Abstract
The B cell-specific transcription factor BACH2 is required for affinity maturation of mature B cells. Here, we show that Bach2-mediated activation of p53 is required for stringent elimination of pre-B cells that failed to productively rearrange immunoglobulin VH-DJH gene segments. Upon productive VH-DJH gene rearrangement, pre-B cell receptor signaling ends negative selection through BCL6-mediated repression of p53. In patients with pre-B ALL, BACH2-mediated checkpoint control is frequently compromised. Low levels of BACH2 expression represent a strong independent predictor of poor clinical outcome. Bach2+/+ pre-B cells resist leukemic transformation by Myc through Bach2-dependent upregulation of p53, and fail to initiate fatal leukemia in transplant recipient mice. ChIP-seq and gene expression analyses reveal that BACH2 competes with BCL6 for promoter binding and reverses BCL6-mediated repression of p53 and other checkpoint control genes. These findings identify Bach2 as a critical mediator negative selection at the pre-B cell receptor checkpoint and a safeguard against leukemogenesis.
Introduction
In mice, bone marrow progenitor cells produce approximately 10 million pre-B cells daily3. Newly formed pre-B cells, however, are destined to die unless they productively rearrange VH-DJH gene segments and are rescued by pre-B cell receptor signals into the long-lived peripheral B cell pool4-5. We recently identified the transcriptional repressor BCL6 as critical survival factor that rescues pre-B cells that productively rearranged VH-DJH gene segments and emerged from the pre-B cell receptor checkpoint6-7. However, the mechanisms leading to clearance of other pre-B cells that failed to productively rearrange VH-DJH gene segments and thus lack pre-B cell receptor expression are poorly understood.
Results
Bach2 induces Arf/p53 downstream of Pax5 during early B cell development
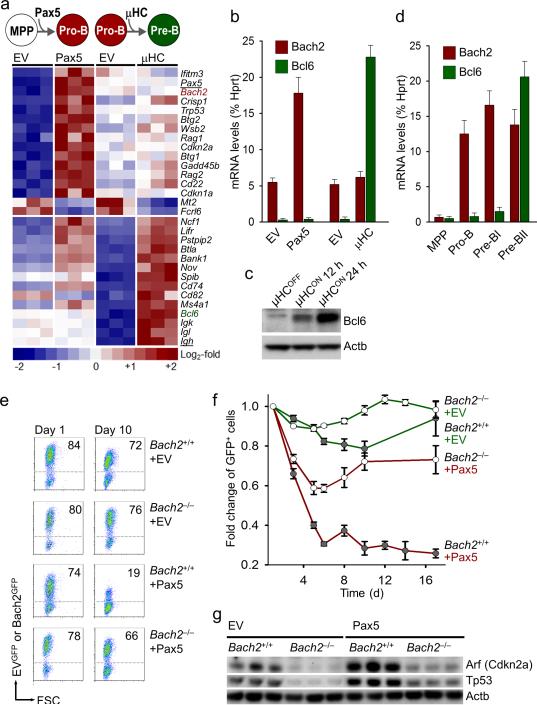
To identify factors that mediate negative selection at the pre-B cell receptor checkpoint in humans, we studied gene expression changes during human B cell development at the pro-B to pre-B cell transition8. We identified 18 genes with specific upregulation at the pre-B cell receptor checkpoint including components of the pre-B cell receptor itself (IGLL1, VPREB1), effectors of VH-DJH recombination (RAG1, RAG2, DNTT), and mediators of survival signaling (Supplementary Fig. 1). We next studied gene expression changes at the pre-B cell receptor checkpoint under two experimental conditions, namely (i) initiation of V(D)J recombination upon inducible activation of Pax59 and, (ii) inducible expression of a productively rearranged VH-DJH encoding a functional immunoglobulin μ heavy chain (μHC), which initiates pre-B cell receptor signaling7 (Fig. 1a). We reason that pre-B cell receptor checkpoint control should occur between these two events. As predicted, Pax5-mediated initiation of V(D)J recombination is reflected by strong upregulation of Rag1, Rag2 and caused upregulation of Arf/p53 and Bach2. By contrast, inducible activation of μHC expression induced expression of Bcl6, a transcriptional repressor of Arf/p53 (Figs. 1a-c). Bcl6-mediated repression of Arf/p53 is needed to rescue μHC+ pre-B cells once they have passed the pre-B cell receptor checkpoint6-7. Consistent with findings in human B cell differentiation (Supplementary Fig. 1), we found high levels of Bach2 prior to (Pro-B and pre-BI cells) and high levels of Bcl6 subsequent to (pre-BII cells) passage of the pre-B cell receptor checkpoint in mice (Fig. 1d). These findings imply that Bach2 potentially contributes to negative selection of pre-B cells that failed to productively rearrange VH-DJH gene segments at the pre-B cell receptor checkpoint. To test this hypothesis, we assayed pre-B cell receptor checkpoint control in pro-B and pre-BI cells from Bach2–/– mice2 and wildtype controls under experimental conditions. Activation of GFP-tagged Pax5 induces accumulation of Arf/p53 and results in rapid elimination of Bach2+/+ but not Bach2–/– pro-B and pre-BI cells (Figs. 1e-g).
Figure 1. Bach2 and Bcl6 maintain balance between negative selection and survival of early B cells at the pre-B cell receptor checkpoint.
(a) Gene expression changes at the pre-B cell receptor checkpoint were studied under two experimental conditions, namely (i) initiation of V(D)J recombination upon inducible activation of Pax5 compared to empty vector control (EV) and (ii) inducible expression of a functional immunoglobulin μ heavy chain (μHC). (b) Quantitative RT-PCR and (c) Western blot depicting regulation of Bach2 and Bcl6 by Pax5 and μHC-induction, respectively. (d) Quantitative RT-PCR for Bach2 and Bcl6 in multi-lineage progenitor cells (MPP), Pro-B, Pre-BI and pre-BII cells sorted from bone marrow of C57BL/6 mice. (e-f) GFP-tagged Pax5 or empty vector controls (EV) were transduced in transformed pro-B and pre-BI cells from Bach2-/- and wildtype mice and GFP+ cells were monitored (day 1 and day 10) by flow cytometry. (g) Bach2+/+ and Bach2-/- pro-B and pre-BI cells transduced with Pax5-GFP or EV were sorted and studied for protein expression of Arf and p53 by Western blotting, using β-actin as loading control.
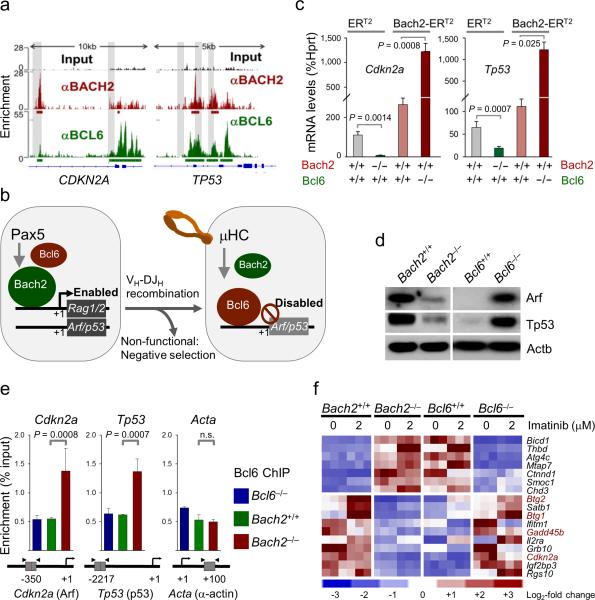
BCL6 reverses Bach2-induced transcriptional activation of Arf/p53
Previous studies by our group identified Bcl6 as a strong transcriptional repressor of Arf/p53 in normal pre-B cells6 and pre-B ALL10. Of note, ChIP-seq analysis and QChIP validation revealed that both BCL6 and BACH2 bind to overlapping promoter regions of CDKN2A (Arf) and TP53 (p53) and other checkpoint regulators (CDKN1A (p21), CDKN1B (p27), GADD45A, GADD45B) (Fig. 2a and Supplementary Figs. 2 and 3). We therefore, tested the hypothesis that BACH2 and BCL6 compete for binding to promoter regions of checkpoint regulator genes, and that the ratio between the two determines negative (Bach2>Bcl6) and positive (Bach2<Bcl6) selection events at the pre-B cell receptor checkpoint (Fig. 2b). Binding of either BCL6 or BACH2 to CDKN2A and TP53 promoters affects gene expression in opposite directions: mRNA and protein levels of Arf and p53 are significantly reduced in the absence of Bach2 but strongly increased in the absence of Bcl6 (Figs. 2d, 2f and Supplementary Fig. 6) or upon inducible overexpression of Bach2 (Fig. 2c and Supplementary Figure 4). Likewise, mRNA levels of the p53-dependent tumor suppressor Btg2 were reduced by >20-fold in the absence of Bach2 but increased by 3-fold in the absence of Bcl6 (Fig. 2f and Supplementary Fig. 4). To test whether Bach2 negatively regulates the ability of Bcl6 to bind to Cdkn2a (Arf) and Tp53 (p53) promoters, we performed Bcl6-ChIP experiments with Bcl6–/– (negative control), Bach2+/+ and Bach2–/– cells. Binding of Bcl6 to Cdkn2a (Arf) and Tp53 (p53) was significantly increased in Bach2–/– compared to Bach2+/+ cells (Fig. 2e and Supplementary Fig. 5). Gene expression analysis for a subset of common Bach2- and Bcl6-target genes revealed that Bcl6 and Bach2 affect gene expression levels of checkpoint regulators including Cdkn2a (Arf), Tp53, Gadd45b, Btg1 and Btg2, in opposite directions (Figs. 2d, 2f and Supplementary Fig. 6). These findings collectively indicate that the balance between Bcl6 and Bach2 determines repression or transcriptional activation of Cdkn2a (Arf), Tp53 (p53) and related checkpoint molecules.
Figure 2. Bach2-dependent activation of Arf/p53 is reversed by Bcl6 upon expression of a functional μ-heavy chain.
(a) ChIP-seq analysis was performed in a lymphoma cell line using antibodies against BACH2 (red) and BCL6 (green) and peaks with significant enrichment relative to background (input; black) are annotated by ChIP-seeqer with bold underlines. Overlapping peaks between BACH2 and BCL6 binding are indicated by shades. (b) The proposed scenario for BACH2-BCL6 interactions at the pre-B cell receptor checkpoint. (c) Effects of presence or absence of Bach2, presence or absence of Bcl6 and inducible overexpression of Bach2 (tamoxifen-inducible Bach2-ERT2 vector) on mRNA levels of Arf and p53 measured by qRT-PCR. (d) Effects of presence or absence of Bach2 and presence or absence of Bcl6 on protein levels of Arf and p53 measured by Western blot using β-actin as loading control. (e) To directly test the hypothesis that Bach2 negatively regulates the ability of Bcl6 to bind to Cdkn2a (Arf) and Tp53 (p53) promoters, Bcl6-ChIP experiments with Bcl6-/- (negative control), Bach2+/+ and Bach2-/- cells were performed. (f) A systematic ChIP-seq analysis revealed that 134 of the 541 BCL6- and 565 Bach2-target genes are shared (not shown). Gene expression analysis (Affymetrix GeneChip) for a subset of common Bach2- and Bcl6-target genes showing that Bcl6 and Bach2 affect gene expression levels of checkpoint regulators like Cdkn2a (Arf), Tp53, Gadd45b, Btg1 and Btg2, in opposite directions.
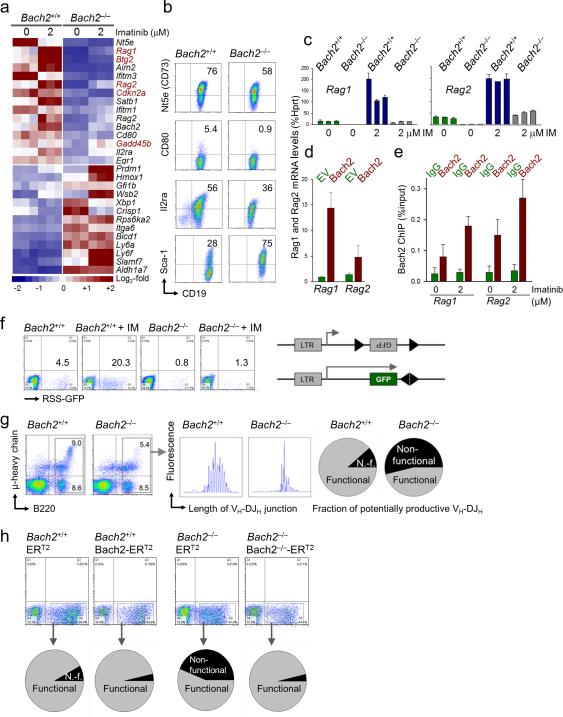
Bach2 mediates expression of Rag1, Rag2 and activates V(D)J recombination
We measured functional consequences of Bach2-deficiency at the pre-B cell receptor checkpoint in Bach2–/– and wildtype mice. Using a classical pre-B cell differentiation model based on tyrosine kinase inhibition (Imatinib; IM) of BCR-ABL1-transformed pre-B cells12-13, we studied gene expression changes upon differentiation of Bach2–/– and Bach2+/+ pre-BI cells (Fig. 3a). In this analysis, Bach2-deficiency was associated with increased expression of the early progenitor antigen Ly6f (Sca-1) and reduced expression of the pre-B cell antigen Il2ra (CD25; Figs. 3a-b). Importantly, mRNA levels of Rag1 and Rag2, critical effectors of V(D)J recombination, were reduced by ~20-fold. Likewise, Imatinib (IM)-induced differentiation induced strong upregulation of Rag1/Rag2 expression in Bach2+/+ pre-BI cells as previously described6,12-13, but failed to upregulate Rag1/Rag2 beyond baseline levels in Bach2–/–cells (Fig. 3c). Inducible overexpression of Bach2 was sufficient to drive Rag1/Rag2 upregulation (Fig. 3d) and single-locus ChIP revealed that BACH2 binds RAG1 and RAG2 promoters, which was enhanced by IM-treatment (Fig. 3e).
Figure 3. Bach2 mediates V(D)J recombination and μ-heavy chain checkpoint control during early B cell development.
(a) Using a classical pre-B cell differentiation model based on tyrosine kinase inhibition (Imatinib; IM) of BCR-ABL1-transformed pre-B cells12,13, gene expression changes upon inducible differentiation of Bach2-/- and Bach2+/+ pre-BI cells were measured by microarray analysis. (b) Increased expression of the early progenitor antigen Ly6f (Sca-1) and reduced expression of the pre-B cell antigen Il2rα (CD25) in Bach2-deficient Pre-BI cells was validated by flow cytometry. (c) Reduced mRNA levels of Rag1 and Rag2 in Bach2-deficient Pre-BI ALL (BCR-ABL1) cells in the presence and absence of IM were validated by qRTPCR. (d) The effect of inducible overexpression of Bach2 on Rag1 and Rag2 mRNA levels was measured by qRT-PCR. (e) Single-locus ChIP analysis using IgG and BACH2-specific antibodies depicting the binding of BACH2 to Rag1 and Rag2 promoters, which is further enhanced by IM-treatment.
(f) To test functional consequences of defective expression of Rag1/Rag2, Bach2+/+ and Bach2-/- pre-BI cells were transduced with a puromycin-selectable V(D)J recombination substrate carrying an inverted GFP flanked by recombination signal sequences (RSS). Recombination activity of the RSS substrate in Bach2+/+ and Bach2-/- pre-BI cells was measured by flow cytometry (inversion of the GFP cassette in the correct orientation; percentages of GFP+ cells are given) in the presence and absence of IM treatment. (g) Comparison of the composition of B cell progenitor populations in the bone marrow of Bach2+/+ and Bach2-/- mice by flow cytometry. Fragment length analysis revealed a size peak distribution of VH-DJH junctions that were indeed selected for multiples of 3 bp among Bach2+/+ compared to random distribution in Bach2-/- pre-B cells.
(h-i) IL7-dependent Bach2+/+ and Bach2-/- pre-B cells were transduced with tamoxifen (4-OHT)-inducible Bach2-ERT2 and ERT2 empty vector controls. Cells were treated with 4-OHT for 24 hours and sequence analysis of VH-DJH junctions was performed. (i) Amino acid sequences of VH-DJH junctions with non-functional VH-DJH rearrangements shaded in gray.
To test if defective expression of Rag1/Rag2 results in impaired V(D)J recombination activity, we transduced Bach2–/– and Bach2+/+ pre-BI cells with a V(D)J recombination RSS substrate. Consistent with the massively increased Rag1/Rag2 expression, IM-treatment resulted in a 6-fold increase of baseline recombination of the RSS substrate in Bach2+/+ pre-B cells. By contrast, IM-induced V(D)J recombination activity was reduced by 20-fold in Bach2–/– pre-BI cells as compared to wildtype counterparts (Fig. 3f and Supplementary Fig.7).
Bach2 is required for clearance of pre-BI cell clones that carry non-functional VH-DJH gene rearrangements
We next studied the composition of B cell progenitor populations in bone marrows and spleens of Bach2+/+ and Bach2–/– mice. Total numbers of CD19+ B220+ B cells were only slightly reduced in the bone marrow and spleen of Bach2–/– mice (Fig. 3g and Supplementary Fig. 8). While pre-B cells from Bach2+/+ bone marrow were stringently selected for productive in-frame VH-DJH gene rearrangements, pre-B cells from Bach2–/– mice lacked selection for VH-DJH junctions with coding capacity (Fig. 3g and Supplementary Fig. 8). Based on random V(D)J recombination, only one of three pre-B cell clones achieve in-frame rearrangements and coding capacity for a functional pre-B cell receptor. Effective negative pre-B cell selection causes clearance of cells with non-functional out-of-frame VH-DJH rearrangements and, hence, results in a length distribution of VH-DJH junctions with size peaks that are spaced by three nucleotides14. Fragment length analysis revealed a peak size distribution of VH-DJH junctions that were selected for multiples of 3 bp among Bach2+/+ as compared to random distribution in Bach2–/– counterparts (Fig. 3g and Supplementary Fig. 8). Sequence analysis of VH-DJH junctions confirmed that out-of-frame VH-DJH rearrangements were stringently cleared from the repertoire in Bach2+/+ pre-B cells as opposed to accumulation of a large fraction of clones with non-functional rearrangements among Bach2–/– pre-B cells (Fig. 3g and Supplementary Fig. 8).
To test if activation of Bach2 downstream of Pax5 is sufficient to clear the pre-B cell repertoire from non-functional VH-DJH rearrangements, we developed a vector system for inducible expression of Bach2. IL7-dependent Bach2+/+ and Bach2–/– pre-B cells were transduced with tamoxifen (4-OHT)-inducible Bach2-ERT2 and ERT2 empty vector controls. Cells were treated with 4-OHT for 24 hours and sequence analysis of VH-DJH junctions was performed (Fig. 3h and Supplementary Figs. 9 and 10). While inducible overexpression of Bach2 had no significant effect on the repertoire composition of Bach2+/+ pre-B cells, inducible Bach2-reconstitution in Bach2–/– pre-B cells resulted in rapid elimination of clones carrying non-functional VH-DJH rearrangements (gray shaded in Supplementary Fig. 10). We conclude that Bach2 is required and sufficient for stringent clearance of pre-B cells that failed to productively rearrange VH-DJH segments at the pre-B cell receptor checkpoint.
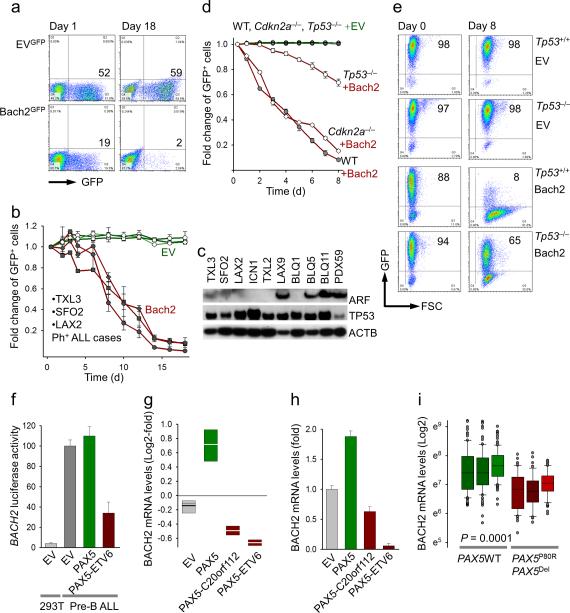
Bach2 mediates PAX5-dependent tumor suppression in pre-B ALL through activation of p53
In a high throughput screen for retroviral integrations in mouse pre-B cell leukemia (RTCGD), we found Bach2 introns 1-3 as common integration sites (Supplementary Fig. 11), raising the possibility that Bach2 functions as tumor suppressor in human pre-B ALL.
We therefore, tested whether Bach2 affects survival and proliferation of human pre-B ALL cells. To this end, we transduced xenografts from clinically-derived Ph+ ALL cells with vectors for either Bach2 or empty vector controls (EV). Primary Ph+ ALL cells carrying GFP-tagged Bach2 were rapidly depleted (Figs 4a-b and Supplementary Fig. 12). While Ph+ ALL cells expressed p53 protein in all cases studied, in 6 of 10 cases Ph+ ALL cells had low expression of ARF (Fig. 4c). Despite low ARF expression, the Ph+ ALL cells underwent cell death upon Bach2 overexpression (Fig. 4b). While Bach2 is required for upregulation of both ARF and TP53, these findings indicate BACH2 induces cell death in Ph+ ALL cells independently of ARF.
Figure 4. BACH2 mediates PAX5-dependent tumor suppression in pre-B ALL through activation of p53.
(a) Patient-derived Ph+ ALL cells were propagated on OP9 stroma and transduced with vectors for Bach2GFP or GFP empty vectors (EV; a), and (b) GFP+ cells were monitored by flow cytometry. (c) Ph+ ALL cases were assayed for ARF and TP53 protein expression by Western blot using β-actin as loading control. (d-e) Flow cytometry for GFP in Cdkn2a-/- and Tp53-/- pre-B ALL cells (BCR-ABL1) transduced with Bach2GFP or EVGFP. (f) Transcriptional regulation of BACH2 by PAX5 assayed by the overexpression of dominant-negative PAX5-ETV6 fusion in 293T cells and Nalm6 pre-B ALL cells transduced with a BACH2 promoter luciferase reporter construct. (g-h) mRNA levels of BACH2 were increased by expression of wildtype PAX5 but decreased by dominant-negative PAX5-C20orf112 and PAX5-ETV6 fusion genes. (i) Gene expression values for three BACH2 probe sets in 160 cases of childhood pre-B ALL, 54 of which carry PAX5 deletions or somatic mutations (221234_s_at, 227173_s_at, 215907_at; COG P9906).
To test the requirements of Arf/p53 downstream of Bach2 in a genetic experiment, Cdkn2a-/- and Tp53-/- pre-B ALL cells were transduced with GFP-tagged Bach2 or empty vector controls (Figs. 4d-e). While Bach2 caused rapid elimination of Cdkn2a-/- and wildtype leukemia cells, the effect of Bach2 on Tp53-/- leukemia cells was delayed and substantially diminished (Fig. 4e and Supplementary Fig. 13). We conclude that p53 is an important effector of Bach2-mediated cell death, whereas Arf is dispensable. Our Arf/p53 Western blot analysis (Fig. 4c) agrees with previous findings that defects of p53 are rare in Ph+ ALL, whereas deletions of ARF are frequent (52%)15.
To identify potential defects in Bach2 function in human leukemia, leukemic blasts from 83 cases of Ph+ ALL (ECOG 2993) and pre-B cells from 12 normal bone marrow samples were analyzed with the HELP assay for promoter methylation (Log HpaII/MspI)16 of the BACH2 gene. The BACH2 promoter was more hypermethylated in Ph+ ALL cells as compared to normal human bone marrow pre-B cells (n=83; P=0.0026). In addition, sequence analysis of the BACH2 coding region of primary Ph+ ALL samples revealed BACH2 BTB domain missense mutations in 2 of 10 cases studied (Supplementary Fig. 14). Given that p53 is intact in most Ph+ ALLs, we conclude that inactivation of Bach2 upstream of p53 represents an important and non-redundant lesion.
Genetic lesions affecting PAX5 result in defective BACH2 expression in B cell lineage ALL
Our earlier experiments showed that forced expression of Pax5 induces upregulation of Bach2 at the pre-B cell receptor checkpoint (Fig. 1a-b). Recent studies demonstrated that deletions and rearrangements of the PAX5 gene resulting in dominant-negative fusion molecules occur in ~20% of cases of pre-B ALL15,19. We therefore tested whether loss of PAX5 function results in transcriptional inactivation of BACH2 in human pre-B ALL cells. While overexpression of wildtype PAX5 increases transcriptional activity of the BACH2 promoter as measured by a luciferase reporter construct, overexpression of the dominant-negative PAX5-ETV6 fusion suppressed BACH2 promoter activity (Fig. 4f). Likewise, mRNA levels of BACH2 were increased by expression of wildtype PAX5 but decreased by dominant-negative PAX5-C20orf112 and PAX5-ETV6 fusion genes19 (Figs. 4g-i). Next, we studied gene expression values for three BACH2 probe sets in 160 cases of childhood pre-B ALL. Compared to 54 cases carrying either PAX5 deletions or somatic mutations of PAX5, BACH2 mRNA levels were significantly higher in 106 wildtype PAX5 pre-B ALL cases (Fig. 4i). We conclude that BACH2 links PAX5 to activation of p53 and, hence, represents an important tumor suppressor in human pre-B ALL.
Loss of BACH2 expression represents an independent predictor of poor outcome in ALL patients
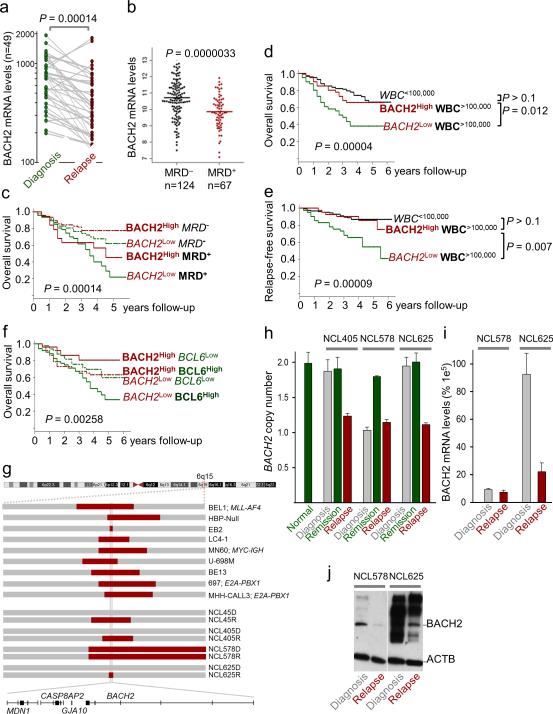
BACH2 levels in ALLs carrying E2A-PBX1 and TEL-AML1 gene rearrangements were similar to those in normal pre-B cells from healthy bone marrow donors. However, Bach2 mRNA levels were significantly lower in MLL-AF4- and Ph+ ALL subtypes (Supplementary Fig. 14). We also studied BACH2 mRNA levels in matched sample pairs from 49 patients that had a relapse of leukemia after initial successful treatment. Comparing matched sample pairs from the time of diagnosis and subsequent relapse of leukemia, BACH2 mRNA levels were substantially reduced in relapse samples (Fig. 5a and Supplementary Fig. 15). In addition, BACH2 expression levels at the time of diagnosis predicted detection of minimal residual disease (MRD) in a bone marrow biopsy taken on day 29 of treatment (Fig. 5b). Importantly, loss of BACH2 expression strongly correlated with poor overall outcome among 207 patients with childhood ALL (P9906 Children's Oncology Group). Patients with higher than median expression levels of BACH2 had a relapse-free survival (RFS) of 77% whereas those with lower than median BACH2 expression levels had a RFS of 47% (Supplementary Fig. 16b).
Figure 5. BACH2 is an independent predictor of poor clinical outcome in ALL patients.
(a) Comparison of BACH2 mRNA levels in matched sample pairs from 49 patients at the time of diagnosis and relapse of leukemia (pair-wise t-test). (b) Comparison of BACH2 mRNA levels in MRD- and MRD+ patients (MRD+; n=67; COG P9906 trial). Multivariate analyses for BACH2 mRNA levels as independent predictors of clinical outcome performed using established predictors of poor clinical outcome, namely, (c) detection of MRD and (d-e) high white blood cell counts (WBC>100,000/μl). (f) Studying BACH2 and BCL6 mRNA levels in a bivariate analysis identified subgroups of patients with particularly favorable (BACH2High-BCL6Low) and particularly poor (BACH2Low-BCL6High) outcome. P values are from logrank test. (g) High resolution SNP analysis identified BACH2 as a region of minimum deletion (RMD) in ALL cell lines (n=9) and primary cases (n=4) with 6q deletions. Longitudinal bone marrow samples at the time of diagnosis, remission and relapse of ALL were studied by (h) genomic PCR using the Taqman probe Hs01527432_cn, (i) qRT-PCR using the Taqman probe Hs00222364_m1 and (j) Western blot to assay allelic status and expression of BACH2.
To study whether Bach2 mRNA levels represent an independent predictor of clinical outcome, we performed multivariate analyses using three established predictors of poor clinical outcome as reference, namely MRD, high white blood cell counts (WBC>100,000/μl) and deletion of the IKZF1 tumor suppressor17. Studying high risk groups of patients on the basis of these three factors, we found that lower than median expression levels of BACH2 defined subgroups of patients with even worse clinical outcome. Lower than median expression levels of BACH2 at diagnosis predicted inferior clinical outcome for patients with positive MRD on day 29, patients with WBC>100,000/μl (Figs. 5c-e and Supplementary Fig. 16), and for patients with IKZF1-deletion (Supplementary Fig. 17).
Inverse relationship between BACH2 and BCL6 in clinical outcome prediction
Our data in Figs. 1 and 2 demonstrated that both Bach2 and Bcl6 bind Cdkn2a and Tp53 promoters but inversely regulate protein levels of Arf and p53 mRNA. Interestingly, the antagonistic function of BACH2 relative to BCL6 in the regulation of ARF/P53 and multiple other checkpoint genes is mirrored by their opposite association with clinical outcome. While higher than median mRNA levels of BACH2 at the time of diagnosis predict favorable clinical outcome, the opposite is true for ALL patients with BCL6 mRNA at higher than median levels (COG P9906). Studying BACH2 and BCL6 mRNA levels in a bivariate analysis identified subgroups of patients with particularly favorable (BACH2High-BCL6Low) and particularly poor (BACH2Low-BCL6High) outcome based on the ratio of BACH2:BCL6 mRNA levels (Fig. 5f).
BACH2 defines a region of minimal deletion at chromosome 6q15 and BACH2 deletion often occurs at the time of leukemia relapse
BACH2 is located at chromosome 6q15 and deletions of 6q encompassing 6q15 were found in 32% of cases of B cell lineage ALL18. We therefore studied B cell lineage ALL cell lines (n=9) and primary cases (n=4) with 6q deletions by high-resolution SNP analysis and found that BACH2 lies within a region of minimal deletion (RMD) on 6q (Fig. 5g). Studying bone marrow samples from primary ALL cases at the time of (i) diagnosis, (ii) clinical remission and (iii) relapse of leukemia, we found that deletion of the BACH2 locus was acquired at the time of leukemia relapse in 3 of 4 cases studied and was pre-existing in the 4th case (Figs. 5g-j).
Bach2-dependent gene expression changes overlap with Stat5- and Myc-regulated genes in Ph+ ALL
Ph+ ALL cells critically depend on tyrosine kinase signaling from (i) the BCR-ABL1 oncogene, (ii) downstream phosphorylation of Stat5 and (iii) transcriptional activation of Myc20. Interestingly, a global analysis of Bach2-dependent gene expression changes revealed that these changes significantly overlap with gene expression changes caused by (i) BCR-ABL1 kinase inhibition, inducible deletion of (ii) Stat5a/b and (iii) Myc (Supplementary Fig. 18). These findings suggest that Bach2-mediated gene expression changes oppose a transcriptional program of survival (Stat5) and proliferation (Myc) downstream of BCRABL1 in Ph+ ALL cells.
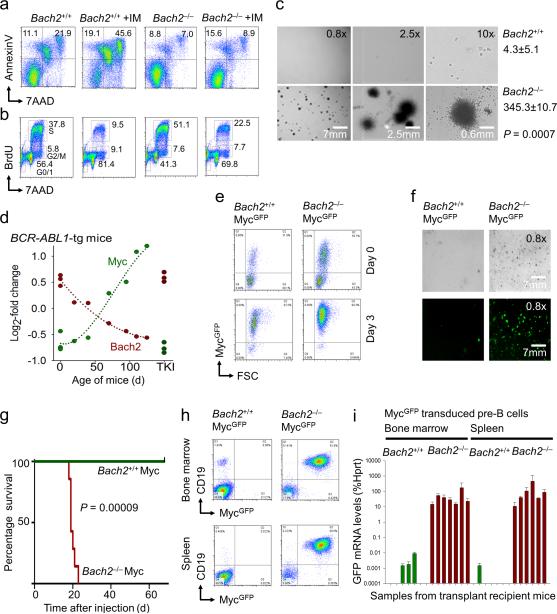
Consistent with this scenario, BCR-ABL1-transformed Bach2–/– pre-B ALL cells are significantly less sensitive to BCR-ABL1 tyrosine kinase inhibition (TKI; IM) than their wildtype counterparts (Figs. 6a-b). In the absence of Bach2, IM-induced apoptosis was reduced by 3-fold (Fig. 6a) and the fraction of cells that remained in cell cycle despite IM-treatment was increased by 2-fold (Fig. 6b). While BCR-ABL1-transformed Bach2+/+ cells are able to form colonies only after a lag phase of ~10 days, colony formation of Bach2–/– pre-B ALL cells was significantly accelerated (Fig. 6c).
Figure 6. Bach2 prevents leukemic transformation by Myc.
(a) Induction of cell death (Annexin V/7AAD staining) and (b) cell cycle arrest (BrdU staining; G0/1, S and G2/M phase distribution) in BCR-ABL1 transformed Bach2+/+ and Bach2-/- pre-B ALL cells before and after imatinib treatment. (c) Comparison of colony forming abilities of Bach2+/+ and Bach2-/- pre-B ALL cells plated in methylcellulose on day 7. Bach2+/+ pre-B ALL cells were able to form colonies after a lag phase of ~10 days (not shown). (d) To study progressive leukemic transformation in vivo, bone marrow pre-B cells were isolated from wildtype mice and BCR-ABL1-tg mice of various ages reflecting different stages of transformation. mRNA levels of Bach2 and Myc were plotted against age of mice [days], TKI denotes in vivo treatment of leukemic mice with the tyrosine kinase inhibitor Nilotinib. (e) Bach2+/+ and Bach2-/- IL-7-dependent pre-B cells were retrovirally transduced with MycGFP or EVGFP and transduction efficiency was monitored by flow cytometry on days 0 and 3. (f) Plating efficiency of MycGFP-transduced Bach2+/+ and Bach2-/- IL-7-dependent pre-B cells in methylcellulose was monitored by fluorescence microscopy. (g) Sorted MycGFP-transduced Bach2+/+ and Bach2-/- IL-7-dependent pre-B cells were also tested for their capacity to initiate fatal leukemia in NOD/SCID recipient mice and Kaplan-Meier analysis was performed to compare overall survival of transplant recipient mice in the two groups. (h) After 70 days, the mice in the surviving Bach2+/+ MycGFP cohort were sacrificed and analyzed together with mice receiving Bach2-/- MycGFP pre-B cells for CD19+ GFP+ cells in bone marrow and spleen (i.e. leukemia) by flow cytometry. (i) Minimal residual disease (MRD) qRT-PCR for leukemic cells infiltrating bone marrow or spleens was performed in triplicates for mice in each group using GFP-specific primers.
Expression of Bach2 and Myc are mutually exclusive during oncogenic transformation of pre-B cells
Overexpression of the Myc proto-oncogene frequently leads to increased proliferation and malignant transformation. However, Myc-driven proliferation can only be increased within certain limits, and Myc alone is not sufficient to cause malignant growth21-22. A number of failsafe mechanisms restrain Myc-induced proliferation and tumorigenesis23. These failsafe mechanisms are critical to terminate pre-malignant clones and, hence, represent a powerful safeguard against malignant transformation. Studying progressive transformation of pre-B cells in BCR-ABL1-tg mice in vivo, we found that mRNA levels of Bach2 and Myc are inversely regulated. In wildtype and young (<30 days, non-leukemic) BCR-ABL1-tg mice, mRNA levels of Bach2 are high whereas mRNA levels of Myc are low. Between ages of 30 and 120 days, BCR-ABL1-tg mice undergo progressive leukemic transformation24. Fully transformed leukemia cells express high levels of Myc and low levels of Bach2, whereas treatment of mice with TKI (Nilotinib) restored normal expression levels for Bach2 (high) and Myc (low; Fig. 6d). We conclude that expression of Bach2 and Myc are mutually exclusive during oncogenic transformation of pre-B cells.
Bach2-dependent activation of p53 represents a critical failsafe mechanism in Myc-mediated pre-B cell transformation
We next tested the hypothesis that Bach2 negatively regulates the susceptibility of pre-B cells to Myc-mediated transformation. In normal pre-B cells, excessively high levels of Myc result in activation of Arf/p53 and oncogene-induced senescence21-23. Retroviral overexpression of Myc was inefficient, induced cell death and caused significant toxicity in Bach2+/+ pre-B cells but not in Bach2–/– pre-B cells (Fig. 6e-f and Supplementary Figs 19 and 20). These findings indicate that Bach2 strongly reduces susceptibility of pre-B cells to malignant transformation by Myc.
Cell cycle analysis revealed that overexpression of Myc in Bach2+/+ pre-B cells increased the fraction of cells in S-phase to a maximum threshold of about 50%. However, Myc overexpression in Bach2–/– pre-B cells caused an increase of proliferation beyond this limit with about 70% of pre-B cells in S-phase (Supplementary Fig. 19b). Consistent with these findings, overexpression of Myc in Bach2+/+ pre-B cells resulted in activation of both Arf and p53. By contrast, p53 levels were very low in Bach2–/– pre-B cells and not increased by Myc-overexpression (Supplementary Fig. 19b). We conclude that in the absence of Bach2, p53-dependent failsafe mechanisms are not activated, obviating the requirement of additional genetic lesions for leukemic transformation.
Loss of Bach2 is sufficient to enable leukemic transformation by Myc
To confirm a critical role of Bach2 in Myc-induced failsafe control in an in vivo setting, IL7-dependent Bach2+/+ and Bach2–/– pre-B cells were transduced with retroviral Myc-GFP. Myc-GFP+ pre-B cells (Fig. 6e) were then injected into sublethally irradiated NSG recipient mice. While Bach2–/– MycGFP pre-B cells readily initiated fatal leukemia that killed the NSG recipient mice within 25 days, all NSG mice receiving Bach2+/+ MycGFP pre-B cells were still healthy at 70 days after injection (Fig. 6g). After 70 days, the mice in the surviving Bach2+/+ MycGFP cohort were sacrificed and analyzed together with mice receiving Bach2–/– MycGFP pre-B cells. In contrast to mice injected with Bach2–/– MycGFP pre-B cells, no CD19+ GFP+ cells (i.e. leukemia) were found in mice receiving Bach2+/+ MycGFP pre-B cells (Fig. 6h and Supplementary Fig. 21). Minimal residual disease (MRD) PCR for GFP revealed small numbers of persisting leukemia cells in most of the surviving mice at 70 days after injection. However, MRD levels were 4-5 log10 orders lower in the wildtype group as compared to mice injected with Bach2–/– MycGFP pre-B cells (Fig. 6i).
Discussion
This study identifies Bach2-mediated activation of p53 as a critical mediator of pre-B cell receptor checkpoint control. During normal B cell development, Bach2 determines stringency of negative selection of pre-B cell clones that failed to productively rearrange productively rearrange VH-DJH gene segments. In addition, Bach2-mediated activation of p53 represents a critical failsafe mechanism to terminate pre-malignant pre-B cell clones before they can complete malignant transformation. Thereby, Bach2 limits pre-B cell proliferation and oncogene signaling through activation of p53. Both in normal pre-B cells and in pre-B ALL, Bach2-mediated activation of p53 is opposed by Bcl6, a potent transcriptional repressor of p53. While inactivation of TP53 in ALL is rare, BACH2 is frequently inactivated in established pre-B ALL clones or at the time of leukemia relapse. Consistent with their opposing role in TP53-regulation and checkpoint control, the ratio of BACH2 (favorable) and BCL6 (poor) expression levels also represents a strong predictor of clinical outcome in patients with ALL (Fig. 5f). While BACH2 function cannot be reinstated in patients with ALL carrying BACH2 deletions, we propose that pharmacological inhibition of BCL6 (e.g. with RI-BPI) will restore the balance of BACH2/BCL6-mediated checkpoint control and relieve BCL6-mediated transcriptional repression of p53 (Supplementary Fig. 22).
Methods
Methods and any associated references are available in the online version of this paper. Sequence data are available from EMBL/GenBank under accession numbers provided in Supplementary Table 7. Microarray data are available from GEO under accession numbers provided in Supplementary Table 8.
Supplementary Material
Acknowledgements
We would like to thank Hilda Ye (Albert Einstein College of Medicine, Bronx, NY) for sharing BCL6-/- mice and wildtype controls with us, Lothar Hennighausen (NIDDK, Bethesda, MD) for Stat5fl/fl mice. Samples used in this research include those obtained from the Newcastle Haematology Biobank-http://www.ncl.ac.uk/nbb/collections/nhb/index.htm). This work is supported by grants from the NIH/NCI through R01CA137060, R01CA139032, R01CA157644, R01CA169458 and (to M.M.), Translational Research Program grants from the Leukemia and Lymphoma Society (grants 6132-09 and 6097-10), a Leukemia and Lymphoma Society SCOR (grant 7005-11, PI Brian J Druker), the William Laurence and Blanche Hughes Foundation and a Stand Up To Cancer-American Association for Cancer Research Innovative Research Grant (IRG00909, to M.M.), the California Institute for Regenerative Medicine (CIRM; TR2-01816 to MM) and Leukaemia and Lymphoma Research (VR and AGH). A.M. and M.M. are Scholars of the Leukemia and Lymphoma Society. VR is a Leukaemia and Lymphoma Research Bennett Fellow.
Abbreviations
- ChIP
chromatin immunoprecipitation
- RFS
relapse-free survival
- EV
empty vector
- HR
hazard ratio
- IL
interleukin
- OIS
oncogene-induced senescence
- ROS
reactive oxygen species
- RSS
recombination signal sequence
- ALL
acute lymphoblastic leukemia
Footnotes
The authors have no conflicting financial interests.
References
- 1.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–95. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–71. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 3.Osmond DG. Proliferation kinetics and the lifespan of B cells in central and peripheral lymphoid organs. Curr Opin Immunol. 1991;3:179–85. doi: 10.1016/0952-7915(91)90047-5. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi N, Melchers F. Lambda 5, a new light-chain-related locus selectively expressed in pre-B lymphocytes. Nature. 1986;324:579–82. doi: 10.1038/324579a0. [DOI] [PubMed] [Google Scholar]
- 5.Kitamura D, Roes J, Kühn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–6. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 6.Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, Schuh W, Jäck HM, Hurtz C, Ramezani-Rad P, Herzog S, Jumaa H, Koeffler HP, de Alborán IM, Melnick AM, Ye BH, Müschen M. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahar R, Ramezani-Rad P, Mossner M, Duy C, Cerchietti L, Geng H, Dovat S, Jumaa H, Ye BH, Melnick A, Müschen M. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood. 2011;118:4174–8. doi: 10.1182/blood-2011-01-331181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Zelm MC, van der Burg M, de Ridder D, Barendregt BH, de Haas EF, Reinders MJ, Lankester AC, Révész T, Staal FJ, van Dongen JJ. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol. 2005;175:5912–22. doi: 10.4049/jimmunol.175.9.5912. [DOI] [PubMed] [Google Scholar]
- 9.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the Ig heavy-chain gene. Genes Dev. 2004;18:411–22. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, Klemm L, Kweon SM, Nahar R, Braig M, Park E, Kim YM, Hofmann WK, Jumaa H, Koeffler HP, Yu JJ, Heisterkamp N, Graeber TG, Wu H, Ye BH, Melnick A, Müschen M. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouault JP, Falette N, Guéhenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, Shaw P, Berger R, Samarut J, Magaud JP, Ozturk M, Samarut C, Puisieux A. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 12.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4:31–7. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 13.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehlich A, Martin V, Müller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr Biol. 1994;4:573–83. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 15.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 16.Geng H, Brennan S, Milne TA, Chen WY, Li Y, Hurtz C, Kweon SM, Zickl L, Shojaee S, Neuberg D, Huang C, Biswas D, Xin Y, Racevskis J, Ketterling RP, Luger SM, Lazarus H, Tallman MS, Rowe JM, Litzow MR, Guzman ML, Allis CD, Roeder RG, Müschen M, Paietta E, Elemento O, Melnick AM. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012;2:1004–23. doi: 10.1158/2159-8290.CD-12-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, Harvey RC, Chen IM, Clifford RJ, Carroll WL, Reaman G, Bowman WP, Devidas M, Gerhard DS, Yang W, Relling MV, Shurtleff SA, Campana D, Borowitz MJ, Pui CH, Smith M, Hunger SP, Willman CL, Downing JR, Children's Oncology Group Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merup M, Moreno TC, Heyman M, Rönnberg K, Grandér D, Detlofsson R, Rasool O, Liu Y, Söderhäll S, Juliusson G, Gahrton G, Einhorn S. 6q deletions in acute lymphoblastic leukemia and non-Hodgkin's lymphomas. Blood. 1998;91:3397–400. [PubMed] [Google Scholar]
- 19.Kawamata N, Pennella MA, Woo JL, Berk AJ, Koeffler HP. Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene. 2012;31:966–77. doi: 10.1038/onc.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA, Fajmann S, Grebien F, Warsch W, Stengl G, Hennighausen L, Poli V, Beug H, Moriggl R, Sexl V. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2:98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drayton S, Rowe J, Jones R, Vatcheva R, Cuthbert-Heavens D, Marshall J, Fried M, Peters G. Tumor suppressor p16INK4a determines sensitivity of human cells to transformation by cooperating cellular oncogenes. Cancer Cell. 2003;4:301–310. doi: 10.1016/s1535-6108(03)00242-3. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–7. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zindy, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signalling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trageser D, Iacobucci I, Nahar R, Duy C, von Levetzow G, Klemm L, Park E, Schuh W, Gruber T, Herzog S, Kim YM, Hofmann WK, Li A, Jäck HM, Groffen J, Martinelli G, Heisterkamp N, Jumaa H, Müschen M. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J Exp Med. 2009;206:1739–1753. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.