Abstract
Diacylglycerols (DAGs) are important intermediates of lipid metabolism and cellular signaling. It is well known that the mass levels of DAG are altered under disease states. Therefore, quantitative analysis of DAGs in biological samples can provide critical information to uncover underlying mechanisms of various cellular functional disorders. Although great efforts on the analysis of individual DAG species have recently been made by utilizing mass spectrometry with or without derivatization, cost effective and high throughput methodology for identification and quantification of all DAG species including regioisomers, particularly in an approach of shotgun lipidomics, are still missing. Herein, we described a novel method for directly identifying and quantifying DAG species including regioisomers present in lipid extracts of biological samples after facile one-step derivatization with dimethylglycine based on the principles of multi-dimensional mass spectrometry-based shotgun lipidomics. The established method provided substantial sensitivity (low limit of quantification at amol/µl), high specificity, and broad linear dynamics range (2,500 folds) without matrix effects. By exploiting this novel method, we revealed a 16-fold increase of total DAG mass in the livers of ob/ob mice compared to their wild type controls at 4 months of age (an insulin-resistant state) vs. a 5-fold difference between 3-month old mice (with normal insulin). These results demonstrated the importance and power of the method for studying biochemical mechanisms underpinning disease states.
Keywords: brain injury, diacylglycerol, dimethylglycine derivatization, lipid metabolism, mass spectrometry, obesity, shotgun lipidomics
Diacylglycerol (DAG) species are glycerol derivatives in which two hydroxyl groups are esterified by fatty acyl (FA) chains. Depending on the position of the two FA chains, they can exist in two stereochemical forms: sn-1,2- and sn-1,3-DAG. The 1,2-DAG species are the cellular intermediates in the biosynthesis and degradation of triacylglycerols, glycerophospholipids, and glyceroglycolipids1. The 1,2-DAG species play important roles in cellular functions, as they serve as second messengers in many cellular processes2. 1,2-DAGs accumulate transiently in membranes where they bind to particular proteins via strong hydrophobic interactions. The accumulated 1,2-DAGs then cause changes in the physical properties of the bilayer, thereby possibly facilitating membrane fusion/fission events, affecting intracellular vesicular trafficking, and influencing the functions of ion channels and pumps3, 4. It has been reported that increased amounts of 1,2-DAG species are associated with numerous disease states, including cardiac hypertrophy5, colonic neoplasia6, diabetes7, obesity8, brain injury9, lipotoxicity10, etc. This suggests that DAGs may play roles in the pathogenesis of these complications and may be used as biomarkers for diagnosis and therapeutic intervention of those diseases. The 1,3-DAG species largely result from the chemical isomerization of 1,2-DAG species11. Ingested 1,3-DAGs are metabolized through different pathways, which is not part of the triacylglycerol pool present in chylomicrons12.
Quantification of DAG species conventionally requires the use of radioactive materials or successive chromatographic separation which is labor intensive and results in the loss of analytes13–15. Alternatively, DAG can be derivatized with chemicals to increase their volatility for gas chromatographic analysis or with chromophores to enhance HPLC detection16, 17. However, DAG species are hardly resolved, which severely affects quantitative analysis. Moreover, these methods only have a limited dynamic range. In addition, neither of these techniques could readily discern the 1,2- and 1,3-DAG regioisomers. Electrospray ionization mass spectrometric (ESI-MS) analysis of DAG species after TLC purification and derivation with N-chlorobetainyl chloride has been developed18. Unfortunately, this method is incapable of identifying acyl chains of DAG species or resolving 1,2- and 1,3-DAG regioisomers. N,N-dimethylglycine (DMG) was used to esterify the hydroxyl group of alcohols including DAG species for faster and more sensitive characterization of those compounds19. However, the information about acyl chains and regioisomers as well as the mass contents of the analyzed DAG species were not determined in that work. High mass resolution MS has been applied for the analysis of DAG species in which quantitative analysis has not been demonstrated and 1,2- and 1,3-DAG regioisomers cannot been distinguished20. LC-MS (including ESI or atmospheric pressure chemical ionization) after pre-fractionation has recently been applied for analysis of DAG species with or without derivatization21–23.
Herein, by applying the principles of multi-dimensional MS-based shotgun lipidomics (MDMS-SL)24–26 and employing an improved method to derivatize the hydroxyl group27, we established an accurate, highly efficient, sensitive, and reproducible method for analysis of DAG species directly from biological lipid extracts. The established method allowed us to identify and quantify all of cellular DAG species, including 1,2- and 1,3-DAG isomers. We believe that this method should extend the MDMS-SL platform to a powerful tool for quantitative analysis of DAG species.
MATERIALS AND METHODS
Materials
All of 1,2-DAG species were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) and 1,3-DAG species were from Nu-Chek Prep, Inc. (Elysian, MN). All these species were used without further purification. Stock solutions of these DAG species were separately prepared in either CHCl3/MeOH (1/1, v/v) or CHCl3 with a concentration of 1 mg/µl, and stored at −20 °C. The derivatization reagents, DMG hydrochloride, N-(3-Dimethylaminopropyl)-N’-ethylcarbodiimide (EDC) hydrochloride, and dimethylaminopyridine (DMAP) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). All solvents used for lipid extraction and sample preparation and for MS analysis were obtained from Burdick and Jackson (Muskegon, MI), except formic acid obtained from Thermo Fisher Scientific, Inc. (Fair Lawn, NJ). All other chemicals were from Sigma-Aldrich.
Animal Experiments
Obese homozygous (ob/ob) and wild type (WT, C57BL/6J) male mice were purchased from the Jackson Laboratories (Bar Harbor, ME), and kept separately under a temperature- (23 °C) and lighting- (12 h of light and dark) controlled house with free access to a standard chow (Teklad Diet 2916, Harlan Laboratories, Indianapolis, IN) and water. At the 3 or 4 mo of age, mice were euthanized by asphyxiation with CO2 followed by cervical dislocation. The livers were excised quickly and perfused with ice-cold PBS to remove blood, blotted with Kimwipes (Kimberly-Clark, Roswell, GA) to remove excess buffer, and then immediately freeze-clamped at the temperature of liquid N2. All of tissue samples were stored at −80 °C until lipid extraction and analysis. All animal procedures were performed in accordance with the “Guide for the Care and Use of Laboratory Animals” (National Research Council of National Academies, 2010) and were approved by the Institutional Animal Care and Use Committee at the Sanford-Burnham Medical Research Institute.
Preparation of Lipid Extracts
Liver wafers were pulverized into a fine powder by a stainless steel Bio-pulverizer (12 wells, capacity 10–100 mg per well, BioSpec Products, Bartlesville, OK) at the temperature of liquid N2. Tissue fine powders (~ 20 mg) were weighed from each liver and homogenized in 10× diluted PBS by using 2.0-ml cryogenic vials (Corning Life Sciences, Tewksbury, MA). Protein assay on the homogenates was performed by using a bicinchoninic acid protein assay kit (Thermo Scientific, Rockford, IL) with bovine serum albumin as standards. All determined lipid levels were normalized to the protein content of individual samples.
Individual homogenate of the liver samples was accurately transferred into a disposable glass culture test tube. Internal standard (1,3-di15:0 DAG) for quantitation of DAG species was added prior to lipid extraction. This DAG species represents < 0.1% of the endogenous DAG mass levels as predetermined by ESI-MS. Lipid extraction was performed by using a modified Bligh and Dyer procedure as described28. Each lipid extract was resuspended in a volume of 100 µl of CHCl3/MeOH (1:1, v/v) per mg of tissue protein, flushed with N2, capped, and stored at −20 °C.
Derivatization of Diacylglycerol with DMG
A certain amount of extract (e.g., equivalent to 0.2 mg of tissue protein content) was transferred to a disposable conical tube, and evaporated completely to dryness under a N2 stream. Two µl of DMG (0.125 M) and DMAP (0.5 M) and 2 µl of EDC (0.25 M) (all in ultra-dry CHCl3) were added into the tube, respectively. After vortexing for 20 s and then centrifuging at 2700 rpm for 1 min, the reaction vessel was flushed with dry N2, capped, and incubated at 45 °C for 90 min or as indicated. The derivatization was terminated by adding 3 ml CHCl3/MeOH (1:1, v/v) and 1.5 ml NH4OH (25 mM) to each of the sample tubes and vortexing for 1 min. The derivative products were extracted using the modified Bligh-Dyer procedure. The CHCl3 layer was collected and the solvent was evaporated to dryness. The residue was re-extracted by using the modified Bligh and Dyer procedure to remove salt content. The extracts were resuspended to a volume of 75 µl CHCl3/MeOH (1:1, v/v), flushed with N2, capped, and stored at −20 °C for MS-ESI analysis.
MS Analysis of DAG derivatives
MS analysis of DAGs was performed on a QqQ mass spectrometer (Thermo TSQ Vantage, San Jose, CA) equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY)29. An ionization voltage of 1.2 kV and gas pressure of 2.0 psi on the NanoMate apparatus were employed for the MS analyses. The device was controlled by Chipsoft 8.3.1 software. All MS or MS/MS analyses were operated under Xcalibur software as described30. Typically, a 1-min period of signal averaging from 1 s/scan in the profile mode was used for each MS spectrum and a 4-min period of signal averaging from 1 s/scan was employed for each MS/MS spectrum. For MS/MS analysis in the neutral-loss scan (NLS), precursor-ion scan (PIS), or product ion mode, collision gas (argon) pressure was set at 1.0 mT or specified, and collision energy was specified. A mass resolution setting of 0.7 Th for Q1 and Q3 was used for both MS and MS/MS analyses. Before ESI analysis in the positive-ion mode through direct infusion, individual DMG-derivatized lipid solution (see above) was further diluted to a final concentration of 100–1000 fmol/µl of the internal standard with 50% of CHCl3/MeOH/isopropanol (1/2/4, v/v/v) plus 50% of CHCl3/MeOH/formic acid (50/50/0.2, v/v/v) for protonated MS analysis or with CHCl3/MeOH/isopropanol (1/2/4, v/v/v) containing 0.025% (v/v) LiOH-saturated MeOH solution for MS analysis of lithiated DMG-DAG.
RESULTS AND DISCUSSION
Derivatization of DAG Species with DMG
Unlike sphingolipids or phospholipids, DAG species do not possess fixed charge moieties, and they show significantly low ionization efficiency by ESI. Through introduction of a fixed charge site to DAGs, they become less volatile and more efficient to be ionized by ESI. Hence, we introduced a tertiary amine to DAG molecules through a facile esterification reaction with DMG (Scheme S1). The tertiary amine in the DAG derivatives can be readily protonated under acidic conditions or adducted with alkali ions, offering a polar group to enhance ionization efficiency in positive-ion ESI-MS.
To examine whether the derivatization efficiency of DAG species depends on the chemical properties of individual DAG species including FA chain length, double bond (DB) numbers, or stereochemical forms (sn-1,2- and 1,3-DAG), we prepared a DAG standard mixture containing 2 nmol of each of 1,2-di14:0, 1,3-di15:0, 1,3-di16:1, 1,2-di16:0, 1-16:0-2-18:1, 1-18:0-2-18:2, 1,3-di18:0, 1-18:0-2-20:4, 1,3-di20:4, 1-18:0-2-22:6, 1,3-di20:2, 1,3-di20:1, 1,3-di21:0, 1,3-di22:0. This DAG mixture was derivatized and the derivatized solution was analyzed by ESI-MS after direct infusion with a concentration of ~1 pmol/µl for each species. The mass spectrum acquired under acidic conditions (Figure S1A) displayed fourteen intense ion peaks at m/z at 598, 626, 650, 654, 680, 706, 710, 730, 750, 754, 758, 762, 794, and 822. These ions showed virtually equal intensities within experimental errors after correction for differential 13C isotope distribution as previously described31 (see discussion below). The results confirmed that introduction of a charge site to DAG species yielded similar ion responses in ESI-MS regardless of the FA chain length, saturation, or stereochemical structures. This is consistent with our previous findings that the head group for polar lipids plays the major role in lipid ionization by ESI-MS and that different molecular species of a polar lipid class possess essentially identical ionization efficiency in the low concentration region25, 32.
In the current study, we also optimized the ratios of DMG vs. DAG for derivatization. Since the hydroxyl groups are common in lipid extracts of biological samples, a large amount of DMG should favor for complete derivatization of DAGs. The amounts of DAGs and other hydroxyl-containing species present in biological samples are varied in a board range that requires a high ratio of DMG to DAG to warrant a complete derivatization of DAG species. However, a very high ratio of DMG to DAG leads to ion suppression resulting from the coexistence of non-reacted DMG and other resultant by-products. To optimize this ratio, we derivatized a DAG standard mixture and a lipid extract from mouse liver, respectively, with varied amounts of DMG from 10 to 10000 times of DAG masses. It was found that when the ratio of DMG/DAG was greater than 1000, the mass spectra showed reduced ionization stability and efficiency. Moreover, the mass spectra showed very strong by-product peaks at m/z 645 and 659, which severely suppressed the ionization of DMG-DAG. It was found that an amount of DMG at the ratio of 20 to 30 times of the total DAG levels gave the best signals of DMG-DAG species with relatively low background noise. This amount of DMG is optimal for the amount of lipids corresponding to 0.2 mg of tissue protein content (i.e., approximately up to 0.3 µmol of total lipids) extracted from the majority of biological samples.
The derivatization time and the reaction temperature were also investigated to optimize the derivatization of DAG species. A mixture of DAG standards with derivative reagents was incubated at the room temperature (23.5), 45, or 70 °C for 2 hrs. The survey mass spectra of the DAG derivatives from the reactions at the room temperature showed lower intensities than the ones from 45 or 70 °C. However, the spectra of the derivatives from the reactions at 70 °C showed many intensive by-product peaks. Therefore, a reaction temperature of 45 °C was selected for the derivative reaction. In another experiment, the reactant mixtures were incubated at 45 °C for 0.75, 1.5, 3, 6, and 15 hrs. The DAG derivatives from the various reaction conditions showed no significant differences in the ESI-MS or MS/MS spectra (data not shown). This result indicated that the dervative reaction was fast and that the reaction conditions examined would not increase the acyl chain migration after derivatization.
Characterization of DMG-DAG species by ESI-MS/MS
ESI-MS/MS analysis of protonated DMG-DAG in the product-ion mode after collision-induced dissociation (CID) (Figure S1B and C) displayed only a predominant fragment ion peak corresponding to the neutral loss of 103 Da (loss of DMG) from the polar group of the derivatives. Moreover, the intensity ratios of this fragment ion vs. the molecular ion in the mass spectra were essentially identical (Figure S1B and C). These observations indicate that NLS of 103 Da from the protonated DMG-DAG could be used for identification and quantification of DMG-DAG species. Unfortunately, the product-ion ESI-MS/MS analysis of protonated DMG-DAG did not provide any information about FA chains and regioisomers of DAG species.
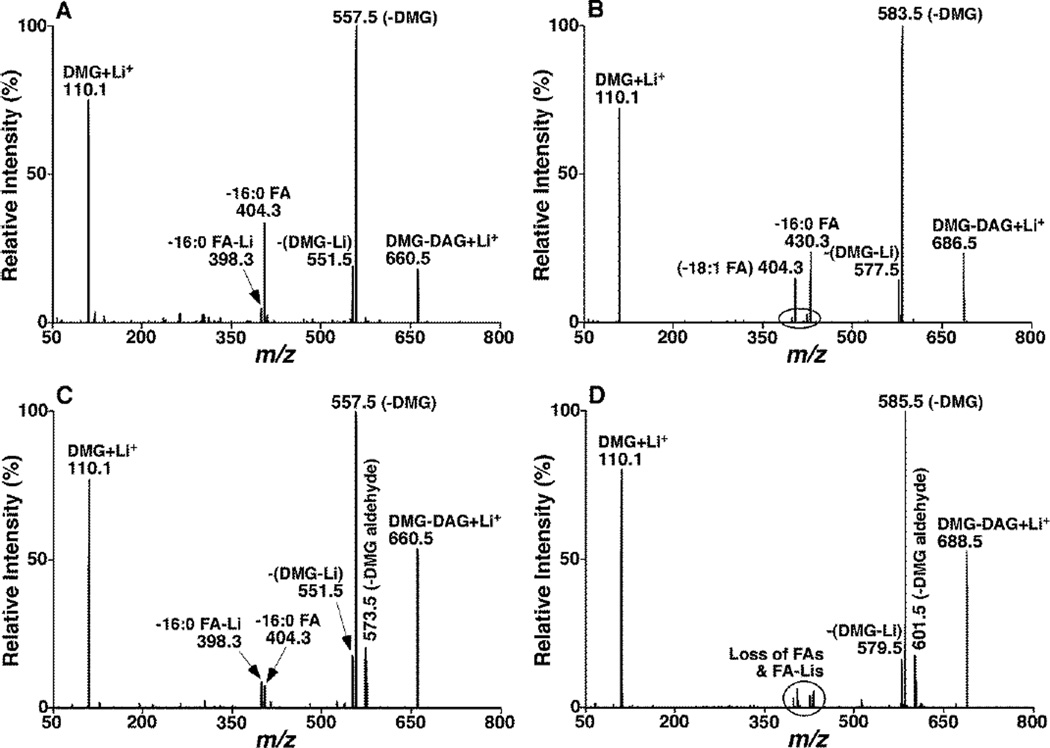
To explore the identification of DAG identities including FA chains and regioisomers, ESI-MS/MS analysis of lithiated DMG-DAG in the product-ion mode after CID was performed, since previous studies have demonstrated that ESI-MS/MS analysis of lithiated lipid species gives great structural information for identification33. We found that there were numerous abundant, informative, and characteristic peaks in the product ion mass spectra of lithiated DMG-DAG (Figure 1). These product ions included an abundant fragment ion at m/z 110 corresponding to lithiated DMG, an abundant product ion corresponding to the neutral loss of 103 Da (i.e., DMG), and a moderate abundance fragment ion corresponding to the neutral loss of 109 Da (i.e., DMG Li-salt) from all examined DMG-DAG species including sn-1,2- and 1,3-DAG species (Figure 1). For example, lithiated molecular ions of both 1,2- and 1,3-di16:0 DAG-DMG at m/z 660 lost DMG head group and yielded product ions at m/z 557 after CID (Figure 1A and C, respectively); lithiated 1-16:0-2-18:1 DMG-DAG at m/z 686 produced a fragment ion at m/z 583 after CID (Figure 1B); a product ion at m/z 585 resulted from the loss of DMG from lithiated 1-16:0-3-18:0 DMG-DAG at m/z 688 (Figure 1D). From these features of DMG-DAG species, it can be concluded that both NLS of 103 Da and PIS of 110 Th can be used to sensitively detect the molecular ions of lithiated DMG-DAG species.
Figure 1.
Representative product ion MS analyses of lithiated DMG-DAG species. Product-ion analyses of 1,2-di16:0 (A), 1-16:0-2-18:1 DAG (B), 1,3-di16:0 (C), and 1-16:0-3-18:0 (D) DMG-DAG species were performed after selection of the corresponding molecular ions by Q1, collision activation in Q2 with collision energy of 35 eV and collision gas pressure of 1.0 mTorr, and product-ion detection by Q3. The majority of the fragment ions in the spectra were assigned.
From the comparison between the product ion mass spectra of lithiated sn-1,2- and 1,3-DMG-DAG species (Figure 1), it was found that the mass spectra of 1,3-DMG-DAG displayed an additional moderate fragment ion peak corresponding to the neutral loss of 87 Da from the molecular ions (i.e., the loss of DMG as an aldehyde). For example, the fragment peak at m/z 573 in the 1,3-di16:0 DMG-DAG spectrum (Figure 1C) was not shown in the 1,2-counterpart (Figure 1A). Identically, the fragment ion peak resulting from the neutral loss of 87 Da did not exist in product-ion mass spectrum of lithiated 1-16:0-2-18:1 DMG-DAG (Figure 1B); whereas such a peak was present in product-ion mass spectrum of lithiated 1-16:0-3-18:0 DMG-DAG (Figure 1D). Collectively, these data indicate that the neutral loss of 87 Da is specific to lithiated 1,3-DMG-DAG species and NLS of 87 Da could be used to discriminate 1,3-DAG from 1,2-DAG isomers.
The product ion mass spectra of lithiated DMG-DAG species also showed a pair of fragment ion peaks corresponding to the loss of an individual FA chain as an acid or lithium salt. For example, the product ion peaks at m/z 398 and 404 yielded from the loss of 16:0 FA lithium salt and 16:0 FA, respectively, from lithiated 1,2- or 1,3-di16:0 DMG-DAG (Figure 1A and C). Similarly, we also detected the product ion peaks corresponding to the loss of 16:0 and 18:1 or 18:0 FAs and their lithium salts in the product ion spectra of lithiated 1-16:0-2-18:1 and 1-16:0-3-18:0 DMG-DAG (Figure 1B and D). It should be recognized that the intensities of the fragment ions corresponding to the loss of FA-Li salt from 1,2-DAG ions were in much lower abundance than that resulted from the loss of FA from the molecular ions. However, this pair of fragment ions was present in similar intensities in product-ion mass spectra of lithiated 1,3-DMG-DAG species (Figure 1C and D). This difference between the 1,2- and 1,3-DAG species is due to the differential fragmentation pathways as previously described33. Accordingly, the acyl chain information of DAG species could be identified by the neutral loss of its corresponding FA or FA lithium salt. From the ratio of the pair of product ion peaks corresponding to the loss of FA and its lithium salt, 1,2-DAG could also readily be discriminated from its 1,3-isomers.
Moreover, product-ion mass spectral analyses of lithiated 1,2-DMG-DAG species showed that the intensity of the fragment ion corresponding to the loss of sn-1 FA is higher than that of sn-2 FA. For example, the peak at m/z 430 corresponding to the loss of 16:0 FA from the sn-1 position was more intensive than that at m/z 404 corresponding to the loss of 18:1 FA from the sn-2 position (Figure 1B). This difference is also consistent with the differential fragmentation patterns resulted from different positions as previously described33. Hence, this difference could be used to identify the FA positions in 1,2-DAG species. It should be pointed out that the different location of FA chains in 1,3-DAG species could result in different chiral isomers. However, product-ion MS analysis is unable to distinguish these isomers.
Taken together, characterization of product-ion mass spectra of lithiated DMG-DAG species indicated that the identities of DAG species could be definitively determined from the lithiated DMG-DAG. The identification includes (1) distinguishing DAG species from other classes of lipids by utilizing NLS103 and PIS110 within mass range of potential DAG species (e.g., m/z 500–850); (2) identifying the DAG acyl chains mostly by using NLS of their corresponding FA (NLS of their FA lithium salt for 1,2-DAG is weak, which was only used for validation of the acyl chain information); (3) resolving 1,2- and 1,3-DAG isomers by using NLS87 as well as the peak intensity ratio of a paired fragment ions resulted from the loss of a FA chain as either FA or FA-Li salt (this ratio is >> 1 for 1,2-DAG whereas ~1 for 1,3-DAG); and (4) determination of the location of FA chains in 1,2-DAG species by exploiting the peak intensity ratio of fragment ions resulted from the loss of the two distinct FA chains (loss of sn-1 FA > loss of sn-2 FA).
Quantification of DAG species by ESI-MS/MS
We first examined the ionization efficiencies of protonated DMG-DAG species by using ESI-MS, which is critical for optimizing the conditions for ESI-MS/MS analysis. ESI-MS analysis of DMG-DAG mixtures at the concentration of ~1 pmol/µl for each species was performed in the presence of 0.1% of formic acid. MS analysis of the equimolar mixture of 14 DAG species showed the protonated DAG peaks in nearly equal intensities (Figure S1A) after considering the differential 13C isotope distribution, which can be corrected as previously described31. The small variation among those peak intensities of the derivatized DAG species after de-isotoping correction likely came from the small and random variation in the concentrations of stock solutions among individual DAG species in the mixture since the ion peak intensities of these species did not show any trends correlated with either the variation of the FA chain lengths or unsaturation in these species. Considering the previous findings that ionization efficiencies of individual molecular species of a polar lipid class are independent of the physical natures of individual species32, we believed that the ionization efficiencies of protonated DMG-DAG species are identical within experimental error (~10%). Similar results were also obtained from the MS analysis of lithiated DMG-DAG in the presence of LiOH (data not shown). These results suggest that the derivatized DAG species can be quantified by using ESI-MS with one internal standard after correction for differential 13C isotope distribution and under a diluted lipid concentration to avoid lipid aggregation as previously described24.
Unfortunately, in biological samples, DAG species are generally present in very low abundance. Moreover, the DMG-DAG ions may overlap or co-exist with those from other lipid classes within the mass region under the experimental condition. These facts, particularly, when the overlapping occurs with the high abundance lipid classes (e.g., phosphatidylcholine), make the quantification of DMG-DAG difficult by utilizing survey scan spectra despite the enhanced ionization after derivatization unless a column separation is employed. Therefore, it is necessary to explore the possibility that MS/MS analysis, which is widely employed in lipidomics due to its enhanced sensitivity and specificity, can be used to quantify the derivatized DAG species.
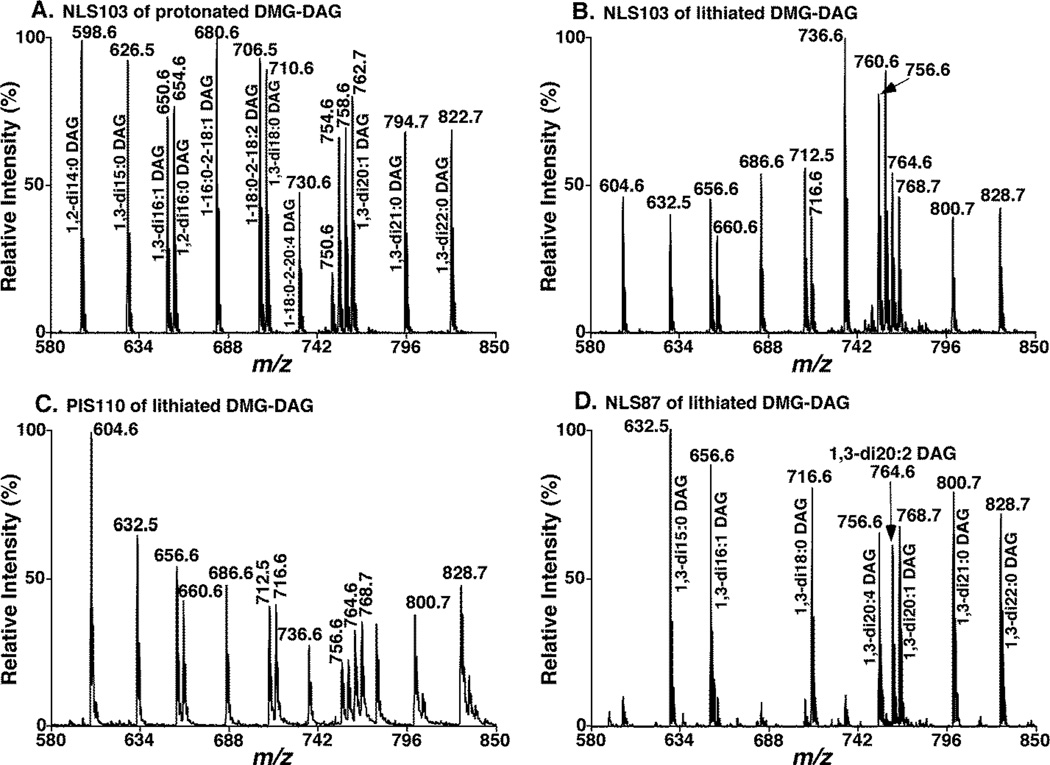
To investigate if MS/MS analysis could be used to quantify the DMG-DAG species, both NLS103 and PIS110 of the same DAG mixture as aforementioned were performed with ramping collision energy under acidic condition or in the presence of LiOH. We found that all of MS/MS spectra including NLS103 of protonated DMG-DAG, and NLS103 and PIS110 of lithiated DMG-DAG, yielded sensitive and specific detection of DMG-DAG ions (Figure 2). At the infusion concentration lower than 0.1 fmol/µl, the spectra from NLS103 of lithiated DAG species showed higher variation and more noisy baselines than the one from protonated adducts. Therefore, we found that NLS103 of protonated DMG-DAG species showed better reproducibility and improved detect limits comparing to that obtained from lithium adducts (Figure S2). We also found that NLS103 spectra of protonated DMG-DAG displayed nearly equal ion intensities of these species except those DAG species containing one or two polyunsaturated FA chains (the number of double bonds (DB) ≥ 3) for which significantly lower intensities were obtained due to the differential fragmentations (Figure 2A). For example, the derivatives of 1-18:0-2-20:4 DAG at m/z 730, 1,3-di20:4 DAG at m/z 750, and 1-18:0-2-22:6 DAG at m/z 754 showed much lower intensities than others. In contrast to NLS103 analysis of protonated DMG-DAG, NLS103 spectra of lithiated DMG-DAG showed the enhanced ion intensities of those DAG species with the DB number ≥ 3 in either of their FA chains (e.g., 1-18:0-2-20:4 DAG at m/z 736, 1,3-di20:4 DAG at m/z 756, and 1-18:0-2-22:6 DAG at m/z 760) relative to the other DAG ions (Figure 2B). We also found that this enhancement did not depend on the concentration of the species when the concentration of the analyzed DMG-DAG mixture was in the range from 0.1 to 1,000 fmol/µl (Figure S2). Based on the peak intensity ratios of DAG species containing polyunsaturated FA chain(s) (DB number ≥ 3) vs. the selected internal standard (i.e., 1,3-di15:0 DAG) under various concentrations, an enhancement factor of 2.1 was determined (Figure S2A).
Figure 2.
Representative MS/MS analysis of DMG-DAG species in the neutral loss and precursor-ion scan modes. Neutral loss scan of DMG (103 Da) (NLS103) from an equimolar mixture of DMG-DAG species at 1 pmol/µl each in the presence of 0.1% formic acid was acquired at collision energy of 27 eV (A), indicating protonated DMG-DAG species. NLS103 (B) and PIS110 (C) of the equimolar DMG-DAG mixture in the presence of LiOH acquired at collision energy of 39 and 76 eV, and collision gas pressure of 1.0 and 0.6 mTorr, respectively, displayed lithiated DMG-DAG species. NLS87 (D) of the equimolar DMG-DAG mixture in the presence of LiOH acquired at collision energy of 39 eV identified 1,3-DMG-DAG species.
Taken together from these determinations, the DAG species could be assessed within experimental error (~10%) by combining NLS103 analyses of protonated and lithiated DMG-DAG because of the enhanced sensitivity for those DAG species with polyunsaturated FA chains (DB number ≥ 3). The analysis of protonated species was used to determine the DAG species with saturated FA chains or DB number < 3 in their FA chains, whereas the analysis of lithiated species was employed for determining the species containing polyunsaturated FA chain(s) (DB number ≥ 3) with an enhanced factor of 2.1 in comparison to the selected internal standard. Another option is to employ an additional internal standard that contains polyunsaturated FA chain (e.g., 1-stearoyl-2-d8-arachidonoyl-sn-glycerol) for polyunsaturated FA-containing DAG species (DB number ≥ 3) quantification.
In order to determine the composition of 1,2- and 1,3-DAG isomers, NLS87 from the DMA-DAG mixture in the presence of LiOH (see above) was performed (Figure 2D). We found that the NLS87 spectrum displayed the very intense ion peaks corresponding to sn-1,3-DAG species and showed nearly equal intensities from the equimolar DAG species including polyunsaturated FA-containing species. Intriguingly, we also detected some minor ion peaks possessing the identical m/z to those of 1,2-DAG isomers in the NLS87 spectrum although the fragment ion corresponding to neutral loss of 87 amu is not observed in the product ion spectra of freshly prepared 1,2-DAG derivatives (Figure 1 A and B). We believe that these ion peaks were yielded from their sn-1,3 DAG counterparts since the chemical isomerization between 1,2-DAGs and their 1,3-isomers easily occurred even though the prepared solutions were stocked at −20 °C. This conclusion is supported by the observation that the intensities of the minor peaks corresponding to the isomerized 1,3-DAGs increased overtime after the non-derivatized 1,2-DAG species were stocked for a period of time at −20 °C. We specifically pointed out that the isomerization would be impossible after the free hydroxyl group at the glycerol backbone of a DAG species is occupied by derivatization. Hence, DMG derivatization could also prevent the chemical isomerization between 1,2-DAGs and their 1,3-isomers. We would like to advise that it is better to derivatize lipid extracts immediately after preparation if identification and quantification of 1,3-DAG species are desired.
The linearity of dynamic range of the established method was determined by spiking different concentrations (5 to 1250 fmol/µl for each DAG species) of the nearly equimolar mixture of DAG species as aforementioned in the selected internal standard (1,3-di15:0 DAG) solution (250 fmol/µl). The DAG mixtures were derivatized with DMG and diluted with a total DAG concentration of < 5 pmol/µl prior to direct infusion for MS analysis. The concentration was measured by comparing the peak intensity corresponding to each individual DAG species to that of the internal standard after 13C de-isotoping and correction for the enhanced factor for the DAG species containing polyunsaturated FA chain(s) (DB number ≥ 3, see above). Linear plots between the measured concentrations and the theoretical concentrations in the mixtures were obtained with linear correlation coefficients between 0.996 and 0.999, slope from 0.94 to 1.12, and intercepts from −0.33 to 0.04 (Figure S2B). The high correlation between the ions corresponding to the individual DAG species and the internal standard indicated that the derivatized DMG head group played a dominant role in the ionization of these derivatized DAG species and that all DAG species present in biological samples could be assessed by using one internal standard under the optimized conditions after appropriate corrections.
Next, we examined the effects of the matrices (i.e., other co-existing lipids in biological sample extracts) on quantification of DAG species by the established method. We spiked various amounts of the nearly equimolar mixture of DAG species and the internal standard (see above) into mouse liver lipid extracts and determined the intensity changes of the ions corresponding to the DAG species. After proper dilution for minimizing potential aggregation (a total lipid concentration of < 5 pmol/µl), the measurement showed a great linear correlation of the spiked amounts of DAG species with those determined by this method with correlation coefficients between 0.991 and 1.000 (Figure S2C).
Collectively, through the dilution experiment and linear correlation analysis of DAG species, we found the established method possessing the limit of quantification at 0.1 fmol/µl, a reproducibility of CV < 5.2% from an identical prepared sample and CV < 10.8% from separately prepared samples, and a linear dynamic range over 2,500 fold.
Determination of DAG Species in Lipid Extracts of ob/ob Mouse Liver Samples
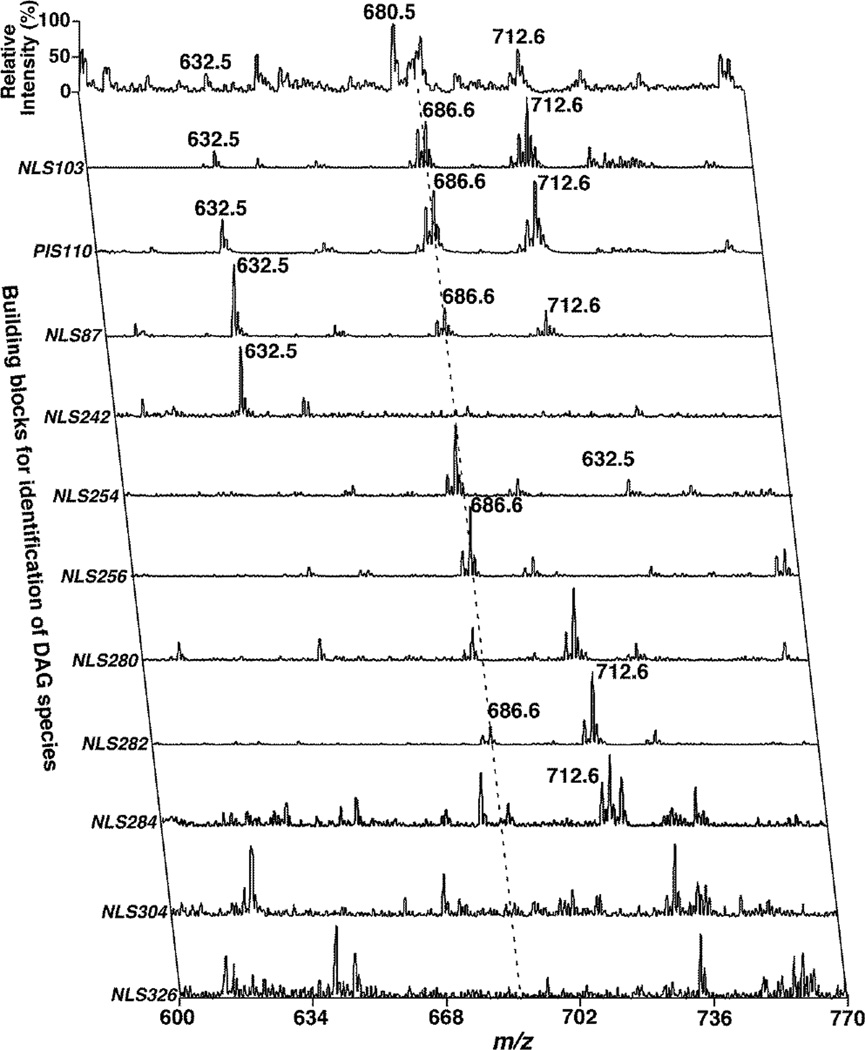
To demonstrate the capability and utility of the current method for DAG analysis, we determined and compared the levels of DAG species including 1,2- and 1,3-DAG isomers present in liver samples from 3- and 4-month old ob/ob mice and their WT controls. We identified the molecular structures of all of the possible DAG species by using 2D-MS (Figure 3). The survey MS scan for the analysis of lithiated DAG species was acquired in the positive-ion mode, which only showed a few abundant DAG species (e.g., m/z 684, 686 and 712). However, NLS103 and PIS110 spectra acquired under the conditions were specific to lithiated DMG-DAG species. These spectra, in combination with the NLS103 spectrum of protonated DMG-DAG (spectrum not shown) showed all of the possible ions corresponding to the DAG species present in the lipid extracts with high sensitivity. From the detected ion masses by these spectra, the numbers of total carbon atoms and double bonds in the aliphatic chains of the DAG species were readily deduced. The NLS87 spectrum was used to identify and quantify 1,3-isomers in the detected DAG species.
Figure 3.
Representative two-dimensional MS analysis of lithiated DMG-DAG species present in lipid extracts of mouse liver samples. All MS/MS scans (i.e., NLS103, PIS110, and NLS87) for characterization of the DMG head group were performed under the same conditions as described in the legend to Figure 2. All NLS of FA chains from lithiated DMG-DAG species were acquired at collision energy of 35 eV.
The FA chains of individual DAG species were determined by NLS of naturally occurring FAs by matching the total number of carbon atoms and the number of double bonds. For example, an ion peak was present at m/z 686 in both NLS103 and PIS110 spectra corresponding to a 34:1 DAG species (34 carbon atoms and one double bond in the FA chains). The NLS87 spectrum indicated the presence of 1,3-DAG from a part or all of the ions. After quantification by using NLS87 for 1,3-DAG species in comparison to the 1,3-di15:0 DAG internal standard, and using NLS103 of protonated DMG-DAG for the total DAG mass of the ion peak, we determined that there was 10% of 1,3-isomer and 90% of 1,2-isomer in this 34:1 DAG. The NLS spectra of naturally occurring FAs indicated that the ion peak at m/z 686 was present in the NLS254 (i.e., 16:1 FA), NLS256 (i.e., 16:0 FA), NLS282 (i.e., 18:1 FA), and NLS284 (i.e., 18:0 FA) spectra. After baseline correction34 and 13C de-isotoping of the intensity of the ion peak at m/z 686 in each of the NLS spectra, it was determined that the 34:1 DAG species contained 2.5% of 18:0-16:1 DAG and 97.5% of 16:0-18:1 DAG species.
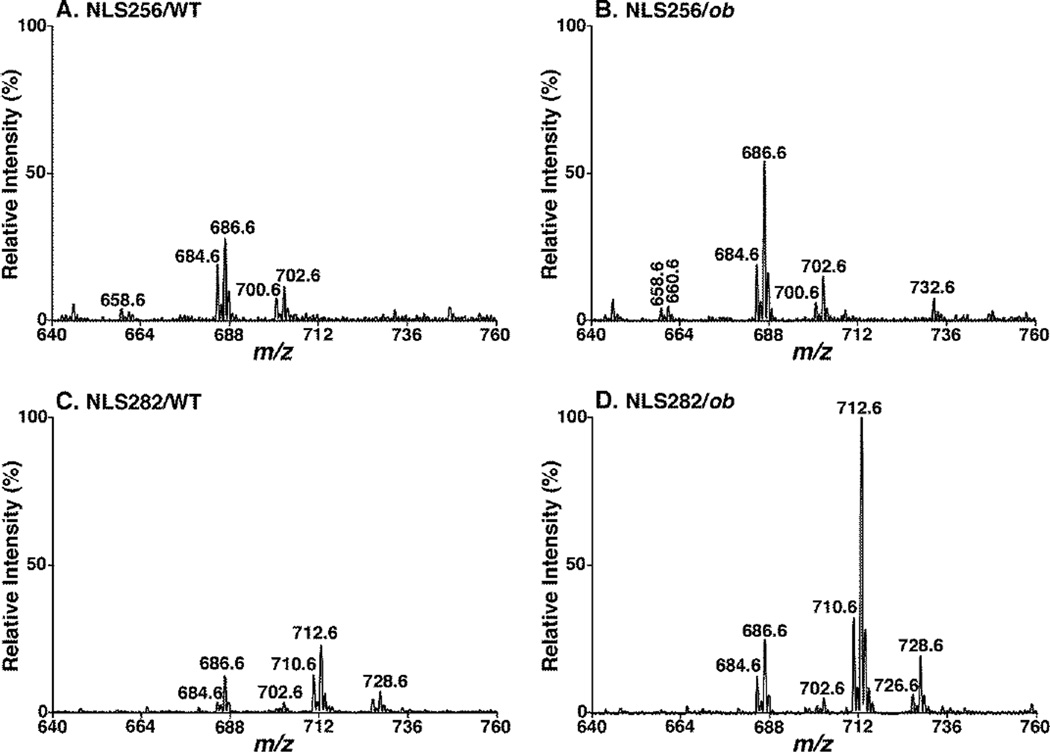
By utilizing the established method, we not only found that the levels of DAG species in the liver of ob/ob mice were much higher than their age-matched controls, but also revealed that the difference of total mass levels of DAG in the livers between ob/ob and WT mice is 16 folds at 4 month (an insulin-resistant state) vs. five folds at 3 months of age (a normal insulin state) (Table 1). Elevated DAG levels are believed to cause the inhibitory action on insulin signaling35. These mass levels of DAG species were also consistent with those of triacylglycerol mass levels, suggesting that DAG species mainly serve as the intermediates for triacylglycerol synthesis or degradation. We also found that the mass levels of all analyzed FA chains increased in the ob/ob mice (data not shown), likely due to the changes of systematic lipid metabolism under ob/ob conditions. However, it is intriguing to note that the mass levels of 18:1 FA was substantially elevated in ob/ob mouse liver, likely due to its increased de novo synthesis (as demonstrated with the increased expression of stearoyl-CoA desaturase 136) to serve as a supply of energy storage through forming of 18:1 FA-containing triacylglycerols. For example, the intensities of ion peaks at m/z 684, 686, 710 and 712 were increased drastically in NLS of 18:1 FA in the ob/ob mice (Figure 4D and Table 1). The ion peak at m/z 712, mainly comprised of di18:1 DAG, was much higher in the livers of ob/ob mice than WT controls: a 10 times higher at 3 months, but over 40 times higher at 4 months (Table 1).
Table 1.
Comparison of the mass levels of DAG species in lipid extracts of liver samples between ob/ob and wild type mice at 3 and 4 months of age (nmol/mg protein).
| DAG species |
3 month | 4 month | ||||||
|---|---|---|---|---|---|---|---|---|
| Wild type | ob/ob | Wild type | ob/ob | |||||
| 1,2-DAG | 1,3-DAG | 1,2-DAG | 1,3-DAG | 1,2-DAG | 1,3-DAG | 1,2-DAG | 1,3-DAG | |
| di14:0 | 0.05±0.02 | 0.04±0.01 | 0.01±0.01 | 0±0 | 0.06±0.01 | 0.02±0.01 | 0.01±0.02 | 0.02±0.03 |
| 14:0-16:1 | 0.02±0.01 | 0.01±0.01 | 0.04±0 | 0.01±0.01 | 0±0 | 0±0 | 0.06±0.01 | 0.01±0.01 |
| di16:1/14:0-18:2 | 0.13±0.04 | 0.01±0.01 | 0.39±0.07 | 0.05±0.02 | 0.03±0 | 0.01±0.01 | 0.75±0.18 | 0.13±0.02 |
| 16:0-16:1/14:0-18:1 | 0.61±0.19 | 0.38±0.05 | 1.92±0.49 | 0.65±0.23 | 0.07±0.01 | 0.39±0.09 | 2.91±0.69 | 0.85±0.17 |
| di16:0 | 0.29±0.09 | 0.1±0.05 | 0.9±0.34 | 0.44±0.25 | 0.08±0.01 | 0.03±0.02 | 1.18±0.22 | 0.5±0.04 |
| 16:0-17:1 | 0.17±0.04 | 0.08±0.03 | 0.3±0.06 | 0.14±0.05 | 0.06±0.01 | 0.04±0.01 | 0.32±0.06 | 0.18±0.03 |
| 16:1-18:2/16:0-18:3 | 0.41±0.13 | 0.05±0.02 | 0.95±0.23 | 0.12±0.03 | 0.14±0.01 | 0.02±0.01 | 2.25±0.55 | 0.23±0.07 |
| 16:1-18:1/16:0-18:2/di17:1 | 3.15±0.71 | 0.36±0.07 | 9.81±2.51 | 1.21±0.45 | 1.33±0.08 | 0.15±0.03 | 14.32±3.53 | 1.82±0.33 |
| 16:0-18:1/18:0-16:1/17:0-17:1 | 3.79±1.22 | 0.45±0.17 | 19±7.2 | 3.02±1.53 | 0.58±0.05 | 0.07±0.01 | 21.99±5.33 | 3.67±0.53 |
| 18:0-16:0/di17:0 | 0.12±0.03 | 0.08±0.02 | 0.39±0.12 | 0.32±0.2 | 0.08±0.04 | 0.05±0.01 | 0.44±0.05 | 0.34±0.09 |
| 18:3-17:0/18:1-17:2/18:2-17:1 | 0.08±0.01 | 0.03±0.02 | 0.04±0.03 | 0.02±0.02 | 0.04±0.01 | 0.02±0 | 0.08±0.01 | 0.04±0.02 |
| 17:1-18:1/17:0-18:2 | 0.2±0.06 | 0.08±0.03 | 0.62±0.14 | 0.2±0.08 | 0.05±0.01 | 0.02±0.02 | 0.55±0.09 | 0.24±0.09 |
| 17:0-18:1/16:0-19:1/17:1-18:0/19:0-16:1 | 0.05±0.01 | 0.04±0.02 | 0.1±0.01 | 0.08±0.05 | 0.02±0 | 0.02±0 | 0.08±0 | 0.11±0.03 |
| di18:2/18:3-18:1/16:0-20:4 | 0.51±0.07 | 0.07±0.01 | 0.7±0.24 | 0.09±0.05 | 0.38±0.08 | 0.04±0.01 | 1.52±0.33 | 0.19±0.06 |
| 18:1-18:2/18:0-18:3/16:0-20:3 | 1.71±0.46 | 0.15±0.06 | 5±1.1 | 0.5±0.15 | 0.62±0.07 | 0.06±0.02 | 8.4±2.11 | 0.64±0.15 |
| di18:1/18:0-18:2/16:0-20:2 | 2.61±0.92 | 0.28±0.11 | 25.01±9.61 | 2.46±1.32 | 0.49±0.03 | 0.06±0.02 | 22.03±5.96 | 2.04±0.41 |
| 18:0-18:1 | 0.29±0.09 | 0.04±0.02 | 1.73±0.6 | 0.39±0.23 | 0.09±0.01 | 0.01±0.01 | 1.47±0.35 | 0.39±0.05 |
| di18:0 | 0.04±0.01 | 0.02±0.01 | 0.01±0.01 | 0.04±0.02 | 0.07±0.04 | 0.04±0.02 | 0.02±0.02 | 0.04±0.03 |
| 16:0-22:6/16:1-22:5/18:1-20:5/18:2-20:4 | 0.46±0.11 | 0.03±0.01 | 2.1±0.8 | 0.11±0.03 | 0.3±0.06 | 0.01±0.01 | 4.07±0.99 | 0.31±0.06 |
| 18:1-20:4/18:2-20:3/16:0-22:5 | 0.35±0.08 | 0.01±0.01 | 0.85±0.37 | 0.06±0.03 | 0.13±0.03 | 0.01±0.01 | 1.03±0.25 | 0.12±0.04 |
| 18:1-20:3/18:2-20:2/18:0-20:4 | 0.39±0.08 | 0.03±0.03 | 1.06±0.42 | 0.15±0.07 | 0.18±0.02 | 0.02±0.01 | 0.94±0.27 | 0.1±0.02 |
| 18:0-20:3/18:1-20:2/16:0-22:3 | 0.27±0.07 | 0.02±0.01 | 1.17±0.51 | 0.1±0.11 | 0.1±0.01 | 0.01±0.01 | 0.89±0.21 | 0.12±0.07 |
| 18:0-20:3/18:1-20:2/16:0-22:4 | 0.28±0.1 | 0.01±0 | 1.96±1.06 | 0.2±0.11 | 0.05±0.01 | 0±0.01 | 1.12±0.28 | 0.11±0.04 |
| 20:0-18:2/18:0-20:2 | 0.07±0.02 | 0.03±0.01 | 0.05±0.02 | 0.08±0.02 | 0.02±0 | 0±0 | 0.03±0.01 | 0.07±0.04 |
| 22:6-18:1 | 0.19±0.02 | 0±0 | 0.63±0.22 | 0.02±0.02 | 0.21±0.03 | 0.01±0 | 1.18±0.43 | 0.05±0.03 |
| 22:5-18:1/22:6-18:0 | 0.18±0.05 | 0.01±0 | 0.42±0.14 | 0.02±0.02 | 0.13±0.03 | 0.01±0.01 | 0.58±0.15 | 0.05±0.05 |
| 22:4-18:1/22:5-18:0 | 0.06±0.01 | 0±0 | 0.14±0.11 | 0±0 | 0±0.01 | 0±0 | 0.11±0.05 | 0.01±0.02 |
| TOTAL | 16.47±4.36 | 2.41±0.74 | 75.31±24.44 | 10.49±4.86 | 5.3±0.36 | 1.12±0.25 | 88.34±21.9 | 12.4±1.79 |
Figure 4.
Comparison of neutral loss scans of fatty acyl chains from lipid extracts of wild type vs. ob/ob liver tissues. Representative MS/MS spectra of NLS256 (i.e., 16:0 FA, A and B) and NLS282 (i.e., 18:1 FA, C and D) were acquired from lipid extracts of WT (A and C) and ob/ob (B and D) mouse liver samples with addition of LiOH at collision energy of 35 eV as described under “MATERIALS AND METHODS”. All the spectra were displayed after separate normalization to the internal standard of each sample and then to the base peak in Panel D. The NLS spectra indicate significantly higher content of DAG species (particularly those containing 18:1 FA) in ob/ob (B and D) compared to WT mouse liver (A and C) at 3 months of age.
In summary, based on the principles of MDMS-SL technology, DAG species including regioisomers in crude lipid extracts of biological samples were characterized and quantified after a facile one-step derivatization with DMG. The derivatization resulted in enhanced ionization efficiency of DAG species by ESI-MS and prevention dynamic migration of FA chains on the hydroxyl groups. By using this method, we revealed the higher mass content of DAG in ob/ob mouse liver compared to that in WT controls, more progressive in the insulin-resistant states. Accordingly, we believe that with the development of this novel methodology, the role of DAG species in biological processes could be effectively investigated, which should provide new insights into the origin of diabetes and obesity development in humans and various animal models.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by the National Institute of General Medical Sciences Grant R01 GM105724 and Intramural institutional research funds.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Goni FM, Alonso A. Prog. Lipid Res. 1999;38:1–48. doi: 10.1016/s0163-7827(98)00021-6. [DOI] [PubMed] [Google Scholar]
- 2.Ohanian J, Ohanian V. J Hum Hypertens. 2001;15:93–98. doi: 10.1038/sj.jhh.1001139. [DOI] [PubMed] [Google Scholar]
- 3.Jun Y, Fratti RA, Wickner W. J. Biol. Chem. 2004;279:53186–53195. doi: 10.1074/jbc.M411363200. [DOI] [PubMed] [Google Scholar]
- 4.Barona T, Byrne RD, Pettitt TR, Wakelam MJ, Larijani B, Poccia DL. J. Biol. Chem. 2005;280:41171–41177. doi: 10.1074/jbc.M412863200. [DOI] [PubMed] [Google Scholar]
- 5.Arimoto T, Takeishi Y, Takahashi H, Shishido T, Niizeki T, Koyama Y, Shiga R, Nozaki N, Nakajima O, Nishimaru K, Abe J, Endoh M, Walsh RA, Goto K, Kubota I. Circulation. 2006;113:60–66. doi: 10.1161/CIRCULATIONAHA.105.560771. [DOI] [PubMed] [Google Scholar]
- 6.Sauter G, Nerlich A, Spengler U, Kopp R, Pfeiffer A. Gut. 1990;31:1041–1045. doi: 10.1136/gut.31.9.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erion DM, Shulman GI. Nat. Med. 2010;16:400–402. doi: 10.1038/nm0410-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudkowska I, Roynette CE, Demonty I, Vanstone CA, Jew S, Jones PJ. Obes Res. 2005;13:1864–1876. doi: 10.1038/oby.2005.229. [DOI] [PubMed] [Google Scholar]
- 9.Nakano T, Iseki K, Hozumi Y, Kawamae K, Wakabayashi I, Goto K. Neurosci. Lett. 2009;461:110–115. doi: 10.1016/j.neulet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Drosatos K, Schulze PC. Curr Heart Fail Rep. 2013;10:109–121. doi: 10.1007/s11897-013-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agren JJ, Kuksis A. Lipids. 2002;37:613–619. doi: 10.1007/s11745-002-0940-0. [DOI] [PubMed] [Google Scholar]
- 12.Reyes G, Yasunaga K, Rothenstein E, Karmally W, Ramakrishnan R, Holleran S, Ginsberg HN. J. Lipid Res. 2008;49:670–678. doi: 10.1194/jlr.P700019-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Preiss J, Loomis CR, Bishop WR, Stein R, Niedel JE, Bell RM. J. Biol. Chem. 1986;261:8597–8600. [PubMed] [Google Scholar]
- 14.Billah MM, Eckel S, Mullmann TJ, Egan RW, Siegel MI. J. Biol. Chem. 1989;264:17069–17077. [PubMed] [Google Scholar]
- 15.Boarder MR, Purkiss JR. Methods Mol. Biol. 1995;41:189–202. doi: 10.1385/0-89603-298-1:189. [DOI] [PubMed] [Google Scholar]
- 16.Lee C, Fisher SK, Agranoff BW, Hajra AK. J. Biol. Chem. 1991;266:22837–22846. [PubMed] [Google Scholar]
- 17.Hubbard WC, Hundley TR, Oriente A, MacGlashan DW., Jr Anal. Biochem. 1996;236:309–321. doi: 10.1006/abio.1996.0172. [DOI] [PubMed] [Google Scholar]
- 18.Li YL, Su X, Stahl PD, Gross ML. Anal. Chem. 2007;79:1569–1574. doi: 10.1021/ac0615910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DW. J. Mass Spectrom. 2001;36:277–283. doi: 10.1002/jms.125. [DOI] [PubMed] [Google Scholar]
- 20.Callender HL, Forrester JS, Ivanova P, Preininger A, Milne S, Brown HA. Anal. Chem. 2007;79:263–272. doi: 10.1021/ac061083q. [DOI] [PubMed] [Google Scholar]
- 21.Leiker TJ, Barkley RM, Murphy RC. Int. J. Mass Spectrom. 2011;305:103–109. doi: 10.1016/j.ijms.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.vom Dorp K, Dombrink I, Dormann P. Methods Mol. Biol. 2013;1009:43–54. doi: 10.1007/978-1-62703-401-2_5. [DOI] [PubMed] [Google Scholar]
- 23.Lee SY, Kim JR, Ha MY, Shim SM, Park TS. Lipids. 2013;48:287–296. doi: 10.1007/s11745-013-3766-6. [DOI] [PubMed] [Google Scholar]
- 24.Han X, Gross RW. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Gross RW. Expert Rev. Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 26.Han X, Yang K, Gross RW. Mass Spectrom. Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Ory DS, Han X. Rapid Commun. Mass Spectrom. 2007;21:141–152. doi: 10.1002/rcm.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christie WW, Han X. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. Fourth ed. Bridgwater, England: The Oily Press; 2010. [Google Scholar]
- 29.Han X, Yang K, Gross RW. Rapid Commun. Mass Spectrom. 2008;22:2115–2124. doi: 10.1002/rcm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang K, Cheng H, Gross RW, Han X. Anal. Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han X, Gross RW. Anal. Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- 32.Han X, Gross RW. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu F-F, Turk J. J. Chromatogr. B. 2009;877:2673–2695. doi: 10.1016/j.jchromb.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang K, Fang X, Gross RW, Han X. J. Am. Soc. Mass Spectrom. 2011;22:2090–2099. doi: 10.1007/s13361-011-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chavez JA, Summers SA. Arch. Biochem. Biophys. 2003;419:101–109. doi: 10.1016/j.abb.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Botolin D, Xu J, Christian B, Mitchell E, Jayaprakasam B, Nair MG, Peters JM, Busik JV, Olson LK, Jump DB. J. Lipid Res. 2006;47:2028–2041. doi: 10.1194/jlr.M600177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.