Abstract
Bacteroides thetaiotaomicron is a prominent member of the human distal gut microbiota that specializes in breaking down diet and host-derived polysaccharides. While polysaccharide utilization has been well studied in B. thetaiotaomicron, other aspects of its behavior are less well characterized, including the factors that allow it to maintain itself in the gut. Biofilm formation may be a mechanism for bacterial retention in the gut. Therefore, we used custom GeneChips to compare the transcriptomes of biofilm and planktonic B. thetaiotaomicron during growth in mono-colonized chemostats. We identified 1154 genes with a fold-change greater than 2, with confidence greater than or equal to 95%. Among the prominent changes observed in biofilm populations were: (i) greater expression of genes in polysaccharide utilization loci that are involved in foraging of O-glycans normally found in the gut mucosa; and (ii) regulated expression of capsular polysaccharide biosynthesis loci. Hierarchical clustering of the data with different datasets, which were obtained during growth under a range of conditions in minimal media and in intestinal tracts of gnotobiotic mice, revealed that within this group of differentially expressed genes, biofilm communities were more similar to the in vivo samples than to planktonic cells and exhibited features of substrate limitation. The current study also validates the use of chemostats as an in vitro ‘gnotobiotic’ model to study gene expression of attached populations of this bacterium. This is important to gut microbiota research, because bacterial attachment and the consequences of disruptions in attachment are difficult to study in vivo.
INTRODUCTION
The adult human gut microbiota is composed of members of all three domains of life and their viruses. This community is dominated by Bacteria, and specifically by members of two bacterial phyla, the Bacteroidetes and Firmicutes. Bacteroides thetaiotaomicron is prominently represented among the Bacteroidetes in the distal gut, where it ferments chemically diverse, complex dietary glycans to short chain fatty acids that can be absorbed by the host (Koropatkin et al. 2012; Martens et al. 2011). B. thetaiotaomicron is also able to utilize host mucus glycans, such as mucin, including mucin O-glycans, as nutrient substrates when polysaccharides are absent from the host diet, giving it a competitive advantage over other, less versatile, simple sugar-fermenting bacteria (Benjdia et al. 2011; Martens et al. 2008; Martens et al. 2011; Sonnenburg et al. 2005).
The saccharolytic capabilities of B. thetaiotaomicron are reflected in its genome. The type strain (VPI-5482) has 88 polysaccharide utilization loci (PULs), composed of 866 genes that comprise 18% of its genome (Martens et al. 2008). Each PUL characterized to date encodes a group of cell envelope-associated proteins collectively known as a Sus-like system, which endows the bacterium with the ability to metabolize a glycan or group of related glycans. Each of B. thetaiotaomicron Sus-like systems contains: (i) a homolog of SusC, which is a TonB-dependent receptor that spans the outer membrane and transports oligosaccharides in an energy dependent manner; and (ii) a homolog of SusD, which is an outer membrane lipoprotein that binds specific glycans and participates in delivering oligosaccharides to the SusC transporter (Koropatkin et al. 2008; Reeves et al. 1996; Reeves et al. 1997). In addition to SusC- and SusD-like proteins, a PUL can include other outer membrane glycan binding proteins, as well as various glycoside hydrolases, polysaccharide lyases, and/or carbohydrate esterases (Koropatkin and Smith 2010). Whole genome transcriptional profiling, targeted gene disruption, characterization of purified Sus proteins, and assays of growth in vitro on glycan arrays (a high-throughput method to directly measure functional interactions with polysaccharides (Blixt et al. 2004; Padler-Karavani et al. 2012; Stevens et al. 2006)) have helped define the carbohydrate recognition and utilization capabilities of B. thetaiotaomicron and the carbohydrate specificities of its PULs (Kitamura et al. 2008; Koropatkin et al. 2009; Koropatkin and Smith 2010). The repertoire of PULs present in the genome, and their patterns of gene expression help define the niches of B. thetaiotaomicron and of other members of Bacteroides in vivo (Martens et al. 2011; Sonnenburg et al. 2010).
A major challenge for members of the gut microbiota is to prevent washout from the gut habitat. The ability to form ‘attached’ populations would provide a competitive advantage to a gut symbiont by increasing retention time, providing access to solid state plant- and human-derived nutrient substrates, and facilitating development of syntrophic (nutrient-sharing) relationships with other members of the microbiota (Sonnenburg et al. 2004). The formation of extracellular matrices composed principally of polysaccharides and other biological polymers with bound/embedded microbes provides an important mechanism for microbes to adhere to each other and to living or non-living surfaces, such as food particles. The intestine is lined by mucus that serves as a microhabitat for members of the microbiota, supplying attachment sites (e.g., O-glycans) and nutrients (Ambort et al. 2012; Ambort et al. 2011; Lindén et al. 2008a; Lindén et al. 2008b; McGuckin et al. 2011). Biofilm formation affects the motility of microbes and also their response to nutrient limitation and other stresses (Beloin and Ghigo 2005; Lazazzera 2005). However, investigation of gut biofilms has been difficult because of the challenges associated with accessing this community in vivo, or replicating the gut environment in vitro (Marzorati et al. 2011).
Studies have shown that B. thetaiotaomicron is prominently represented on the surfaces of mixed food particles isolated from human feces (Macfarlane and Dillon 2007; Macfarlane and Macfarlane 2006), and undigested plant material in the gut lumen and within the mucus layer of gnotobiotic mice colonized by B. thetaiotaomicron and fed simple-sugar or polysaccharide-rich diets (Sonnenburg et al. 2005). Because both plant materials and host-derived mucus can serve as carbon and energy sources for B. thetaiotaomicron, attachment is likely to be mediated, at least in part, by Sus-like proteins involved in nutrient binding (Shipman et al. 2000). When cells bind to a nutrient that is part of a solid surface they, in essence, attach to that surface. However, B. thetaiotaomicron also attaches to glass surfaces, indicating that nutrient binding is not its only attachment mechanism (Macfarlane et al. 2005). One study demonstrated that attachment of this bacterial species to glass was regulated by the availability of soluble substrate (i.e., it occurred only under conditions where glucose concentrations were high), indicating that nutrient binding and attachment are tightly linked in this organism (Macfarlane et al. 2005).
In the present study, we use GeneChip-based whole genome transcriptional profiling to explore how biofilm formation impacts gene expression in B. thetaiotaomicron. To do so, we sampled mono-colonized chemostats, examining biofilm as well as planktonic populations. The results are compared to the transcriptional profiles of B. thetaiotaomicron obtained in monocolonized gnotobiotic mice as well as during in vitro culture as planktonic cells under defined limiting and non-limiting nutrient conditions. Specifically, we investigated the links between attachment, nutrient binding and uptake, and capsule formation to generate hypotheses on how attachment of this organism may affect human health, and to compare biofilm and planktonic populations as in vitro models of gene expression in vivo.
RESULTS AND DISCUSSION
Growth in chemostats and differential expression analysis
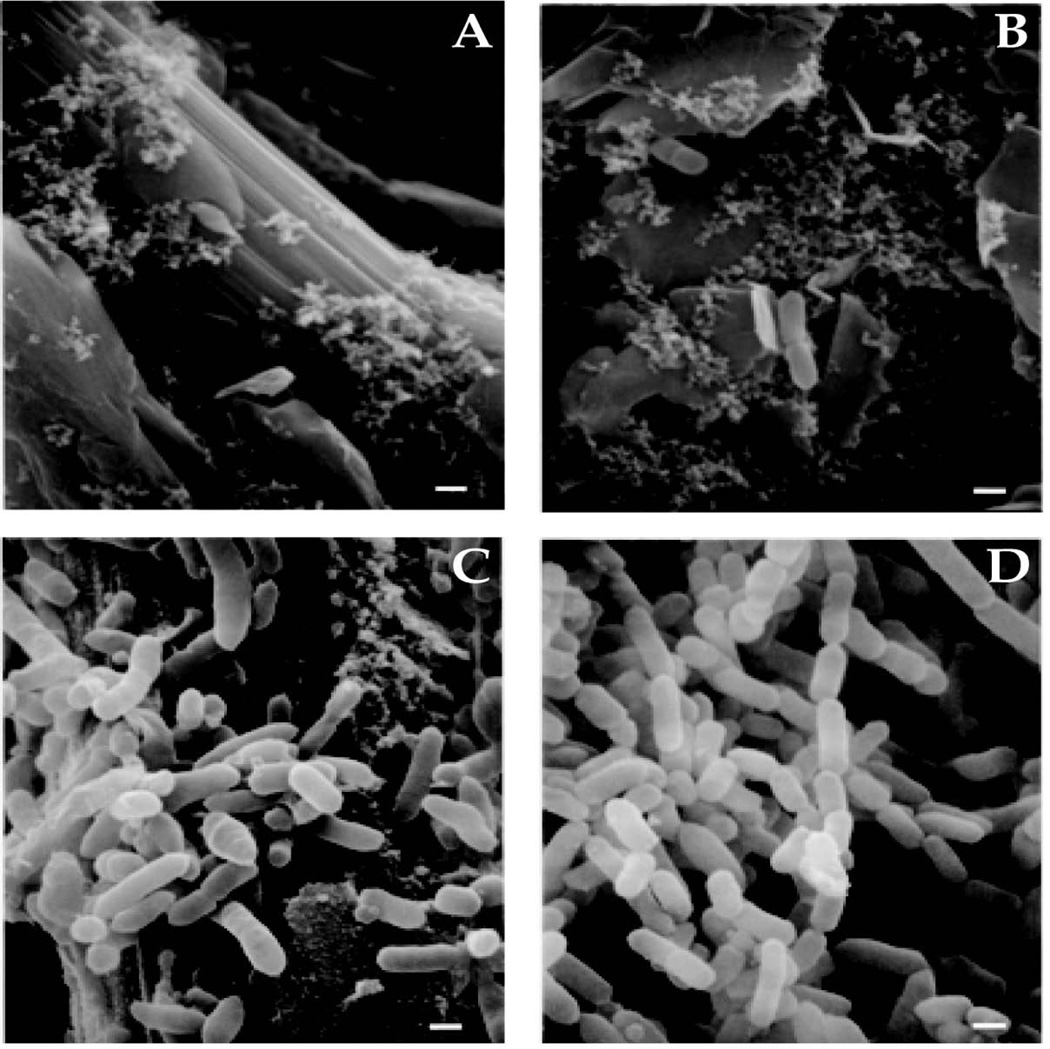
The sequenced type strain, B. thetaiotaomicron VPI-5482, was inoculated into six sterile chemostats and fed continuously with sterile TYG (tryptone, yeast extract, glucose) medium. Growth proceeded at 37°C under an atmosphere of N2 and CO2 (80%/20%). Each chemostat contained a carbon paper growth surface to allow for biofilm formation. During the first 24 h, dense planktonic growth occurred (OD600 ~ 0.6). Biofilm was not detected by scanning electron microscopy (SEM) of the carbon paper surface at 8 h but was clearly visible by the naked eye and SEM at 8 d (Figure 1A-D).
FIGURE 1. Attachment of B. thetaiotaomicron to carbon paper surfaces in the chemostat (A-D).
Scanning electron micrographs of the growth surface sampled at 8 h (panels A and B) and 8 days (panels C and D) after inoculation. Few bacteria are shown in panels A and B, illustrating the rarity of their association with the growth surface at this early time point. Bars, 0.5 µm.
RNA was extracted from planktonic cells harvested from the chemostats after 8 h of growth. Biofilm cells were removed from the carbon paper surface after 8 days of growth. Because the experiments were performed in chemostats, both samples were in a steady-state growth phase and not in stationary or exponential growth phases. The samples were harvested at different operating periods and HRTs to ensure that the planktonic sample was not contaminated with biofilm cells and vice versa. Gene expression was compared for biofilm (n=6) and planktonic (n=6) samples using the LEMMA (Laplace EM Microarray Analysis) method implemented as a package in R (Bar et al. 2010). Using a false discovery rate of 0.05, 1584 genes were detected as differentially expressed between the two groups. Of these, we defined 1145 genes as “differentially expressed” for the rest of the study (base 2 logarithm of the fold-change in their expression was greater than 1 [i.e., fold-change was greater than 2]). Transcripts were categorized based on a variety of annotation schemes: COG category, KEGG orthology group (KO) (KEGG database), KEGG enzyme commission (EC) number, carbohydrate active enzyme (CAZyme) family (CAZy database), and peptidase family (MEROPS database). The results are provided in Table S1. Using these annotations, we determined which functional groups were significantly enriched within the differentially expressed genes compared to the genome (determined using the hypergeometric distribution; p<0.05) (see Table S2). This analysis indicates that groups that are significantly enriched within the differentially expressed genes are specifically involved in changes between biofilm and planktonic samples, but does not take into account up- or down-regulation. Further functional insights about the significance of these observed differences in gene expression came from a follow-up analysis that placed them in the context of B. thetaiotaomicron’s PULs.
Substrate acquisition and utilization
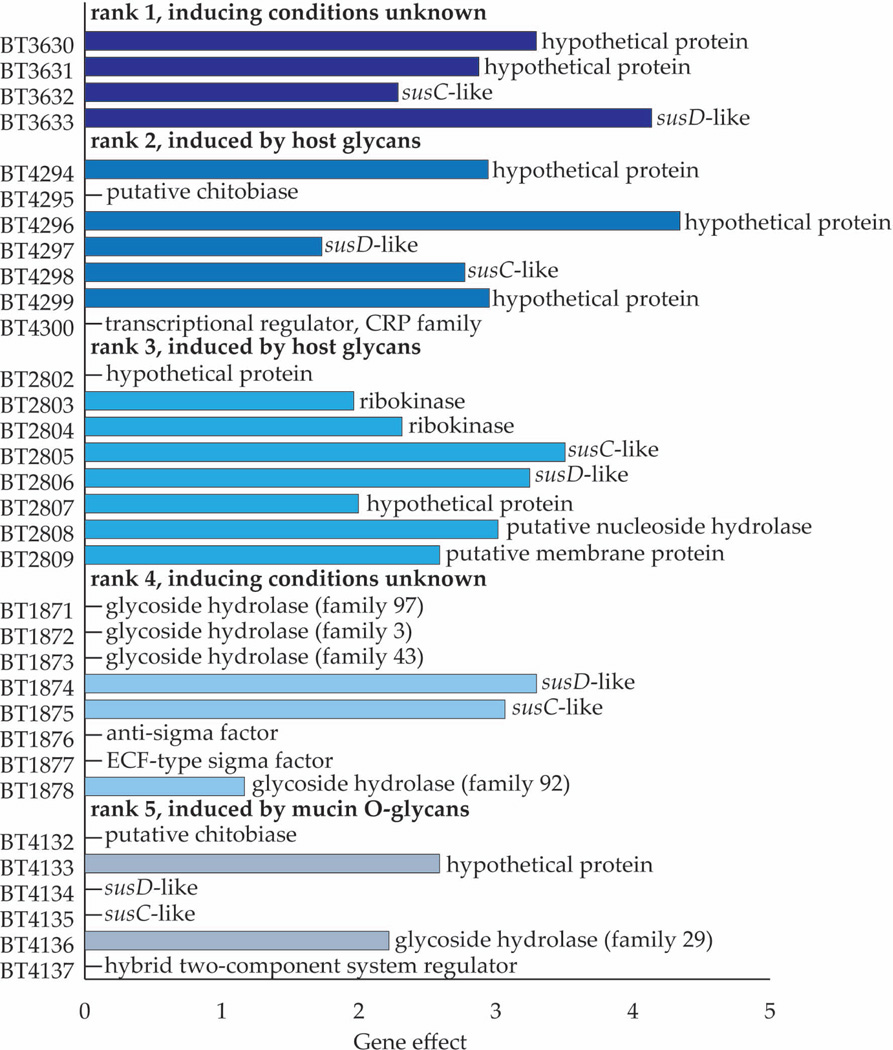
As noted in the Introduction, B. thetaiotaomicron VPI-5482 has 88 PULs, containing a total of 866 genes. Two hundred seventy eight of these genes were differentially expressed in the biofilm cells, with 80 PULs being represented (although not all genes in each of these 80 PULs were differentially expressed). Among the 866 PUL genes are 209 susC/susD homologs, 93 of which (51 SusC homologs and 42 SusD homologs) were differentially expressed. With the exception of two susC homologs and two susD homologs from 3 different PULs all of the differentially expressed susC/susD homologs showed increased expression in the biofilm compared to planktonic populations. Table S3 provides: (i) a rank ordering of PULs based on the magnitude of the average difference in expression of their constituent genes between biofilm versus planktonic populations; and (ii) includes information about the differences in expression of their other genes (e.g., CAZyme family members, hypothetical proteins). Figure 2 shows the change in expression for all genes in the 5 PULs with the largest differences (ranking based on their susC/susD responses) and annotation of the genes that comprise these PULs. Interestingly, two of these 5 PULs, BT4294-4300, and BT2802-2809 are known to be induced in response to host-derived glycans (Sonnenburg et al. 2005) and five sulfatases, including a mucin-degrading sulfatase, were expressed at significantly higher levels in biofilm compared to planktonic cell populations (Table S3). One possible reason for this is that starvation during biofilm growth causes the cell to upregulate Sus-like systems to “surveillance levels” that prime the cell to gather any nutrients that are available. Alternatively, attachment per se may prime B. thetaiotaomicron cells for degradation of host mucins. Mono- and co-colonization studies in gnotobiotic mice have established that sulfatases are important fitness factors for B. thetaiotaomicron, especially when the mice were fed a simple sugar diet that requires adaptive foraging on host glycans (Benjdia et al. 2011).
FIGURE 2. Differential patterns of PUL gene expression.
Gene effect (base 2 logarithm of the fold expression change) of each gene within the 5 most differentially expressed PULs (based on change in susC/susD expression) and annotations for each gene. Bars are not shown for genes that were not detected as differentially expressed. Inducing conditions for each PUL were determined in reference (3).
The B. thetaiotaomicron genome contains 8 capsular polysaccharide synthesis (CPS) loci, each comprised of 15–32 genes (Martens et al. 2009). A total of 74 genes, distributed among all 8 CPS loci, exhibited significant differences in their expression between biofilm and planktonic communities, including 13 genes that were upregulated in CPS locus 8 (BT0037-68), and 24 genes that were downregulated in CPS locus 1 (BT0375-402). This indicates that specific changes in the capsule are required for attachment or life in the biofilm (see Table S4 for a complete list of the genes present in each CPS locus and the magnitude of their differential expression in biofilm versus planktonic populations). Our findings show that attachment to a carbon surface in a chemostat not only regulates expression of PULs involved in adaptive forging of mucus glycans in vivo but also regulates expression of capsular biosynthetic loci. We have not defined how these changes impact capsular glycan composition. It is possible that changes in the capsule are involved in interactions between biofilm community members, whether at the level of attachment or nutrient sharing/harvest.
Comparison of transcriptional profiles of the biofilm community to profiles obtained in vitro under defined growth conditions and in gnotobiotic mice
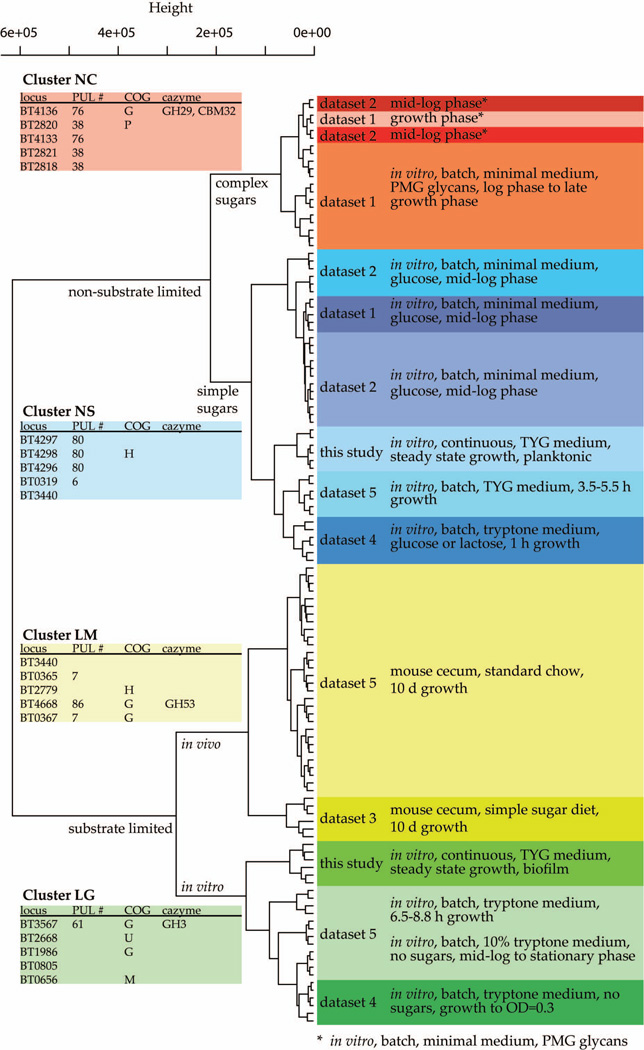
To gain additional perspective about the response of B. thetaiotaomicron to attachment and biofilm community formation, we compared the GeneChip datasets we generated in other studies to the datasets from biofilm and planktonic communities in our chemostats. We previously used the custom B. thetaiotaomicron GeneChip employed in the present study to characterize the transcriptome of this organism under a variety of conditions, including during in vitro growth in defined minimal medium containing a range of potential substrates, and in vivo in mono-associated gnotobiotic mice consuming a plant polysaccharide-rich diet, or a diet devoid of complex polysaccharides and rich in simple sugars (see Table S5 for an annotated list of these 5 datasets and their GEO accession numbers). The 6 datasets were subjected to unsupervised hierarchical clustering analysis with the dist and hclust functions in R. The results of this analysis were visualized as a dendrogram, which was labeled with relevant experimental information, as well as the dataset of origin (Figure 3).
FIGURE 3. Hierarchical clustering of transcriptional profiles of B. thetaiotaomicron grown under different environmental conditions.
The analysis is based on genes that are differentially expressed between biofilm and planktonic samples. Their expression under other environmental conditions was used to perform the unsupervised clustering shown, using the hclust function in R. Color code: red labels, ‘non-limited with complex sugars’ (NC) cluster; blue, ‘non-limited with simple sugars’ (NS) cluster; yellow, ‘substrate limited grown in mouse’ (LM) cluster; green, the ‘substrate limited grown in glass or tube’ (LG) cluster. Labels on the right indicate the datasets, and essential information describing them. Tables on the left show loci and annotations for the five most discriminatory genes for each cluster. (PMG, porcine gastric mucin)
When all genes were used in the clustering analysis, the biofilm and planktonic cells clustered together. However, when the differentially expressed genes identified by the chemostat experiment were used to perform the clustering analysis, the first branch point showed a clear division based on ‘substrate availability’ (where available substrate is defined as sugars or polysaccharides given within the previous 6 h of growth). Using only the differentially expressed genes highlighted differences between biofilm and planktonic samples and allowed us to interpret these differences in terms of other growth conditions. Within the substrate limited cluster, there was a division between cultures grown in vitro versus those harvested from the distal gut (cecum) of mono-colonized gnotobiotic mice fed various diets (Figure 3). Non-limited samples broke into two groups depending on whether complex or simple sugars were fed (Figure 3). Thus, overall, samples could be classified into four major groups based on this clustering pattern: (i) substrate limited, grown in mouse (LM); (ii) substrate limited, grown in glass or tube (LG); (iii) non-limited with complex sugars (NC); and (iv) non-limited with simple sugars (NS).
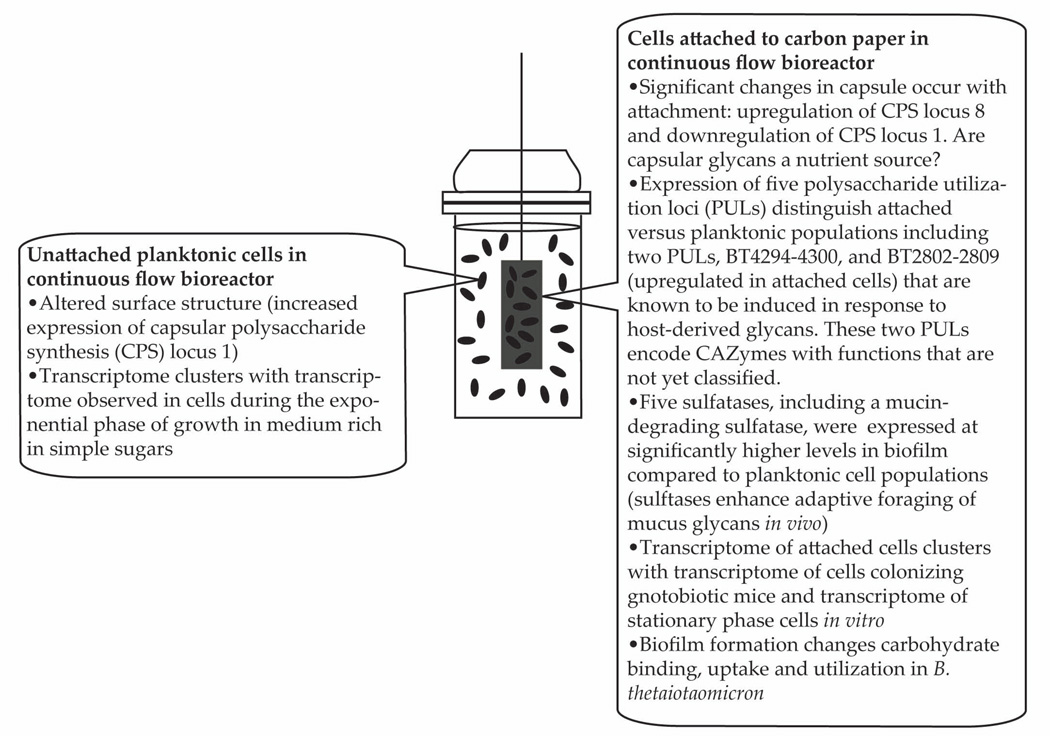
To observe which of the genes used in the clustering analysis were predictive for the sample cluster, we used a machine learning approach implemented in the PAMr package for R (Tibshirani et al. 2002). With a threshold of 5.0, 278 “key clustering genes” were required to accurately predict which of the four major clusters each sample belonged to (Table S6). These genes represent the core transcriptomic changes among the four groups and their potential phenotypic differences. The key clustering genes were enriched in 5 COG functional categories compared to the genome and the differentially expressed genes: (i) cell envelope biogenesis, outer membrane (M); (ii) inorganic ion transport and metabolism (P); (iii) carbohydrate transport and metabolism (G); (iv) amino acid transport and metabolism (E); and (v) coenzyme metabolism (H), with the largest enrichment in categories M and G. When combining the annotated dendrogram with the machine learning results, it becomes clear that the essential differences between the four sample groups lie in carbohydrate uptake and utilization; dendrogram clustering occurred mainly on the basis of substrate type and availability and the key clustering genes were enriched in carbohydrate utilization functions (Table S6). This indicates that there were widespread differences in carbohydrate utilization function between the biofilm and planktonic groups, although both were grown under the same experimental conditions, suggesting further that biofilm formation changes carbohydrate binding, uptake and utilization in B. thetaiotaomicron (Figure 4).
FIGURE 4.
Summary of major transcriptional differences that distinguish the expressed functional features of biofilm and planktonic B. thetaiotaomicron populations present in the chemostat.
Prospectus
Mechanisms that mediate and regulate attachment of gut bacteria to various living and non-living surfaces represent a key area that needs further investigation. Attachment is likely key to harvesting nutrients present in partially digested food. Direct attachment to other bacterial cells and/or gaining proximity to these cells via attachment to common nutrient platforms could be an important step in establishing syntrophic relationships, as in water columns within aquatic ecosystems. Bacterial attachment and the consequences of ‘attachment disorders’ are difficult to study in vivo, but the current study shows that chemostats can be used as an in vitro ‘gnotobiotic’ model to study how a prominent human saccharolytic bacterium attaches to a defined surface and its response to attachment. In terms of gene expression, chemostat-grown cells were more similar in vivo-grown cells than cells grown in batch-fed conditions were, making chemostats a good choice for in vitro experiments with this organism. In this system, we see that biofilm formation occurs in a reproducible fashion and that attachment to a carbon surface ‘primes’ the organism to induce expression of polysaccharide utilization loci (PULs) involved degradation of mucus glycans while also affecting the expression of capsular polysaccharide biosynthetic genes involved in decorating the surface of the bacterial cell with carbohydrates. Both types of responses will change the interactions of this organism with nutrient foundations, including those derived from the host or capsular glycans of other attached cells.
Hierarchical clustering of transcriptional profiles of B. thetaiotaomicron populations studied under a wide variety of environmental conditions indicate that populations in the gut of mono-colonized gnotobiotic mice fed various diets, and biofilm communities elicit transcriptional responses resembling those seen under nutrient limiting conditions, making them more similar to each other than to high-nutrient growth conditions. This study suggests that future genetic and biochemical/metabolic analyses of B. thetaiotaomicron during its assembly into biofilm communities within continuous flow chemostats may provide new ways for defining and testing hypotheses about how this mutualist attaches to various surfaces present in the gut, acquires and processes various nutrients in an attached state, and how it adjusts to various perturbations.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions
B. thetaiotaomicron strain VPI-5482 (ATCC 29148) was used in all experiments. Tryptone, yeast extract, glucose (TYG) medium was used for bacterial growth in the chemostats, containing (per liter deionized water): tryptone, 10 g; yeast extract, 5 g; glucose, 4 g; L-cysteine, 0.5 g; KH2PO4, 4 g; K2HPO4, 9 g; TYG salt solution, 40 mL; CaCl2·2H2O, 8 mg; FeSO4, 0.4 mg; hematin, 1.2 mg. TYG salt solution consists of (per liter deionized water): MgSO4·7H2O, 0.5 g; NaHCO3, 10 g; NaCl, 2 g. All chemicals were used as purchased. Prior to inoculation, the bacterial culture was pre-grown in TYG medium overnight. Pre-cultures were inoculated directly from frozen stocks prior to each experiment.
Chemostat Design and Operation
Two identical chemostats were constructed from glass with a ~210 mL liquid volume. A water jacket around each chemostat maintained an operating temperature of 37°C. A 54 cm2 carbon paper growth surface for biofilm attachment was inserted into each chemostat, (P50, Ballard Material Products, Lowel, MA, USA). Before operation, the chemostats, medium storage tanks, and all connections/pump tubing were autoclaved at 121°C for 60 min. Glucose and hematin solutions were filter-sterilized (0.22-µm pore diameter), and added to the medium storage tank after autoclaving. To inoculate, 3 mL of overnight-grown B. thetaiotaomicron culture (~ 6 × 109 CFU) were injected into the chemostats. The chemostats were continuously fed with TYG medium at a hydraulic retention time (HRT) of 13 h (unless stated otherwise). A N2/CO2 (80%/20%) gas mixture was constantly sparged into the working chamber to remove the potential oxygen flux from the feeding solution and to compensate for the pressure loss due to the difference of flow rates between influent and effluent. Cell density was obtained by measuring optical density of 100 µL aliquots of the culture at a wavelength of 600 nm (Synergy HT microplate reader; Bio-TEK Instruments Inc., Winooski, VT, USA).
Sample Collection and RNA Extraction
Planktonic cells were harvested during the exponential growth phase (optical density of 0.50 – 0.55 at 600 nm, 8 h after inoculation). A sample of the working chamber culture was collected in RNAprotect bacteria reagent (Qiagen Inc., Valencia, CA, USA) at a volumetric ratio of 1:2. After being centrifuged at 5000 × g for 30 min, the supernatant was decanted and the cells were stored at −80°C prior to extraction of total RNA using an RNeasy kit (Qiagen).
Biofilm samples were collected after an 8-day operating period. During the last two days of the operating period, the HRT was shortened gradually until it was ~ 30 min. In this way, planktonic cells were largely removed by fast replacement of the medium. We do not anticipate that this change in HRT would cause a significant impact on gene expression. Indeed, the biofilm experienced substrate limitations (as shown by our transcriptomic data) due to substrate diffusion limitations even though the substrate concentration in the bulk liquid was high at an HRT of 30 min. In addition, shortening the HRT from 13 h to 30 min would not have an overarching effect on turbulence because of the presence of a magnetic stir bar. To minimize RNA degradation in the biofilm during sampling, the growth surface was immediately frozen in liquid nitrogen after its removal from the chemostat. The growth surface was cut into small pieces with sterile scissors and distributed into 2 mL centrifuge vials containing 500 µL of extraction buffer (200 mM Tris, pH 8.0 / 200 mM NaCl/ 20 mM EDTA), 210 µL of 20% SDS, 500 µL of phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.5, Ambion, Foster City, CA, USA) and 150 µL of 0.1-mm silica beads. Biofilm and planktonic samples were mechanically disrupted with a mini-beadbeater (BioSpec Products Inc., Bartlesville, OK, USA) on instrument setting “high” for 5 min at room temperature. After centrifugation at 10,000 × g for 3 min, the supernatant was extracted once more with phenol:chloroform:isoamyl alcohol and RNA was precipitated by adding 60 µL of sodium acetate (3 M) and 600 µL of ethanol (−20°C). The extracted RNA was stored in 100 µL of water at −80 °C.
GeneChip Analysis
Total cellular RNA was purified using a Qiagen RNA Easy mini kit according to the manufacturer's directions. Following extraction of RNA, samples were treated with DNAse I (Ambion) and purified again with a Qiagen RNA Easy column. Due to the large initial concentration of contaminating DNA, biofilm samples were subjected to an additional DNAse I treatment and purification. All samples were monitored for DNA contamination by PCR using primers specific for B. thetaiotaomicron genes. cDNA targets were prepared as previously described according to a standard Affymetrix protocol (Santa Clara, CA, USA) and applied to custom B. thetaiotaomicron GeneChips (Sonnenburg et al. 2005). These GeneChips contain probe pairs derived from 4719 of the 4779 predicted B. thetaiotaomicron genes. GeneChip data was processed using Microarray Suite 5 (Affymetrix). Each array was normalized to an arbitrary mean value of 500. The data generated for this study have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE38534 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38534) (Edgar et al. 2002).
Statistical Analysis
Differential expression analysis was performed using the LEMMA package for R, with a false discovery rate of 0.05 (Bar et al. 2010). Expression values were normalized using the quantile normalization function in R prior to the differential expression analysis. Genes with a gene effect (base 2 logarithm of the fold expression change) less than 1 were not considered. Hierarchical clustering was also performed in R, using the dist function with the Euclidean method to calculate the distance matrix and the hclust function with the Ward method to generate the tree. Plots were drawn using the plotColoredClusters function in the ClassDiscovery package, which is part of the Object-Oriented Microarray and Proteomic Analysis (OOMPA) suite (http://bioinformatics.mdanderson.org/Software/OOMPA). GeneChip data for B. thetaiotaomicron grown under different conditions used in hierarchical clustering analyses was downloaded from the NCBI Gene Expression Omnibus (GEO) database. These datasets consisted of publicly available data collected from a variety of previous studies on B. thetaiotaomicron grown under widely varying conditions. In general, all samples were grown either in standard culture flasks with minimal or standard rich media (e.g., TYG), or in mono-colonized gnotobiotic mice, and sampled at defined time points for RNA extraction. More details are given in Table S5, and more information about each study can be found by looking up the relevant GEO accession number at http://www.ncbi.nlm.nih.gov/geo/. All data were ranked prior to analysis to normalize between different experiments (Folsom et al. 2010). The significance of the enrichment of different gene groups in the differentially expressed genes was checked using the hypergeometric distribution using the dhyper function in R, and a p-value cutoff of 0.05. Machine learning to determine key genes in the clustering analysis was performed using the PAMr package for R, with a threshold value of 5.0 (Tibshirani et al. 2002).
Scanning Electron Microscopy
Small pieces of the carbon paper were fixed overnight in a solution containing 2% glutaraldehyde at 4°C, followed by washing with deionized water for 10–20 min. Secondary fixation was conducted in 1% osmium tetroxide at 4°C for 2 h. The carbon paper pieces were washed with deionized water for 10–20 min, dehydrated in a series of ethanol solutions (50, 70, 90, and 100%), and critical point dried in CO2. The samples were coated with gold and viewed using a scanning electron microscope (Hitachi S-450, Hitachi Ltd., Tokyo, Japan). To examine biofilm formation during the exponential growth phase (at 8 h, when planktonic cells were harvested for RNA extraction), the growth surface was gently rinsed with sterile phosphate buffer to remove any adsorbed planktonic cells.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the National Science Foundation (CAREER grant 0939882 to LTA) and the NIH (DK30292 to JIG).
REFERENCES
- Ambort D, Johansson MEV, Gustafsson JK, Nilsson HE, Ermund A, Johansson BR, Koeck PJB, Hebert H, Hansson GC. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(15):5645–5650. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambort D, van der Post S, Johansson MEV, MacKenzie J, Thomsson E, Krengel U, Hansson GC. Function of the CysD domain of the gel-forming MUC2 mucin. Biochemical Journal. 2011;436:61–70. doi: 10.1042/BJ20102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar H, Booth J, Schifano E, Wells MT. Laplace Approximated EM Microarray Analysis: An empirical Bayes approach for comparative microarray experiments. Statistical Science. 2010;25(3):388–407. [Google Scholar]
- Beloin C, Ghigo J-M. Finding gene-expression patterns in bacterial biofilms. Trends in Microbiology. 2005;13(1):16–19. doi: 10.1016/j.tim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-Adenosyl-l-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron . Journal of Biological Chemistry. 2011;286(29):25973–25982. doi: 10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(49):17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom J, Richards L, Pitts B, Roe F, Ehrlich G, Parker A, Mazurie A, Stewart P. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiology. 2010;10(1):294. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura M, Okuyama M, Tanzawa F, Mori H, Kitago Y, Watanabe N, Kimura A, Tanaka I, Yao M. Structural and Functional Analysis of a Glycoside Hydrolase Family 97 Enzyme from Bacteroides thetaiotaomicron. Journal of Biological Chemistry. 2008;283(52):36328–36337. doi: 10.1074/jbc.M806115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin N, Martens EC, Gordon JI, Smith TJ. Structure of a SusD Homologue, BT1043, Involved in Mucin O-Glycan Utilization in a Prominent Human Gut Symbiont. Biochemistry. 2009;48(7):1532–1542. doi: 10.1021/bi801942a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature Reviews Microbiology. 2012;10(5):323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16(7):1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Smith TJ. SusG: A unique cell-membrane-associated α-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18(2):200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Lazazzera BA. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Current Opinion in Microbiology. 2005;8(2):222–227. doi: 10.1016/j.mib.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Lindén SK, Florin THJ, McGuckin MA. Mucin dynamics in intestinal bacterial infection. PLoS ONE. 2008a;3(12):e3952. doi: 10.1371/journal.pone.0003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindén SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunology. 2008b;1(3):183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. Journal of Applied Microbiology. 2007;102(5):1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Applied and Environmental Microbiology. 2006;72(9):6204–6211. doi: 10.1128/AEM.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Woodmansey EJ, Macfarlane GT. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Applied and Environmental Microbiology. 2005;71(11):7483–7492. doi: 10.1128/AEM.71.11.7483-7492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host & Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biology. 2011;9(12):e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. Journal of Biological Chemistry. 2009;284(27):18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzorati M, Van den Abbeele P, Possemiers S, Benner J, Verstraete W, Van de Wiele T. Studying the host-microbiota interaction in the human gastrointestinal tract: basic concepts and in vitro approaches. Annals of Microbiology. 2011;61(4):709–715. [Google Scholar]
- McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nature Reviews Microbiology. 2011;9(4):265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Padler-Karavani V, Song XZ, Yu H, Hurtado-Ziola N, Huang SS, Muthana S, Chokhawala HA, Cheng JS, Verhagen A, Langereis MA, et al. Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. Journal of Biological Chemistry. 2012;287(27):22593–22608. doi: 10.1074/jbc.M112.359323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, D'Elia JN, Frias J, Salyers AA. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. Journal of Bacteriology. 1996;178(3):823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, Wang GR, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. Journal of Bacteriology. 1997;179(3):643–9. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. Journal of Bacteriology. 2000;182(19):5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141(7):1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leipi DD, Chen C-H, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. Journal of Molecular Biology. 2006;355(5):1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.