Abstract
Introduction
Identification of some somatic molecular alterations in non-small-cell lung cancer (NSCLC) has become evidence-based practice. The success and failure rate of using commercially-available tumor genotyping techniques in routine day-to-day NSCLC pathology samples is not well described. We sought to evaluate the success and failure rate of EGFR mutation, KRAS mutation, and ALK FISH in a cohort of lung cancers subjected to routine clinical tumor genotype.
Methods
Clinicopathologic data, tumor genotype success and failure rates were retrospectively compiled and analyzed from 381 patient-tumor samples.
Results
From these 381 patients with lung cancer, the mean age was 65 years, 61.2% were women, 75.9% were white, 27.8% were never smokers, 73.8% had advanced NSCLC and 86.1% had adenocarcinoma histology. The tumor tissue was obtained from surgical specimens in 48.8%, core needle biopsies in 17.9%, and as cell blocks from aspirates or fluid in 33.3% of cases. Anatomic sites for tissue collection included lung (49.3%), lymph nodes (22.3%), pleura (11.8%), bone (6.0%), brain (6.0%), among others. The overall success rate for EGFR mutation analysis was 94.2%, for KRAS mutation 91.6% and for ALK FISH 91.6%. The highest failure rates were observed when the tissue was obtained from image-guided percutaneous transthoracic core-needle biopsies (31.8%, 27.3%, and 35.3% for EGFR, KRAS, and ALK tests, respectively) and bone specimens (23.1%, 15.4%, and 23.1%, respectively). In specimens obtained from bone, the failure rates were significantly higher for biopsies than resection specimens (40% vs 0%, p=0.024 for EGFR) and for decalcified compared to non-decalcified samples (60% vs 5.5%, p=0.021 for EGFR).
Conclusions
Tumor genotype techniques are feasible in most samples, outside small image-guided percutaneous transthoracic core-needle biopsies and bone samples from core biopsies with decalcification, and therefore expansion of routine tumor genotype into the care of patients with NSCLC may not require special tissue acquisition or manipulation.
Keywords: lung cancer, non-small-cell lung cancer, never smokers, epidermal growth factor receptor, EGFR, anaplastic lymphoma kinase, ALK, KRAS, tumor genotype, failure, bone specimen, core biopsy, molecular testing
INTRODUCTION
In the United States in 2013, the expected number of new cases of and deaths from lung cancer will exceed 220,000 and 159,000, respectively (1). The overall, of all stages combined, five-year survival for the most prevalent form of lung cancer - non-small-cell lung cancer (NSCLC) - does not exceed 15% despite use of surgical resection, radiotherapy and systemic chemotherapy (1). The last decade of research in lung cancer has yielded important advances in the development of targeted therapies that target driver oncogenes (2). The most prevalent mutated or rearranged oncogenes identified in non-small cell lung cancers (NSCLCs) are v-ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), ROS1, BRAF, ERBB2 and RET (3). Specifically, mutations in EGFR and rearrangements (either inversions or translocations) involving ALK are part of the pathogenesis of some lung adenocarcinomas, predominantly in never and/or light smokers, and predict for improved outcomes with tyrosine kinase inhibitors (TKIs), such as erlotinib and crizotinib, respectively, that target these aberrant kinases (4–6). As such, molecular testing of lung cancer specimens has become part of routine clinical practice in the care for patients with advanced NSCLC.
Currently, the proposed testing guidelines from the College of American Pathologists, the International Association for the Study of Lung Cancer, the Association for Molecular Pathology, and the National Comprehensive Cancer Network recommend testing all advanced NSCLCs with an adenocarcinoma component at the time of diagnosis for EGFR mutation and for ALK fluorescence in situ hybridization (FISH) analysis (7; 8). These specimens are more frequently encountered as small biopsy or cytology specimens derived from either the primary tumor or from lymph node or distant metastatic sites, and processed as formalin-fixed, paraffin embedded tissue samples.
Limited tumor cellularity in small biopsy or cytology specimens can lead to molecular testing failure. Additionally, it is recognized that different tissue processing techniques, including acid decalcification or heavy metal fixatives, can cause DNA degradation and impede molecular analysis (9). These issues can influence the selection and success rates of specimens submitted for mutational analysis when considering sampling of bony lesions. Practically, this can pose a dilemma for the clinician and pathologist alike, in determining whether an initial biopsy used to establish the diagnosis of NSCLC is sufficient for additional molecular testing, or whether another specimen must be procured for such a purpose.
As molecular testing of NSCLCs has become a standard of clinical practice, various institutions have reported their testing protocol and experience (10–13). From these studies, one can occasionally glean the failure rates of their specimens, though to our knowledge a systematic study of genetic testing failure rates for NSCLC using commercially-available testing in a typical clinical practice setting has not been published to date. Here, we present the molecular testing efficiency for EGFR, KRAS, and ALK FISH analysis of clinical specimens from NSCLC patients from our institution over a five year time period, reflecting real-life clinical practice experience.
MATERIALS AND METHODS
Patient selection
Patients with a diagnosis of lung cancer who were seen by our providers and whose tumors were genotyped for at least EGFR mutations were identified through an ongoing Institutional Review Board (IRB) approved protocol at Beth Israel Deaconess Medical Center (BIDMC2009-P-000182). Patients and tumor pairs were excluded if genotyping was not performed. There were 381 patient-tumor specimens that were submitted to a commercial vendor for tumor genotype techniques between 2007 and 2012. The data cut off for analyses was December 19th, 2012. Study data were collected and managed using REDCap electronic data capture tools hosted at BIDMC.
Tumor processing and genotype
Surgical (i.e., either incisional or excisional biopsies that required a surgical procedure) and core needle biopsies were processed using standard techniques: 10% neutral buffered formalin fixation and paraffin-embedding. For any bone specimens that could not be cut with a scalpel at the grossing bench,, an acid decalcification was performed using RDO rapid decalcifier solution (Apex Engineering Products Corporation, Aurora, Illinois) following formalin fixation. Core needle and small biopsy specimens typically underwent a brief 15–30 minute decalcification, whereas larger surgical resection specimens were decalcified for 2–6 hours depending on the amount of calcified bone in the chosen sections. Cell aspirates or cell concentrates from fluid samples were collected into a methanol-water fixative (CytoLyt, Hologic Corp., Marlborough, MA) and a single ThinPrep slide prepared, with residual material used to create a cell block using a plasma-thrombin method prior to formalin-fixation and paraffin embedding. Once a diagnosis was established on histologic and/or immunohistiologic staining profiles as per evidence-based recommendations (8), the residual material in the formalin-fixed paraffin-embedded (FFPE) tissue blocks were submitted for molecular analysis. When multiple tissue blocks were available, the one with the highest tumor cellularity was chosen, without additional tumor microdissection or enrichment. Molecular analysis of tumor specimens was performed by a commercial vendor, Integrated Oncology (LabCorp, Esoterix Genetic Laboratories, LLC). EGFR mutation analysis was performed using standard DNA sequencing techniques with exons 18 to 21 sequenced (14; 15). For KRAS mutation analysis, DNA from exon 2 was amplified and subjected to single nucleotide primer extension to detect mutations at codons 12 and 13. ALK translocation status was analyzed using the Vysis ALK Break-Apart fluorescence in situ hybridization (FISH) probe (Abbott Molecular, Inc., Des Plaines, IL), as previously described (16). Failure of the assays was defined as insufficient/unusable material to isolate DNA or inability to perform/complete sequencing for EGFR and KRAS mutations, and lack of hybridization signals after two attempts for ALK FISH.
Data collection
Clinical, pathologic, radiographic and tumor genotyping information was collected from chart extraction. The site of biopsy (lung, lymph node, pleura, bone, brain, liver, pericardium, or adrenal) and the type of biopsy (surgical specimen [both excisional and incisional], core needle biopsy, or cell block from aspirate/fluid) were extracted from the medical record. Slides from all specimens that failed molecular testing, as well as a subset of the successfully genotyped cases were re-reviewed by a pathologist (PAV), with data compiled on tumor cellularity, use of ancillary studies, histopathologic features, and specimen processing including decalcification.
Statistical methods
Logistic regression was used for the univariable and multivariable analyses to capture the influence of each clinical predictive factor on the failure rate of tumor genotyping. The clinically relevant predictive factors were included into the multivariable analysis regardless of their statistical significance in the univariable analysis. All categorical variables were dichotomized in the regression analyses. We report odds ratio (OR) and 95% confidence interval (95%CI) to each predictive factor. Fisher’s exact test was performed to compare categorical variables. p-value < 0.05 was considered as statistically significant. All p-values we reported were two-sided. We performed our statistical analyses with STATA version 12 (STATA Corp, College Station, TX).
RESULTS
Patient and tumor characteristics
Table 1 summarizes the clinical and pathological characteristics of the 381 patient-tumor pairs that were included in our cohort.
Table 1.
Baseline characteristics of patients and tumors for which a sample was submitted for somatic genotyping technologies
| All patients(n=381) | |
|---|---|
| Age at the time of biopsy mean(SD) | 65.0(11.4) |
| Women n (%) | 233(61.2) |
| Race n (%) | |
| White | 289(75.9) |
| Asian | 50(13.1) |
| Black | 25(6.56) |
| Others | 17(4.46) |
| Smoking status n (%) | |
| Current smoker | 66(17.3) |
| Former smoker | 209(54.9) |
| Never smoker | 106(27.8) |
| Stage n (%) | |
| I | 24(6.30) |
| II | 22(5.77) |
| III | 47(12.1) |
| IV | 288(73.8) |
| Histology n (%) | |
| Adenocarcinoma | 328(86.1) |
| Squamous cell carcinoma | 11(2.89) |
| NSCLC (NOS) | 39(10.2) |
| Others | 3(0.78) |
| Anatomic site of biopsy n (%) | |
| Bone | 23(6.04) |
| Brain | 23(6.04) |
| Liver | 14(3.67) |
| Lung | 188(49.3) |
| Lymph node | 85(22.3) |
| Pleura | 45(11.8) |
| Others | 3(0.79) |
| Type of biopsy n (%) | |
| Core needle biopsy | 68(17.9) |
| Surgical | 186(48.8) |
| Cell block from FNA | 98(25.7) |
| Cell block from fluid | 29(7.61) |
|
EGFR mutation analysis Submitted samples n (%) |
381(100) |
|
KRAS mutation analysis Submitted samples n (%) |
226(59.31) |
|
ALK FISH analysis Submitted samples n (%) |
275(72.17) |
Success and failure rates of tumor genotype techniques
Out of the 381 samples, all were sent for EGFR mutation analysis, 226 for KRAS mutation analysis, and 275 for ALK FISH (Table 1). In 207 tumors (54.3%), all three tests were ordered simultaneously. Table 2 details the success and failure rates for EGFR, KRAS, and ALK analyses techniques. The success rate for EGFR mutation analysis was 94.2% and the failure rate 5.8%; for KRAS mutation 91.6% and 8.36%, respectively; and for ALK FISH 91.6% and 8.41%, respectively. In the 207 tumors in which the three tests were ordered, the success rate for EGFR was 92.3%, for KRAS 91.8% and for ALK FISH 89.9% (Table 2). Out of the 25 failure cases of the latter cohort, 11 failed EGFR, KRAS and ALK concurrently, 4 failed ALK FISH alone, 3 each failed EGFR/ALK or KRAS/ALK pairs, 2 failed KRAS alone, and 1 EGFR/KRAS or EGFR mutation analysis alone
Table 2.
Success and failure rates for EGFR mutation, KRAS mutation and ALK FISHtechniques in the submitted samples
| All samples(n=381) | |||
|---|---|---|---|
| EGFR mutation | ALK FISH | KRAS mutation | |
| Success n(%)* | 359(94.2) | 252(91.6) | 207(91.6) |
| Positive/Mutated | 86(22.6) | 23(8.36) | 71(31.4) |
| Negative/Wild-type | 273(71.7) | 229(83.3) | 136(60.2) |
| Failure n(%)* | 22(5.8) | 23(8.36) | 19(8.41) |
| Not done n(%)** | 0(0) | 106(27.8) | 155(40.7) |
| Samples who underwent all tests(n=207) | |||
| EGFR mutation | ALK FISH | KRAS mutation | |
| Success n(%)* | 191(92.3) | 186(89.9) | 190(91.8) |
| Positive/Mutated | 32(15.5) | 11(5.31) | 65(31.4) |
| Negative/Wild-type | 159(76.8) | 175(84.5) | 125(60.4) |
| Failure n(%)* | 16(7.73) | 21(10.1) | 17(8.21) |
| Not done n(%)** | 0(0) | 0(0) | 0(0) |
Percentage out of the attempted assay
Percentage out of the number of all patients
Tumor characteristics that correlate with increased failure rate
To determine the possible clinical or tumor characteristics that were associated with an increased incidence of failure of tumor genotyping techniques, we performed both univariable and multivariable logistic regressions in the 381 samples and focused on EGFR mutation analysis (due to its higher number of patient-tumor samples). Table 3 indicates the results. The patient’s sex, ethnicity, smoking history, tumor stage, and histologic subtype were not associated with an increase in failure of EGFR mutation analysis. However, non-surgical forms of tumor acquisition (core needle biopsies or cytologic specimens with cell blocks), with an odds ratio (OR) of 28.45 (p=0.002), and bone as the site of tumor acquisition, with an OR of 6.16 (p=0.010), were significantly associated with increase failure rates for EGFR mutational analysis (Table 3). Similarly, non-surgical forms of tumor acquisition, with an OR of 4.32 (p=0.006), and bone as the site of tumor acquisition, with an OR of 3.58, p=0.03, were significantly associated with increase failure rates for ALK FISH analysis (data not shown).
Table 3.
Characteristics of failure cases for EGFR mutation detection and their potential predictive factors by multivariable logistic regression
| Predictive factors | No. of patients(%) | Multivariable regression |
||
|---|---|---|---|---|
| Odds ratio |
95% CI* | p value | ||
| Male | 148(39) | 1.00 | 0.57 to | 0.45 |
| Female | 233(61) | 1.42 | 3.58 | |
| White | 289(76) | 1.21 | 0.39 to | 0.74 |
| Others | 92(24) | 1.00 | 3.64 | |
| Non-smokers | 106(28) | 1.00 | 0.45 to | 0.51 |
| Smokers | 275(72) | 1.50 | 5.00 | |
| Stage I-III | 93(24) | 1.11 | 0.29 to | 0.88 |
| Stage IV | 288(76) | 1.00 | 4.22 | |
| Adenocarcinoma | 328(86) | 1.00 | 0.63 to | 0.22 |
| Others | 53(14) | 2.15 | 7.34 | |
| Bone | 23(6) | 6.17 | 1.56 to | 0.01 |
| Others | 358(94) | 1.00 | 24.5 | |
| Surgical | 186(49) | 1.00 | 3.58 to | 0.002 |
| Non-surgical | 195(51) | 28.5 | 226.2 | |
95%CI*: 95% Confidence interval.
To better define these tumor characteristics associated with genotype failure, we analyzed the cohort for differences in success rate by site and type of tissue acquisition (Tables 4 and 5).
Table 4.
Failure rates of each test stratified by the anatomic site sampled in the complete cohort
| EGFR Mutation analysis (n=381) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Bone | Brain | Liver | Lung | Lymph node | Pleura | Others | Total | |
| Failure n | 4 | 0 | 2 | 11 | 2 | 2 | 1 | 22 |
| %Total* | 18.2 | 0 | 9.09 | 50.0 | 9.09 | 9.09 | 4.55 | 100.0 |
| %Site** | 17.4 | 0 | 14.3 | 5.85 | 2.35 | 4.44 | 33.3 | 5.77 |
| Total | 23 | 23 | 14 | 188 | 85 | 45 | 3 | 381 |
| ALK FISH analysis (n=275) | ||||||||
| Bone | Brain | Liver | Lung | Lymph node | Pleura | Others | Total | |
| Failure n | 4 | 0 | 1 | 12 | 4 | 1 | 1 | 23 |
| %Total* | 17.4 | 0 | 4.35 | 52.2 | 17.4 | 4.35 | 4.35 | 100.0 |
| %Site** | 22.2 | 0 | 12.5 | 9.1 | 6.25 | 3.33 | 50.0 | 8.36 |
| Total | 18 | 21 | 8 | 132 | 64 | 30 | 2 | 275 |
|
KRAS mutation analysis (n=226) | ||||||||
| Bone | Brain | Liver | Lung | Lymph node | Pleura | Others | Total | |
| Failure n | 2 | 0 | 2 | 9 | 4 | 2 | 0 | 19 |
| %Total* | 10.5 | 0 | 10.53 | 47.4 | 21.1 | 10.53 | 0.00 | 100.0 |
| %Site** | 14.3 | 0 | 50.0 | 8.11 | 7.41 | 8.00 | 0.00 | 8.41 |
| Total | 14 | 17 | 4 | 111 | 54 | 25 | 1 | 226 |
Percentage out of total success or failure
Percentage out of the number of attempted biopsy on the each anatomic site
Table 5.
Failure rates of each test stratified by the anatomic site sampled in the core needle biopsy group
|
EGFR Mutation analysis (n=68) | |||||||
|---|---|---|---|---|---|---|---|
| Bone | Liver | Lung (bronchos copy- guided) |
Lung (image- guided percutaneous transthoracic) |
Pleura | Others | Total | |
| Failure n | 4 | 2 | 2 | 7 | 0 | 1 | 16 |
| %Total* | 25.0 | 12.5 | 12.5 | 43.8 | 0 | 6.25 | 100.0 |
| %Site** | 44.4 | 14.3 | 11.1 | 31.8 | 0 | 50.0 | 23.5 |
| Total | 9 | 14 | 18 | 22 | 3 | 2 | 68 |
|
ALK FISH analysis (n=53) | |||||||
| Bone | Liver | Lung (bronchos Copy- guided) |
Lung (image- guided percutaneous transthoracic) |
Pleura | Others | Total | |
| Failure n | 4 | 1 | 2 | 6 | 0 | 1 | 14 |
| %Total* | 28.6 | 7.14 | 14.3 | 42.9 | 0 | 7.14 | 100.0 |
| %Site** | 44.4 | 12.5 | 12.5 | 35.3 | 0 | 100 | 26.4 |
| Total | 9 | 8 | 16 | 17 | 2 | 1 | 53 |
|
KRAS mutation analysis (n=41) | |||||||
| Bone | Liver | Lung (bronchos Copy- guided) |
Lung (image- guided percutaneous transthoracic) |
Pleura | Others | Total | |
| Failure n | 2 | 2 | 1 | 3 | 0 | 1 | 9 |
| %Total* | 22.2 | 22.2 | 11.1 | 33.3 | 0 | 11.1 | 100.0 |
| %Site** | 25.0 | 50.0 | 6.67 | 27.3 | 0 | 100 | 22.0 |
| Total | 8 | 4 | 15 | 11 | 2 | 1 | 41 |
Percentage out of total success or failure
Percentage out of the number of attempted biopsy on the each anatomic site
Type of tissue acquisition and failure rate of tumor genotype
Out of the 207 patients with concurrent testing, the highest failure rates were observed when the tissue was obtained from core needle biopsies (30.8%, 20.5%, and 30.8% for EGFR, KRAS, and ALK tests, respectively) when compared to larger surgical resection specimens (0%, 4.21%, and 3.16% for EGFR, KRAS and ALK tests, respectively) or cell block preparations (5.47%, 8.21% and 6.84% for EGFR, KRAS and ALK tests, respectively. In the latter group, cell block preparations from aspirates or fluid had similar failure rates [data not shown]).
We further on determined that out of the core needle biopsies, the failure rates were higher when the tissue of origin was either bone or lung when compared to all other sites (Table 5).
In particular, the failure rate of image-guided percutaneous transthoracic core-needle biopsies was high at 31.8% for EGFR mutations while the failure rate for bronchoscopy-guided lung transbronchial biopsies was much lower at 11.1% (Table 5). We compared the two aforementioned types of lung biopsies in relation to estimated tumor cellularity, tumor area, use of touch preparation for rapid on-site cytologic evaluation, presence of extensive desmoplastic stromal response, and number of slides cut from the paraffin block used for diagnostic immunohistochemical and ancillary studies. The most significant differences observed in the retrievable specimens were in the use of a touch preparation (image-guided percutaneous transthoracic core-needle biopsies 17/19 cases versus 0/15 cases for bronchoscopy-guided lung transbronchial biopsies, p<0.0001) and the number of slides obtained (image-guided percutaneous transthoracic core-needle biopsies had 9/14 cases with ≥ 10 slides versus 2/12 cases for bronchoscopy-guided lung transbronchial biopsies, p=0.0214). These findings may infer increased tumor tissue use or loss in image-guided percutaneous transthoracic core-needle biopsies prior to tumor genotyping.
Bone specimens and failure of tumor genotype
Table 5 shows the success and failure rate by site of tumor acquisition. The highest failure rates for tumors with more than ten specimens sent for molecular analysis were observed in samples derived from bone (Tables 4 and 5). The failure rate for EGFR mutation analysis of tumors obtained from bone was significantly higher than in all other sites combined (4/23 vs. 18/340, p=0.0357). Out of the 207 patients with concurrent testing, the failure rate for bone-derived specimens was 23.1%, 15.4%, and 23.1% for EGFR, KRAS, and ALK tests, respectively (data not shown).
We further went on to examine factors that might explain the higher failure rate in specimens originating from bone. The failure rate was significantly different based on the type of tissue acquisition, with a higher failure rate from non-surgical biopsy or cytologic cell block specimens from bone (Table 4). The failure rate was significantly higher in non-surgical biopsies versus surgical resections for EGFR mutations (40% [4/10] vs. 0% [0/13], respectively, p=0.024), ALK FISH (44.4% [4/9] vs. 0% [0/9], p= 0.082) and KRAS mutations (2/9 [22.2%] vs. 0% [0/5], p=0.505).
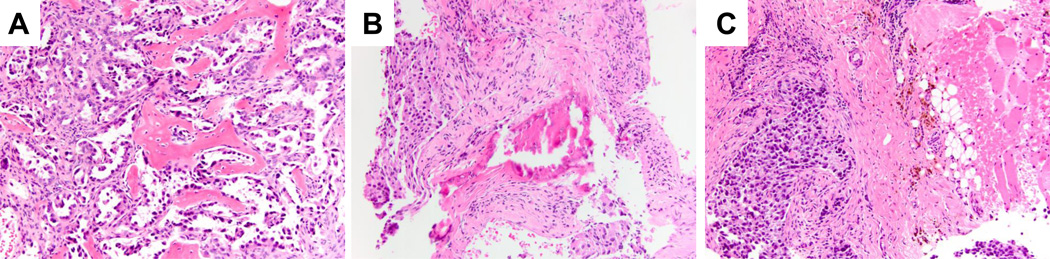
In addition, decalcification of the tumor tissue prior to preparation of the paraffin sample correlated with a higher rate of genotype failure. For EGFR mutation analysis, the failure rate for decalcified versus non-decalcified samples was significantly higher (60% [3/5] vs. 5.5% [1/18], respectively, p=0.021). Representative examples of the histologic features of bone-derived specimens that either were successful for or failed subsequent genotyping are provided in Figure 1.
FIGURE 1.
Tumor specimens obtained from bony tissue. A: Failed genotyping of a core needle biopsy from a glenoid metastasis. Although the specimen demonstrates high tumor cellularity, the extensive infiltration of bone trabeculae necessitated decacification prior to processing. Subsequent genotyping from a second specimen (cell block from a transbronchial FNA) successfully demonstrated a deletion mutation in exon 19 of the EGFR gene. B: Successful tumor genotyping of a core needle biopsy from a L3 spinal metastasis in a 64 year old female never smoker. The poorly differentiated adenocarcinoma (5 and 9 o’clock) is seen in a background of fibrosis with only rare bony trabeculae, thus decalcification was unnecessary for specimen processing. Compare the appearance of the calcified bone trabeculae with the decalcified osteoid in part A; the former demonstrates shattering or fracturing artifact, and had a more basophilic hue. C: Successfully genotyped core needle biopsy specimen of a sacral metastasis. The poorly differentiated adenocarcinoma can be seen extending into the periosseous soft tissues (adipose tissue and skeletal muscle on the right). The lack of calcified tissue in this bony metastasis obviated the need for decalcification, leading to successful tumor genotyping. All images were taken at 200x original magnification, hematoxylin and eosin stain.
DISCUSSION
We have shown that our institutional success rate for EGFR, KRAS, and ALK testing of clinical lung cancer specimens exceeds 90% using routine tissue samples and a commercial vendor. As molecular testing of non-squamous, non-small cell lung carcinomas has become the standard of care for patients with advanced stage lung cancer, numerous studies by high-volume academic centers have been published outlining mutation frequency in different patient populations (17). Less has been published regarding the actual testing success or failure of tumor specimens in clinical practice. Overall tumor genotyping success rates extracted from various studies demonstrate a range from 81–94% following initial pathologist screen for sufficient tumor cell in the specimen (10; 13; 18; 19). Comparable results have also been described more specifically for small biopsy and cytology-derived specimens, with success rates ranging from 83–100% in published studies (12; 20; 21). A large proportion of these studies perform molecular/FISH testing in-house, whereas here we describe a large cohort of patient samples with molecular/FISH testing performed by an independent commercial lab. It is encouraging to find that the overall molecular testing success rate of 90–94%, as well as the success rate for small biopsy or cytology derived specimens of 85–88%, demonstrated in this study are right in line with the reported testing success rates in the literature. Thus, comparable success rates can be achieved in a send-out manner for institutions or medical centers that either do not have in-house molecular testing capabilities, or otherwise choose to outsource such tests.
After identifying the overall tumor genotyping success rates; we performed a detailed analysis using multivariate statistics to identify those factors that might predict testing failure, and found that while clinical characteristics (patient sex, ethnicity, smoking status, tumor stage, histologic subtype) had no impact on testing failure, the method of tumor tissue acquisition and the anatomic site from which that tumor came did correlate with tumor testing success rates. The finding that smaller biopsy or cytology-derived specimens had a higher chance of testing failure than surgical specimens is clearly linked to the amount of tumor cells obtained for analysis: larger specimens generally have ample tissue for diagnostic workup and testing, whereas the tumor tissue present in small biopsy/cytology specimens may be largely used for the diagnostic work-up limiting the material available for subsequent testing. Along these lines, recommendations for limited immunohistochemical work-up and tissue conservation strategies have been put forth to both maximize the diagnostic accuracy as well as preserve as much tissue as possible for molecular testing in these small lung cancer specimens (22).
The majority of tumor genotyping failures from lung specimens were from image-guided percutaneous transthoracic core-needle biopsies. These biopsies had a high frequency of rapid on-site cytologic evaluation in the form of touch-imprint cytologic slides prepared from the core biopsies. This method of cytologic on-site assessment transfers tumor cells from the tissue core to the cytologic slide, in doing so reduces the number of tumor cells that remain in the core that is used for diagnosis and molecular testing. Additionally, it was found that more slides on average were cut from the tissue blocks of image-guided percutaneous transthoracic core-needle biopsies than other forms of lung biopsies (such as transbronchial biopsies). More slides cut from the paraffin block mean less material remaining in the block for subsequent testing. When considering other institution’s experience, a recent large-scale study for the feasibility of image-guided percutaneous transthoracic core-needle biopsies for lung tumor genotyping reports slightly lower testing failure rates (approximately 20%), though it appears as if cytologic on-site evaluation was not utilized (20).
Tumor cells obtained from bony metastasis is the other specimen characteristic that was associated with a higher rate of molecular testing failure. The routine processing of these specimens incorporating an acid decalcification step so that histologic sections can be cut poses problems for downstream molecular testing. Acid decalcification of bone tissue extensively fragments DNA and also interferes with subsequent FISH analysis (9), and should be avoided if possible for metastatic lung cancer specimens. Even when ample tumor cellularity is present, acid decalcification often causes subsequent molecular testing failure (Figure 1A). If the amount of calcified bone tissue is small and the specimen can be cut without a decalcification step, one greatly increases the chances of successful molecular testing (Figure 1B). Along these lines, a clinically or radiologically characterized “bone lesion” can in reality have little or no bone tissue in the biopsy specimen and obviates the need for tissue decalcification (Figure 1C). Therefore, it is important for pathologists and clinicians alike to recognize that just because a specimen is labeled as a bone biopsy or a bony lesion, this should not preclude molecular testing.
From the clinical standpoint, there are many factors that go into deciding what putative tumor site to biopsy, and how to obtain tissue for diagnosis and ancillary testing. For patients that initially present with clinically or radiologically apparent stage IV disease, the principal goal in obtaining a tissue diagnosis should be to both diagnosis and pathologically stage the patient’s lung cancer simultaneously, using the safest, least invasive, and least costly test available (23). Although open surgical biopsies are the gold standard for obtaining ample diagnostic tissue, the associated morbidity/mortality and cost that can be associated with this modality has motivated the shift towards less invasive small biopsy or fine needle aspiration of lesions in establishing a pathologic diagnosis of cancer. As has been shown in this report as well as others, the use of these minimally invasive small biopsy or cytologic-derived specimens - when adequate tumor cellularity is present and decalcification has not been performed - are also more than adequate for additional molecular testing in lung cancer (12; 20; 21).
In conclusion, we have presented a five-year academic institutional experience relating the feasibility and success rates for lung cancer molecular testing of tissue obtained from a broad range of specimen types and modes of tissue acquisition. Importantly, since the molecular testing was not performed in-house, but rather by an independent commercial vendor, the results and conclusions drawn from this study are broadly applicable to other institutions or practices that also send out molecular testing of lung cancer specimens. Attention paid to specific specimen characteristics can help ensure optimal selection of tumor biopsies for successful genotyping.
ACKNOWLEDMENTS
We would like to thank all current and former members of the Thoracic Oncology Clinic at Beth Israel Deaconess Medical Center, and our patients.
Funding/Grant Support: This work was funded in part through a fellowship from the American Society of Clinical Oncology Conquer Cancer Foundation (DBC), an American Cancer Society grant (RSG 11-186 to DBC), a Lung Cancer Foundation of America-International Association for the Study of Lung Cancer grant (to DBC), and National Institutes of Health (NIH) grant CA090578 (to DBC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
Daniel B. Costa has received consulting fees from Pfizer, Roche and AstraZeneca. Paul A. VanderLaan, Erik Folch, David H. Boucher, Michael S. Kent, Sidharta P. Gangadharan, Adnan Majid, Olivier N. Kocher, Michael A. Goldstein, Mark S. Huberman have no conflicts to disclose. No other conflict of interest is stated.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 3.Cheng L, Alexander RE, Maclennan GT, Cummings OW, Montironi R, Lopez-Beltran A, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25(3):347–369. doi: 10.1038/modpathol.2011.215. [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 5.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced nonsmall-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29(15):2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 7.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013 doi: 10.1016/j.jmoldx.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10(10):1236–1271. doi: 10.6004/jnccn.2012.0130. [DOI] [PubMed] [Google Scholar]
- 9.Baloglu G, Haholu A, Kucukodaci Z, Yilmaz I, Yildirim S, Baloglu H. The effects of tissue fixation alternatives on DNA content: a study on normal colon tissue. Appl Immunohistochem Mol Morphol. 2008;16(5):485–492. doi: 10.1097/PAI.0b013e31815dffa6. [DOI] [PubMed] [Google Scholar]
- 10.Cardarella S, Ortiz TM, Joshi VA, Butaney M, Jackman DM, Kwiatkowski DJ, et al. The introduction of systematic genomic testing for patients with non-small-cell lung cancer. J Thorac Oncol. 2012;7(12):1767–1774. doi: 10.1097/JTO.0b013e3182745bcb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2(5):146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rekhtman N, Brandt SM, Sigel CS, Friedlander MA, Riely GJ, Travis WD, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol. 2011;6(3):451–458. doi: 10.1097/JTO.0b013e31820517a3. [DOI] [PubMed] [Google Scholar]
- 13.Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22(12):2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15(16):5267–5273. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa DB, Nguyen KS, Cho BC, Sequist LV, Jackman DM, Riely GJ, et al. Effects of erlotinib in EGFR mutated non-small cell lung cancers with resistance to gefitinib. Clin Cancer Res. 2008;14(21):7060–7067. doi: 10.1158/1078-0432.CCR-08-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27(26):4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dogan S, Shen R, Ang DC, Johnson ML, D'Angelo SP, Paik PK, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smouse JH, Cibas ES, Janne PA, Joshi VA, Zou KH, Lindeman NI. EGFR mutations are detected comparably in cytologic and surgical pathology specimens of nonsmall cell lung cancer. Cancer. 2009;117(1):67–72. doi: 10.1002/cncy.20011. [DOI] [PubMed] [Google Scholar]
- 19.Pang B, Dettmer M, Ong CW, Dhewar AN, Gupta S, Lim GL, et al. The positive impact of cytological specimens for EGFR mutation testing in non-small cell lung cancer: a single South East Asian laboratory's analysis of 670 cases. Cytopathology. 2012;23(4):229–236. doi: 10.1111/j.1365-2303.2012.01000.x. [DOI] [PubMed] [Google Scholar]
- 20.Tam AL, Kim ES, Lee JJ, Ensor JE, Hicks ME, Tang X, et al. Feasibility of image-guided transthoracic core-needle biopsy in the BATTLE lung trial. J Thorac Oncol. 2013;8(4):436–442. doi: 10.1097/JTO.0b013e318287c91e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billah S, Stewart J, Staerkel G, Chen S, Gong Y, Guo M. EGFR and KRAS mutations in lung carcinoma: molecular testing by using cytology specimens. Cancer Cytopathol. 2011;119(2):111–117. doi: 10.1002/cncy.20151. [DOI] [PubMed] [Google Scholar]
- 22.Travis WD, Brambilla E, Noguchi M, Nicholson A, Geisinger K, Yatabe Y, et al. Diagnosis of Lung Cancer in Small Biopsies and Cytology: Implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society Classification. Arch Pathol Lab Med. 2012 doi: 10.5858/arpa.2012-0263-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yung RC. Tissue diagnosis of suspected lung cancer: selecting between bronchoscopy, transthoracic needle aspiration, and resectional biopsy. Respir Care Clin N Am. 2003;9(1):51–76. doi: 10.1016/s1078-5337(02)00083-7. [DOI] [PubMed] [Google Scholar]