with modifications (refer to manuscript).
GST or GST tagged cytoplasmic tails of SYG-1(aa575–727) were expressed in BL21 (DE3) T1R cells at 37°C. Proteins were purified using glutathione sepharose beads (GE Healthcare), followed by a Source Q15 column.
Protein concentrations were calculated using absorption extinction coefficients calculated by the ProtParam website (ref to Expasy) using protein primary sequences.
Pull-down Assays
GST pull-down was performed by mixing 1nmol of GST or GST-Syg1-CT with 0.4 nmol of (MBP)2-WRC and 20mL of glutathione sepharose beads in 1mL of binding buffer (20mM HEPES, 50 mM NaCl, 5% (w/v) glycerol and 5mM beta-mercaptoethanol, pH7). 250nmol of WIRS peptides (WT or mutant, synthesized by Abgent) were also included in the reactions as competitors. After continuous mixing at 4°C for 30min, the beads were centrifuged and washed three times using the binding buffer. Bound proteins were eluted with GST elution buffer (100mM Tris-HCl, 120mM NaCl, 5%(w/v) glycerol, 1mM EDTA, 5mM beta-mercaptoethanol and 30mM reduced glutathione, pH8.5) and examined by SDS-PAGE.
MBP pull-down was performed similarly, using 60pmol of (MBP)2 tagged WRC as bait, 5 fold excess GST-Syg1-CT as prey, 500nmol of WIRS peptides where indicated as competitors, and 15mL of amylose beads in 1mL of binding buffer at 4°C for 30min, followed by three washes. Bound proteins were eluted with 0.5%(w/v) maltose added in binding buffer, and examined by SDS-PAGE.
For mouse brain lysate pull-down, frozen adult mouse brain (Pel-Freez Biologicals) was lysed on ice using a dounce homogenizer in 10 fold (v/w) lysis buffer (50mM Tris-HCl, 150mM NaCl, 5%(w/v) glycerol, 1%(w/v) NP40, 1mM EDTA, 1mM PMSF, 5mM Leupeptin, 5mM Antipain and 5mM Benzamidine, pH7.6), followed by rotary mixing at 4°C for 1hr and centrifugation at 50krpm (18kg) in a Ti70 rotor at 4°C for 1hr. In GST pull-down reactions, the clarified lysate (containing 1mg total protein measured by the BCA method) was mixed with 0.5nmol of purified GST or GST-Syg1-CT, 125nmol of WIRS peptide competitors where indicated, and 20mL of glutathione sepharose beads in 0.6mL of lysis buffer at 4°C for 1hr. The beads were washed 3 times and eluted in GST elution buffer. The bound proteins were resolved by SDS-PAGE and examined by Western Blot for WAVE1 (Neuromab, clone K91/36) or Sra1 (Upstate, 07–531).
Supplementary Material
Highlights.
An adhesion molecule, SYG-1 specifies both synapse formation and axon branching
F-actin networks at presynaptic sites coordinate synapse formation and axon branching
SYG-1 interacts with the WAVE Regulatory Complex through a receptor sequence (WIRS)
The SYG-1 WIRS/WRC interaction is required to pattern synaptic F-actin
Acknowledgements
This work was funded by the Howard Hughes Medical Institute. P. H. Chia is supported by the Agency for Science, Technology, and Research, Singapore. We thank the International Caenorhabditis Genetic Center and the Japanese National Bioresource Project for strains. We also thank C. Gao for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Besse F, Mertel S, Kittel RJ, Wichmann C, Rasse TM, Sigrist SJ, Ephrussi A. The Ig cell adhesion molecule Basigin controls compartmentalization and vesicle release at Drosophila melanogaster synapses. The Journal of Cell Biology. 2007;177:843–855. doi: 10.1083/jcb.200701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckert A, Photowala H, Alford S. Dual pools of actin at presynaptic terminals. J Neurophysiol. 2012;107:3479–3492. doi: 10.1152/jn.00789.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom O, Evergren E, Tomilin N, Kjaerulff O, Löw P, Brodin L, Pieribone VA, Greengard P, Shupliakov O. Colocalization of synapsin and actin during synaptic vesicle recycling. The Journal of Cell Biology. 2003;161:737–747. doi: 10.1083/jcb.200212140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Tari PK, She K, Haas K. Neurexin-Neuroligin Cell Adhesion Complexes Contribute to Synaptotropic Dendritogenesis via Growth Stabilization Mechanisms In Vivo. Neuron. 2010a;67:967–983. doi: 10.1016/j.neuron.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature. 2010b;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Chia PH, Patel MR, Shen K. NAB-1 instructs synapse assembly by linking adhesion molecules and F-actin to active zone proteins. Nature Neuroscience. 2012;15:234–242. doi: 10.1038/nn.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline H, Haas K. The regulation of dendritic arbor development and plasticity by glutamatergic synaptic input: a review of the synaptotrophic hypothesis. The Journal of Physiology. 2008;586:1509–1517. doi: 10.1113/jphysiol.2007.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle IP, Koh Y-H, Lee W-CM, Slind J, Fergestad T, Littleton JT, Ganetzky B. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The Growth Cone Cytoskeleton in Axon Outgrowth and Guidance. Cold Spring Harbor Perspectives in Biology. 2011;3:a001800–a001800. doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Kalil K. Axon branching requires interactions between dynamic microtubules and actin filaments. Journal of Neuroscience. 2001;21:9757–9769. doi: 10.1523/JNEUROSCI.21-24-09757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Gautreau A. Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. BioEssays. 2010;32:119–131. doi: 10.1002/bies.200900123. [DOI] [PubMed] [Google Scholar]
- Doussau F, Augustine GJ. The actin cytoskeleton and neurotransmitter release: an overview. Biochimie. 2000;82:353–363. doi: 10.1016/s0300-9084(00)00217-0. [DOI] [PubMed] [Google Scholar]
- Dwivedy A, Gertler FB, Miller J, Holt CE, Lebrand C. Ena/VASP function in retinal axons is required for terminal arborization but not pathway navigation. Development (Cambridge, England) 2007;134:2137–2146. doi: 10.1242/dev.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-Molecule Discrimination of Discrete Perisynaptic and Distributed Sites of Actin Filament Assembly within Dendritic Spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G. The cytoskeletal and signaling mechanisms of axon collateral branching. Developmental Neurobiology. 2011;71:201–220. doi: 10.1002/dneu.20852. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. The Journal of Cell Biology. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Duan R, Luo F, Zhang G, Hong SN, Chen EH. Competition between Blown Fuse and WASP for WIP Binding Regulates the Dynamics of WASP-Dependent Actin Polymerization In Vivo. Dev Cell. 2011;20:623–638. doi: 10.1016/j.devcel.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketschek A, Gallo G. Nerve Growth Factor Induces Axonal Filopodia through Localized Microdomains of Phosphoinositide 3-Kinase Activity That Drive the Formation of Cytoskeletal Precursors to Filopodia. Journal of Neuroscience. 2010;30:12185–12197. doi: 10.1523/JNEUROSCI.1740-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu S, Takenawa T. The WASP and WAVE family proteins. Genome Biology. 2009;10:226. doi: 10.1186/gb-2009-10-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Molecular Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau P. Actin in axons: stable scaffolds and dynamic filaments. Cell Biology of the Axon. 2009:65–90. doi: 10.1007/400_2009_15. [DOI] [PubMed] [Google Scholar]
- Lucido AL, Suarez Sanchez F, Thostrup P, Kwiatkowski AV, Leal-Ortiz S, Gopalakrishnan G, Liazoghli D, Belkaid W, Lennox RB, Grutter P, et al. Rapid assembly of functional presynaptic boutons triggered by adhesive contacts. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:12449–12466. doi: 10.1523/JNEUROSCI.1381-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manitt C, Nikolakopoulou AM, Almario DR, Nguyen SA, Cohen-Cory S. Netrin participates in the development of retinotectal synaptic connectivity by modulating axon arborization and synapse formation in the developing brain. Journal of Neuroscience. 2009;29:11065–11077. doi: 10.1523/JNEUROSCI.0947-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MP, Smith SJ. Evidence from in vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. Journal of Neuroscience. 2006;26:3604–3614. doi: 10.1523/JNEUROSCI.0223-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Current biology : CB. 2011;21:R560–R569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. The EMBO journal. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ, Hong W, Dani VS, Favaloro V, Luo L. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 2012;484:237–241. doi: 10.1038/nature10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy VN, De Camilli P. Cell biology of the presynaptic terminal. Annual review of neuroscience. 2003;26:701–728. doi: 10.1146/annurev.neuro.26.041002.131445. [DOI] [PubMed] [Google Scholar]
- Nelson JC, Colon-Ramos DA. Serotonergic Neurosecretory Synapse Targeting Is Controlled by Netrin-Releasing Guidepost Neurons in Caenorhabditis elegans. Journal of Neuroscience. 2013;33:1366–1376. doi: 10.1523/JNEUROSCI.3471-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK. Physical Mechanisms of Signal Integration by WASP Family Proteins. Annu Rev Biochem. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Lehrman EK, Poon VY, Crump JG, Zhen M, Bargmann CI, Shen K. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nature Neuroscience. 2006;9:1488–1498. doi: 10.1038/nn1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson C, Eaton BA, Davis GW. Formin-dependent synaptic growth: evidence that Dlar signals via Diaphanous to modulate synaptic actin and dynamic pioneer microtubules. Journal of Neuroscience. 2008;28:11111. doi: 10.1523/JNEUROSCI.0833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage J, Bulat V, Zuchero J, Fetter R. Hts/Adducin Controls Synaptic Elaboration and Elimination. Neuron. 2011 doi: 10.1016/j.neuron.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annual review of biophysics and biomolecular structure. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of Formins in Actin Assembly: Nucleation and Barbed-End Association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Motola-Barnes RN, Littleton JT. Nervous Wreck and Cdc42 Cooperate to Regulate Endocytic Actin Assembly during Synaptic Growth. Journal of Neuroscience. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan S, Atluri PP, Ryan TA. Actin has a molecular scaffolding, not propulsive, role in presynaptic function. Nature Neuroscience. 2003;6:127–135. doi: 10.1038/nn1002. [DOI] [PubMed] [Google Scholar]
- Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, Parachini L, Chen EH. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. The Journal of Cell Biology. 2010;191:1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir MA, Jiang K, Struckhoff EC, Demarco RS, Patel FB, Soto MC, Lundquist EA. The Arp2/3 Activators WAVE and WASP Have Distinct Genetic Interactions With Rac GTPases in Caenorhabditis elegans Axon Guidance. Genetics. 2008;179:1957–1971. doi: 10.1534/genetics.108.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–630. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- Spillane M, Ketschek A, Jones SL, Korobova F, Marsick B, Lanier L, Svitkina T, Gallo G. The actin nucleating Arp2/3 complex contributes to the formation of axonal filopodia and branches through the regulation of actin patch precursors to filopodia. Developmental Neurobiology. 2011;71:747–758. doi: 10.1002/dneu.20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AKH, Nelson JC, Martínez-Velázquez LA, Klein M, Samuel ADT, Colón-Ramos DA. Synaptic vesicle clustering requires a distinct MIG-10/Lamellipodin isoform and ABI-1 downstream from Netrin. Genes & development. 2012;26:2206–2221. doi: 10.1101/gad.193409.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Bamji SX. Pix Modulates Actin-Mediated Recruitment of Synaptic Vesicles to Synapses. Journal of Neuroscience. 2011;31:17123–17133. doi: 10.1523/JNEUROSCI.2359-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP–WAVE protein network: connecting the membrane to the cytoskeleton. NATURE REVIEWS MOLECULAR CELL BIOLOGY. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tang H, Li A, Bi J, Veltman DM, Zech T, Spence HJ, Yu X, Timpson P, Insall RH, Frame MC, et al. Loss of Scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Current biology : CB. 2013;23:107–117. doi: 10.1016/j.cub.2012.11.059. [DOI] [PubMed] [Google Scholar]
- Vaughn JE. Fine structure of synaptogenesis in the vertebrate central nervous system. Synapse (New York, NY) 1989;3:255–285. doi: 10.1002/syn.890030312. [DOI] [PubMed] [Google Scholar]
- Waites CL, Leal-Ortiz SA, Andlauer TFM, Sigrist SJ, Garner CC. Piccolo Regulates the Dynamic Assembly of Presynaptic F-Actin. Journal of Neuroscience. 2011;31:14250–14263. doi: 10.1523/JNEUROSCI.1835-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, Spectrin, and Associated Proteins Form a Periodic Cytoskeletal Structure in Axons. Science (New York, NY) 2012 doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Benson DL. Stages of synapse development defined by dependence on F-actin. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:5169–5181. doi: 10.1523/JNEUROSCI.21-14-05169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Benson DL. Developmentally regulated changes in cellular compartmentation and synaptic distribution of actin in hippocampal neurons. Journal of neuroscience research. 2002;69:427–436. doi: 10.1002/jnr.10313. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kubiseski TJ. Caenorhabditis elegans wsp-1 Regulation of Synaptic Function at the Neuromuscular Junction. J Biol Chem. 2010;285:23040–23046. doi: 10.1074/jbc.M109.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.