Abstract
Wild species of Oryza are extremely valuable sources of genetic material that can be used to broaden the genetic background of cultivated rice, and to increase its resistance to abiotic and biotic stresses. Until recently, there was no sequence information for the BBCC Oryza genome; therefore, no special markers had been developed for this genome type. The lack of suitable markers made it difficult to search for valuable genes in the BBCC genome. The aim of this study was to develop microsatellite markers for the BBCC genome. We obtained 13,991 SSR-containing sequences and designed 14,508 primer pairs. The most abundant was hexanuclelotide (31.39%), followed by trinucleotide (27.67%) and dinucleotide (19.04%). 600 markers were selected for validation in 23 accessions of Oryza species with the BBCC genome. A set of 495 markers produced clear amplified fragments of the expected sizes. The average number of alleles per locus (Na) was 2.5, ranging from 1 to 9. The genetic diversity per locus (He) ranged from 0 to 0.844 with a mean of 0.333. The mean polymorphism information content (PIC) was 0.290, and ranged from 0 to 0.825. Of the 495 markers, 12 were only found in the BB genome, 173 were unique to the CC genome, and 198 were also present in the AA genome. These microsatellite markers could be used to evaluate the phylogenetic relationships among different Oryza genomes, and to construct a genetic linkage map for locating and identifying valuable genes in the BBCC genome, and would also for marker-assisted breeding programs that included accessions with the AA genome, especially Oryza sativa.
Introduction
The Oryza genus comprises more than 22 species with 10 recognized genomic types, six of which are diploid genome sets (2n = 24, AA, BB, CC, EE, FF, and GG) and four of which are tetraploid (2n = 4x = 48, BBCC, CCDD, HHJJ, and HHKK) [1]. According to their genome constitution, species in this genus can be classified into four main complexes [2]: Oryza ridleyi complex (including the HHJJ genome); Oryza granulate complex (including the GG genome); Oryza officinalis complex (including the BB, CC, BBCC, CCDD, and EE genomes); and Oryza sativa complex (including the AA genome). There are two cultivated Oryza species, referring to Oryza sativa and Oryza glanerrima. Asian cultivated rice (Oryza sativa) is one of the most important food crops in the world, and serves as a primary food source for more than half of the world's population [3]. In the field, cultivated rice plants are continuously damaged by various biotic and abiotic factors. The planting of modern varieties with resistance and/or tolerance genes is one of the best strategies to control pests in rice production. Some populations of wild species of Oryza have been identified as extremely valuable resources that can be used to broaden the genetic background of cultivated rice to increase its resistance to adverse factors.
The BBCC Oryza genome (2n = 4x = 48) is characteristic of allotetraploid wild species with two homologous genomes, B and C. Three species have this genome type: Oryza malampuzhaensis, which is found in India; Oryza minuta, which is endemic to Philippines and Papua New Guinea; and Oryza punctata (tetraploid, 2n = 48), which is widely distributed in Africa. The BBCC genome is related to the BB and CC genomes [1]. Only Oryza punctata (2n = 24) has the BB genome [4], [5], while Oryza officinalis, Oryza rhizomatis and Oryza eichingeri have the CC genome. These species are regarded as donors of genes that promote resistance to rice blast, bacterial leaf blight, brown planthopper, and white backed planthopper [6], [7].
However, the transfer of valuable genes from these wild species to Oryza sativa via crossing has been proved to be extremely difficult because of low seed set, hybrid sterility, and the lack of chromosome recombination [8]. There is no doubt that appropriate gene identification technologies will promote the use of genetic material from these wild species. The traditional method to identify the genomes of Oryza was to observe chromosome pairing behavior at meiotic metaphase-I in interspecific hybrids [9], [10]. However, this process was affected by genetic and environmental factors [11], [12]. Subsequently, genomic in situ hybridization (GISH) was used to identify genomes [13], followed by multicolor genomic in situ hybridization (McGISH), an improved method that used two different genomic probes simultaneously [14]. Both GISH and McGISH were complex methods with highly technical requirements. More recently, DNA molecular techniques, especially simple sequence repeat (SSR) markers, have been proved to be simple and highly effective methods for genetic analysis. A large number of SSR markers have been developed for Oryza sativa [15], [16]. While some of the SSRs developed for Oryza sativa could be amplified from other AA genomes in the Oryza genus, they were not suitable for cross-amplifications from Oryza species with different genome types [17], as preceding cross-amplifications by Miscanthus sinensis (Poaceae) and its relative [18] and Narcissus papyraceus (Amarillydaceae) and its relatives [19]. Since there had being no sequence information available for the BBCC genome, no special markers have been developed for it. This made it difficult to explore the BBCC genome to find valuable genes, and to study the phylogenetic relationships among diverse members of the Oryza genus.
Hence, the goal of this study was to develop the first set of microsatellite markers for the BBCC Oryza genome using next generation sequencing (NGS) technology. These microsatellite markers could be used to evaluate the phylogenetic relationships among different Oryza genomes, and to construct a genetic linkage map for locating and identifying valuable genes in the BBCC genome, and would also for marker-assisted breeding programs that include accessions with the AA genome, especially Oryza sativa.
Materials and Methods
Plant materials and DNA extraction
We chose seven Oryza species including 48 accessions (Table S1) in this study, referring to different ploidy levels, genomic constitutions, and genome origins. 38 accessions were obtained from the Germplasm Resource Center of the International Rice Research Institute (Los Banos, Philippines), including 23 accessions with the BBCC genome, 1 with the BB genome, and 14 with the CC genome. The other 10 accessions of Oryza sativa were obtained from the National Mid-term Genebank for Rice (Hangzhou, China).
Total genomic DNA was extracted from fresh leaves using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA).
Microsatellite loci search and SSR primer development
Genome libraries were constructed from the accession W303 (Oryza minuta) based on shotgun method, and then sequenced using the Illumina Hi Seq 2000 sequencer (Illumina Inc., San Diego, CA, USA). The genome of W303 (European Bioinformatics Institute; Accession number: PRJEB5091) was assembled using Phusion2 [20] and Phrap [21]. The N50 length of the entire assembly was calculated for the initial contigs with small contigs <1000 bp excluded.
The SSRs were identified by the software MISA (Microsatellite identification tool, http://pgrc.ipk-gatersleben.de/misa/). The primers for each unique SSR were designed using the Primer 3.0 (http://sourceforge.net/projects/primer3/). The primer design parameters were as follows: length from 18 bp to 23 bp with 21 bp as the optimum; annealing temperature between 55°C and 63°C with 60°C as the optimum; GC content from 40% to 60% with 50% as the optimum; and PCR product size between 80 bp and 250 bp.
SSR genotyping
The PCR amplifications were carried out with a 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA) in 10 μL reaction mixtures. Each reaction contained 1.0 μL 10× buffer, 1.0 μL 2 mmol/L dNTPs, 1.0 μL 25 mmol/L MgCl2, 0.6 μL each of forward and reverse primer (10 μmol/L), 0.1 μL 5 U/μL Taq polymerase, and 20 ng template DNA. The PCR cycling profile was as follows: 94°C, 2 min; 35 cycles of 94°C, 30 s, 60°C with a increase/decrease of 1°C, 30 s, and 72°C, 1 min; and 72°C, 8 min. The amplification products were analyzed by an Applied Biosystems 3130xl DNA analyzer (Applied Biosystems), and the data were processed using GeneScan and GeneMapper software (Applied Biosystems).
Statistical analysis
The average number of alleles per locus (Na), the genetic diversity per locus (He), and the polymorphic information content (PIC) were calculated with the Powermarker Software [22]. All 48 accessions were clustered using the Neighbor-Joining (NJ) tree implemented in the TreeView program [23] according to the Nei's unbiased genetic distance [24] with 100 bootstrap replications, using the Oryza sativa as an out-group.
Results
Data from sequencing and microsatellite loci detected
As shown in Table 1, a total length of the assemble sequences >1000 bp was 480,470,380 bp (n = 225,883) (http://www.ricedata.cn/down/W303_fasta.rar). The average length of the read sequences was 2,128 bp, with a maximum length of 41,615 bp and no sequences shorter than 1,000 bp.
Table 1. The statistics about the sequence assembly.
| Category | Length (bp) | Number |
| sum | 480,470,380 | 225,833 |
| ave | 2,128 | |
| largest | 41,615 | |
| N50 | 2,329 | 65,627 |
| N90 | 1,203 | 182,019 |
In total, 16,197 SSR loci were identified with discrete repeats accounting for 97% and compound repeats (C* type and C type) accounting for only 3%. We obtained 13,991 SSR-containing sequences, and 1,814 sequences contained more than one SSR. There were 503 SSRs present in compound formation (Table 2). Finally, 14,508 primer pairs were designed.
Table 2. Occurrence of the sequence analysis and microsatellites in the genome survey.
| Category | Numbers |
| Total number of sequences examined | 225,833 |
| Total size of examined sequences (bp) | 480,470,380 |
| Total number of identified SSRs | 16,197 |
| Number of SSR containing sequences | 13,991 |
| Number of sequences containing more than 1 SSR | 1,814 |
| Number of SSRs present in compound formation | 503 |
Distribution of identified microsatellite motifs and classified repeat types
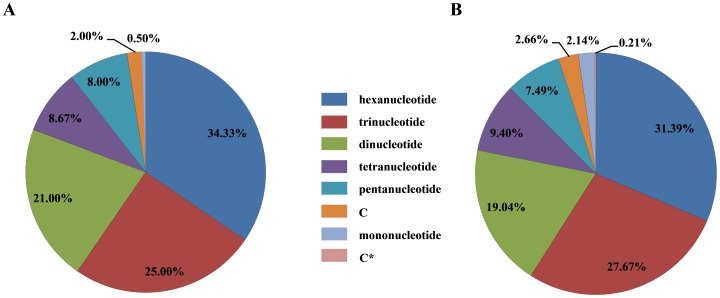
We set the following minimum length criteria in MISA to extract repeated units (unit size/minimum number of repeats): (1/18), (2/9), (3/6), (4/5), (5/4), and (6/3). The SSR motif of hexanucleotide repeats (5,090, 31.4%) was the most abundant class, followed by trinucleotide (4,529, 28.0%), dinucleotide (3,131, 19.3%), tetranucleotide (1,603, 9.9%), pentanucleotide (1,182, 7.3%) and mononucleotide repeats (662, 4.1%) (Figure 1a); the SSR motif detected at the highest frequency in each class was ATCTTT, CGC, CT, TATG, AATCT, and G, respectively. The most abundant SSR repeat type in each class was AAAAAG/CTTTTT (4.0%), AGG/CCT and CCG/CGG (16.3%), AG/CT (74.6%), ACAT/ATGT (13.7%), AGAGG/CCTCT (9.7%) and C/G (64.7%), respectively.
Figure 1. Frequencies of different classes of nucleotide repeats.
(A) 14508 primer pairs; (B) 600 selected primer pairs.
Characterization of microsatellite markers for the BBCC genome
We designed 14,508 primer pairs, and selected a set of 600 SSR markers based on proportional distribution (Figure 1). We tested the ability of the 600 primer sets to amplify SSRs from 23 accessions with the BBCC genome. Of the 600 primer pairs, 50 did not produce amplicons, probably because of mutations at the SSR locus. 55 did not amplify fragments of the expected size, probably because of In/Del mutations at the SSR locus. Of the remaining 495 microsatellite markers (Table S2, http://www.ricedata.cn/down/SSR_data.xlsx), 156 were monomorphic, and 339 were polymorphic. There were 223 single copy and 272 multi-copy markers. The mean Na value was 2.5 with a range from 1 to 9. The He value varied from 0 to 0.844 with a mean of 0.333. The mean PIC was 0.290, and ranged from 0 to 0.825. Among these markers, 46 were unique to Oryza minuta, five were unique to Oryza punctata, and none were specific to Oryza malampuzhaensis. The genetic diversity of Oryza minuta was lower than that of Oryza punctata (Table 3; Na = 1.4 vs. 1.4; He = 0.093 vs. 0.125; PIC = 0.081 vs. 0.102).
Table 3. Details of 46 and 5 microsatellites specific to Oryza minuta and Oryza punctata, respectively.
| Locus | Repeat motif | Forward primer (5'-3') | Reverse primer (5'-3') | Tm(°C) | Expected product size (bp) | Oryza minuta genetic characterization | Oryza punctata genetic haracterization | ||||||
| Especially | Na | He | PIC | Especially | Na | He | PIC | ||||||
| CN001 | (CAGCT)4 | TACAAGTGGGCTTAGGGTGG | GTCGAGCCAGTTCGTTATCC | 60 | 100 | √ | 2 | 0.298 | 0.253 | ||||
| CN019 | (CTTTTC)4 | ATCCACATGGCAAACTACCC | ACATCTTTTGCCACACATCG | 60 | 115 | √ | 1 | 0.000 | 0.000 | ||||
| CN026 | (AATT)5 | AATGTGGATTAGGCACGAGG | ACGGGCATACTAATCAACGC | 60 | 120 | √ | 2 | 0.444 | 0.346 | ||||
| CN028 | (CTTTAT)3 | CGCACGTTAATATCACCTCG | GAAGACAATTCTGGTCGATGG | 60 | 120 | √ | 1 | 0.000 | 0.000 | ||||
| CN032 | (CTCCGT)3 | GATCGATCCTTCTGGAACCC | CAGTCGGAGGAGAAAAGTGC | 60 | 122 | √ | 2 | 0.180 | 0.164 | ||||
| CN036 | (ATCTAT)3 | ATAGATCCCACGTGTCAGCC | GTCTTGGACTCGGATTTTGC | 60 | 125 | √ | 1 | 0.000 | 0.000 | ||||
| CN040 | (AAAC)5 | GCAGTCATCGAGTCCCTAGC | TGCTTACTCATCATCCTGCG | 60 | 128 | √ | 1 | 0.000 | 0.000 | ||||
| CN049 | (TC)14 | TACACGCCTTTTGTTCTTCG | CAACGATGATTATGATGCGG | 60 | 133 | √ | 1 | 0.000 | 0.000 | ||||
| CN066 | (AAGATA)3 | ACCTGCATCCTACACTTGCC | TATTGTCACCTCGTTTTGCG | 60 | 146 | √ | 3 | 0.292 | 0.272 | ||||
| CN070 | (GAATCG)3 | TAAGGATGAAAACCGCTTGG | CCGTATTTGCTCAGTTTTCG | 58 | 148 | √ | 1 | 0.000 | 0.000 | ||||
| CN079 | (AGAGGG)3 | AATCTGTCAATGGGCAGTCC | CGCAACTCACATAGAAACGG | 59 | 153 | √ | 3 | 0.512 | 0.444 | ||||
| CN081 | (CTG)6 | AATGCACAACAAGTCTCCCC | ATCTGGAAGGAGCAATGACG | 60 | 154 | √ | 1 | 0.000 | 0.000 | ||||
| CN082 | (AAAATG)3 | ATTTTGTTCCGATGGTCTCG | GTTAGGGATGAAAACGGTCG | 59 | 154 | √ | 1 | 0.000 | 0.000 | ||||
| CN093 | (CAAC)5 | TTGGTGATCGAGCACATAGC | AGATTGATTCACATGCGTGG | 60 | 159 | √ | 1 | 0.000 | 0.000 | ||||
| CN099 | (AAAGG)4 | TCTGTGGATCACAAGCAAGC | ATAAAAAGGGAAGGCATCCG | 60 | 161 | √ | 1 | 0.000 | 0.000 | ||||
| CN102 | (ATATAC)3 | TGGAGGGTTATAATCAGCGG | AAGACATTGGAGCTTGACCG | 60 | 161 | √ | 2 | 0.444 | 0.346 | ||||
| CN108 | (TTTAT)5 | GGGGAGAAATACCGGTAAGC | ACCTCACATCTCAACCCTGC | 60 | 165 | √ | 1 | 0.000 | 0.000 | ||||
| CN124 | (TTGTG)4 | ATTCAGATTCACCTCCGACG | ACCCACGAAAAGGTGTATCG | 60 | 169 | √ | 1 | 0.000 | 0.000 | ||||
| CN150 | (CGC)8 | TAATCCGAGGACCAAAGTGC | CTGAGCGTAGGATGAGGAGG | 60 | 178 | √ | 4 | 0.597 | 0.552 | ||||
| CN156 | (AGG)6 | ACGTCGACCTCTTCACAAGC | CTTCCTCCACAGCTCACTCC | 60 | 179 | √ | 2 | 0.298 | 0.253 | ||||
| CN171 | (ATGAAG)3 | GGAGCACATGGAAGAGAAGC | AATGGATTTCTCGTTTTGCG | 60 | 184 | √ | 1 | 0.000 | 0.000 | ||||
| CN174 | (T)18 | TACCAGCTCCTTCTGATGGC | ACTTGTTAATCCAGTGGCGG | 60 | 185 | √ | 1 | 0.000 | 0.000 | ||||
| CN181 | (CAACGG)3 | TCTGACGATGCAATCAAAGC | TTGTCGTAAGCAGCAACTCG | 60 | 185 | √ | 1 | 0.000 | 0.000 | ||||
| CN189 | (CCTCGT)3 | ATCGATGCACACTCAACTGC | ACAATGATGGAGAGGAACGC | 60 | 188 | √ | 2 | 0.153 | 0.141 | ||||
| CN206 | (CCAAAT)3 | TGCCATATTTGAGCTGATGC | CAAGAGATGGAGGAGCAAGG | 60 | 194 | √ | 1 | 0.000 | 0.000 | ||||
| CN207 | (CGGTCA)3 | GGCTTAAAACCAAACCCTCC | TTGTGTAGTGAGGCGAGTGC | 60 | 194 | √ | 1 | 0.000 | 0.000 | ||||
| CN208 | (TC)9 | AACCCTAGTTTTCCCATCCG | AGGAGCCGATCTAGAGGTCC | 60 | 195 | √ | 3 | 0.542 | 0.460 | ||||
| CN210 | (ACAT)5acacacat(ATAC) 7atatatatgtgtgtg(TATGTA)3 | ATCGGTATCATATGCAGCGG | TTTGCTACATCCAACATGTGC | 60 | 198 | √ | 1 | 0.000 | 0.000 | ||||
| CN212 | (GCGAG)4 | TATGTCTCGTCACAGCTGGC | GAGACGGGTAGGTAGGGAGG | 60 | 198 | √ | 1 | 0.000 | 0.000 | ||||
| CN217 | (CCAGCG)3 | GGCTCCTGAAAACAATCTGC | TTCCAATCTCTCCCATCTCG | 60 | 199 | √ | 1 | 0.000 | 0.000 | ||||
| CN253 | (TCTTC)4 | GGACGAAAAACCTAATCCCC | TAACACTGATCCGCACAACG | 61 | 208 | √ | 2 | 0.165 | 0.152 | ||||
| CN275 | (CAACGG)3 | CTCTTATGCCAAATCCGACG | GCATTTTGGTATTTCCACCG | 60 | 211 | √ | 2 | 0.153 | 0.141 | ||||
| CN281 | (GCGA)5 | CATGATTGAACTGGTGACCG | GCGTGGGTAGAGAGAGATGG | 60 | 213 | √ | 1 | 0.000 | 0.000 | ||||
| CN283 | (CT)9 | GATGAGGGTGACAGAGAGGC | AGTGTATCTTGCTCCACCGC | 60 | 214 | √ | 1 | 0.000 | 0.000 | ||||
| CN287 | (CTTTAT)3 | CGCACGTTAATATCACCTCG | TTAAGAAGGCAAATCGGAGC | 59 | 214 | √ | 1 | 0.000 | 0.000 | ||||
| CN300 | (CAATGG)3 | GAGACAGCCAACTCCTACGC | TCGGCTACATTGTGTGAAGC | 60 | 218 | √ | 1 | 0.000 | 0.000 | ||||
| CN310 | (CCG)6 | TGGGAATGAGAAGGAAGACG | TGTCCGCTACTACTGATGCG | 60 | 221 | √ | 1 | 0.000 | 0.000 | ||||
| CN330 | (AGTGCT)3 | CCTCCTGCTTCACAAACTCC | ACGATATGCTCCCATGTTCC | 60 | 225 | √ | 1 | 0.000 | 0.000 | ||||
| CN348 | (CGCCCG)3 | ACCTTCCTCCTCAACTTCCC | CTTGAAAATTCGGGTTAGCG | 60 | 229 | √ | 1 | 0.000 | 0.000 | ||||
| CN356 | (CTTCAG)3 | CTCAACAGTTCAAATGGATTGC | TTTGTGCTGTGAAAGCAAGG | 60 | 230 | √ | 1 | 0.000 | 0.000 | ||||
| CN379 | (ATCC)5 | ATCGCTTCTCTCCCTTAGCC | AAATGCTCAGTGGGTTTTGG | 60 | 235 | √ | 1 | 0.000 | 0.000 | ||||
| CN387 | (CTC)7 | TGTCGTTGTCACCTTCTTCG | GCGAGAATAAGCTGTTTGGC | 60 | 237 | √ | 1 | 0.000 | 0.000 | ||||
| CN391 | (AGA)9 | AGTGGGCTACATGAGATGGC | GCATCCTGTTCTTGACCACC | 61 | 238 | √ | 2 | 0.153 | 0.141 | ||||
| CN418 | (CGGAAA)4 | TCTGCTGTGGTAAAAACCCC | TTTCGAAAAGTTTCCAACCG | 60 | 241 | √ | 2 | 0.153 | 0.141 | ||||
| CN434 | (CTAGCT)3 | GGAAGCTCATGACAACCTCC | CTCTTGTTGAGCTTGCCTCC | 60 | 245 | √ | 1 | 0.000 | 0.000 | ||||
| CN453 | (GTC)6gctcccttgctcctcgccat gcttcgggtgaagctggcgagg(GTG)6 | CTGATCACCATGTACCACGC | GAACCTGTCACCGATCATGG | 61 | 251 | √ | 3 | 0.500 | 0.449 | ||||
| CN467 | (ATCTAA)3(TCTAAA)4 | TGCAAAAGAAAAGCTAATCGG | GGAAAAGAAACGTGCAAAGC | 60 | 267 | √ | 1 | 0.000 | 0.000 | ||||
| CN476 | (GA)11 | AAAAATTGTCTGCCGTCTGG | AACCCTAGAATTCCCATCCG | 60 | 271 | √ | 1 | 0.000 | 0.000 | ||||
| CN478 | (AATAGG)3 | ACCAATCTTGGGAAGACACG | TCCCTTCTCATATCGCATCC | 60 | 271 | √ | 1 | 0.000 | 0.000 | ||||
| CN486 | (AAAAGT)3 | AAACCTTGGAAAAGGCTTGG | CTTGAGGAGTCCACTCTCCG | 60 | 276 | √ | 1 | 0.000 | 0.000 | ||||
| CN490 | (TG)9 | GAATGGCTGTGTCAATGTGG | ATGACTCATCTCAATCGGGC | 60 | 279 | √ | 1 | 0.000 | 0.000 | ||||
| Mean | 1.4 | 0.093 | 0.081 | 1.4 | 0.125 | 0.102 | |||||||
Cross-amplification from other related genomes
Next, we evaluated the suitability of these 495 markers for use in other closely related species. Of the 495 markers, only 12 (2.4%) were specific to the BB genome, 173 (34.9%) were specific to the CC genome, and 299 (60.4%) were common to the BB, CC, and BBCC genomes. Eleven markers (2.2%) were neither in the BB nor the CC genome. Most interestingly, 198 markers (40.0%) were also present in the AA genome.
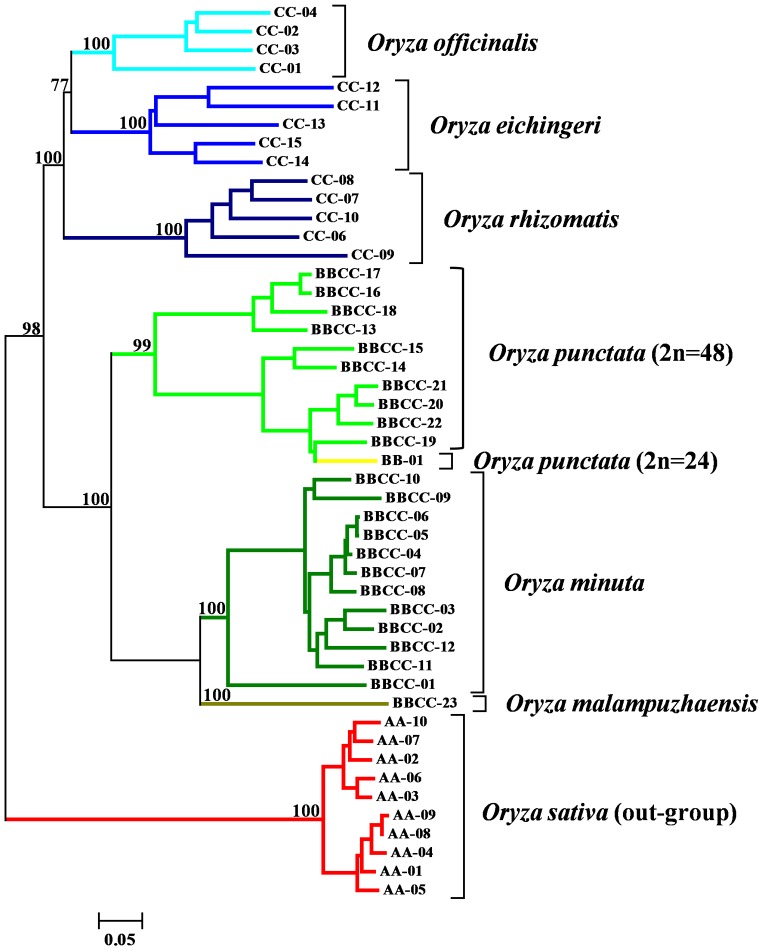
The phylogenetic tree (Figure 2) grouped the 48 accessions into two significant, distinct clusters. Cluster I consisted of the BB, CC, and BBCC genome species; and cluster II consisted of the AA genome species. Cluster I was further divided into two groups, one consisting of species with the BBCC and BB genomes, and the other consisting of species with the CC genome. Within the BBCC genome, Oryza minuta and Oryza punctata formed different subgroups. Oryza malampuzhaensis was more closely related to Oryza minuta than to Oryza punctata. Among the species with the CC genome, Oryza eichingeri was more closely related to Oryza officinalis than to Oryza rhizomatis. In cluster II, Oryza sativa indica and Oryza sativa japonica were clearly divided into two groups. The groups in the NJ tree were consistent with the intrinsic relationships among Oryza species [17], and further confirmed the usefulness of the new developmental microsatellite markers in genetic analyses.
Figure 2. Neighbor-Joining tree of 48 accessions based on Nei's unbiased genetic distance from 495 SSR markers.
Bootstrap values (out of 100) are indicated at the branch points.
Discussion
We developed the first set of microsatellite markers for the BBCC Oryza genome. The SSRs were located in both coding and non-coding regions, and therefore, they would be useful for genetic and evolutionary analyses, high-throughput mapping, and marker-assisted plant improvement strategies. In this study, 82.5% of selected markers produced clear amplified fragments of the expected sizes. This was similar to the success rate of 60–90% amplification reported elsewhere [25]. Among these markers, 12 were specific to the BB genome and 173 were unique to the CC genome. Thus, these unique microsatellite markers could be developed as probes to identify different species and various genomes. We evaluated the transferability rates of the markers in different Oryza species. The transferability rate between Oryza minuta and Oryza punctata was 89.7%. This was higher than that for Oryza species with the BB, CC, and BBCC genomes (60.4%), and that between AA and BBCC genomes (40.0%). These high transferability rates suggested that different species or genomes within the Oryza genus were closely related.
Our results showed that hexanucleotide repeat motif (31.4%) was the most abundant repeat type, followed by trinucleotide (28.0%) and dinucleotide (19.3%). These findings differed from those of previous studies in which dinucleotide or trinucleotide repeats were reported to be the most abundant motifs in genomes of cultivated rice [16], [26], and pentanucleotide repeats (30.5%) were the most abundant type in Gossypium raimondii [17]. The nature of the microsatellites obtained was related not only to the thresholds used to define the microsatellites, but also to genome organization, since heterogeneity could lead to differences in microsatellite size [27]. The most common hexanucleotide motif was AAAAAG/CTTTTT (4.0%), which made up a much lower proportion than that of the most common motif in faba bean, ACACGC/CGTGTG (49.5%) [28]. The main trinucleotide repeats were AGG/CTT and CCG/CGG, representing 16.3% of all of the trinucleotide repeats analyzed. The most common trinucleotide repeats were AGG/CTT in Amorphophallus [25], and CGG/GCC in cultivated rice [16], [26]. These results provided further evidence that the CCG/CGG motif was very common in monocots [29]. This reflected the strong conservation of synteny among genomes of diverse monocots, and could result from a high GC content and codon bias [30], [31].
In previous studies, mitochondrial restriction fragment length polymorphisms (RFLPs) [32] and inter simple sequence repeat (ISSR) [33] markers had been used to study genetic relationships among members of the Oryza genus. However, these analyses could only distinguish the AA genome from other types, and could not separate other related genomes, such as the BB, CC, and BBCC genomes. In contrast, the SSR markers developed from the BBCC genome were able to differentiate the AA, BB, CC, and BBCC genomes, and also distinguished the BB and CC genomes from the BBCC genome, even identified various species within the AA, CC, and BBCC genomes. Thus, the relationships predicted from analyses using these markers were consistent with the established evolutionary relationships among members of the Oryza genus [17]. Despite this, a new marker, SNP (Single Nucleotide Polymorphism), is now on the scene and has gained increasing popularity. In terms of genetic information provided, as simple bi-allelic co-dominant markers, they can be considered as a step backwards when compared to the highly informative multi-allelic microsatellites [34].
The NJ tree further revealed that the BB genome species were more closely related to species with the BBCC genome than to those with the CC genome, demonstrating that the BB genome was the maternal parent of the BBCC genome [35], [36] and CC species evolved later [37]. Oryza malampuzhaensis and Oryza officinalis, both of which had the BBCC genome, shared similar morphologies; in fact, Oryza malampuzhaensis was considered to be a subspecies of Oryza officinalis [38]. There were clear differences in the panicle and spikelet between these two species [14]. Our results showed that Oryza malampuzhaensis was more closely related to Oryza minuta than to Oryza officinalis, consistent with the fact that Oryza malampuzhaensis was an allotetraploid with the BBCC genome [39] while Oryza officinalis was a diploid with the CC genome.
Conclusions
We present the first set of microsatellite markers from the nuclear BBCC Oryza genome. Our results showed that the high-throughput approach for sequencing was useful for obtaining many high quality SSR markers. These markers can be used to study the origins and evolutionary relationships among members of the Oryza genus, and could also be used to construct physical maps and for map-based gene cloning from the BBCC genome to identify valuable genes. Furthermore, they could be used for marker-assisted trait selection in cultivated rice breeding programs. By using the pre-existing sequence information, the further analysis will focus on the SNPs development which is known as a new marker.
Supporting Information
Oryza species and accessions used in this study.
(XLS)
Characteristics of 495 microsatellite markers producing clear amplified fragments of the expected sizes.
(XLS)
Acknowledgments
The authors thank Dr. Qiang Zhao and Dr. Qi Feng (National Center for Gene Research, Chinese Academy of Sciences) for assembling genome sequences. We also thank Dr. Bin Han (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for critical comments in the manuscript. We thank the International Rice Research Institute (Los Banos, Philippines) for providing wild seeds.
Funding Statement
This work was supported by the National Key Technology R&D Program in China (2013BAD01B02-14). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ge S, Sang T, Lu B, Hong D (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proceedings of the National Academy of Sciences 96: 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Current Opinion in Plant Biology 6: 139–146. [DOI] [PubMed] [Google Scholar]
- 3.Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Oryza: From Molecule to Plant: Springer. pp. 25–34. [PubMed]
- 4. Tateoka T (1962) Taxonomic studies of Oryza. I. O latifolia complex. II. Several species complexes. Bot Mag, Tokyo 75: 418–427. [Google Scholar]
- 5. Tateoka T (1962) Taxonomic studies of Oryza II. Several species complexes. Bot Mag Tokyo 75: 455–461. [Google Scholar]
- 6.Heinrichs E, Medrano F, Rapusas H (1985) Genetic evaluation for insect resistance in rice: IRRI (free PDF download).
- 7. Amante-Bordeos A, Sitch L, Nelson R, Dalmacio R, Oliva N, et al. (1992) Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa . Theoretical and Applied Genetics 84: 345–354. [DOI] [PubMed] [Google Scholar]
- 8. Sitch L, Dalmacio R, Romero G (1989) Crossability of wild Oryza species and their potential use for improvement of cultivated rice. Rice Genet Newsl 6: 58–60. [Google Scholar]
- 9.Ho KC, Lic H (1961) Cytogenetical studies of Oryza sativa L. and its related sepecies
- 10. Katayama T (1977) Cytogenetical studies on the genus Oryza . Japanese Journal of Genetics 52: 301–307. [Google Scholar]
- 11. Gupta P, Fedak G (1985) Genetic control of meiotic chromosome pairing in polyploids in the genus Hordeum. Canadian journal of genetics and cytology 27: 515–530. [Google Scholar]
- 12.Jauhar P, Joppa L (1996) Chromosome pairing as a tool in genome analysis: merits and limitation. Methods of genome analysis in plants CRC Press, Boca Raton 21.
- 13. Fukui K, Shishido R, Kinoshita T (1997) Identification of the rice D-genome chromosomes by genomic in situ hybridisation. Theoretical and Applied Genetics 95: 1239–1245. [Google Scholar]
- 14. Li C, Zhang D, Ge S, Lu B, Hong D (2001) Identification of genome constitution of Oryza malampuzhaensis, O. minuta, and O. punctata by multicolor genomic in situ hybridization. Theoretical and Applied Genetics 103: 204–211. [Google Scholar]
- 15. Harushima Y, Yano M, Shomura A, Sato M, Shimano T, et al. (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA research 9: 199–207. [DOI] [PubMed] [Google Scholar]
- 17. Zou X, Zhang F, Zhang J, Zang L, Tang L, et al. (2008) Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol 9: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou H, Li S, Ge S (2011) Development of microsatellite markers for Miscanthus sinensis (Poaceae) and cross-amplification in other related species. American Journal of Botany 98: e195–e197. [DOI] [PubMed] [Google Scholar]
- 19. Simón VI, Picó FX, Arroyo J (2010) New microsatellite loci for Narcissus papyraceus (Amarillydaceae) and cross-amplification in other congeneric species. American journal of botany 97: e10–e13. [DOI] [PubMed] [Google Scholar]
- 20. Mullikin JC, Ning Z (2003) The phusion assembler. Genome research 13: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bastide M, McCombie WR (2007) Assembling genomic DNA sequences with PHRAP. Current Protocols in Bioinformatics: 11.14. 11–11 14: 15. [DOI] [PubMed] [Google Scholar]
- 22. Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129. [DOI] [PubMed] [Google Scholar]
- 23.Page RD (2001) TreeView. Glasgow University, Glasgow, UK.
- 24. Nei M (1973) Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng X, Pan C, Diao Y, You Y, Yang C, et al. (2013) Development of microsatellite markers by transcriptome sequencing in two species of Amorphophallus (Araceae). BMC genomics 14: 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akagi H, Yokozeki Y, Inagaki A, Fujimura T (1996) Microsatellite DNA markers for rice chromosomes. Theoretical and Applied Genetics 93: 1071–1077. [DOI] [PubMed] [Google Scholar]
- 27. Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nature Reviews Genetics 5: 435–445. [DOI] [PubMed] [Google Scholar]
- 28. Yang T, Bao S, Ford R, Jia T, Guan J, et al. (2012) High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC genomics 13: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z, Li J, Luo Z, Huang L, Chen X, et al. (2011) Characterization and development of EST-derived SSR markers in cultivated sweetpotato (Ipomoea batatas). BMC plant biology 11: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nature genetics 30: 194–200. [DOI] [PubMed] [Google Scholar]
- 31. La Rota M, Kantety RV, Yu J, Sorrells ME (2005) Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. Bmc Genomics 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abe T, Edanami T, Adachi E, Sasahara T (1999) Phylogenetic relationships in the genus Oryza based on mitochondrial RFLPs. Genes & genetic systems 74: 23–27. [DOI] [PubMed] [Google Scholar]
- 33. Joshi S, Gupta V, Aggarwal R, Ranjekar P, Brar D (2000) Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza . Theoretical and Applied Genetics 100: 1311–1320. [Google Scholar]
- 34. Vignal A, Milan D, SanCristobal M, Eggen A (2002) A review on SNP and other types of molecular markers and their use in animal genetics. Genetics Selection Evolution 34: 275–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dally A, Second G (1990) Chloroplast DNA diversity in wild and cultivated species of rice (Genus Oryza, section Oryza). Cladistic-mutation and genetic-distance analysis. Theoretical and Applied Genetics 80: 209–222. [DOI] [PubMed] [Google Scholar]
- 36. Kanno A, Hirai A (1992) Comparative studies of the structure of chloroplast DNA from four species of Oryza: cloning and physical maps. Theoretical and Applied Genetics 83: 791–798. [DOI] [PubMed] [Google Scholar]
- 37. Nishikawa T, Vaughan DA, Kadowaki K (2005) Phylogenetic analysis of Oryza species, based on simple sequence repeats and their flanking nucleotide sequences from the mitochondrial and chloroplast genomes. Theoretical and Applied Genetics 110: 696–705. [DOI] [PubMed] [Google Scholar]
- 38. Tateoka T (1963) Taxonomic studies of Oryza. III. Key to the species and their enumeration. Bot Mag, Tokyo 76: 165–173. [Google Scholar]
- 39. Krishnawamy N, Chandrasekharan P (1957) Note on a naturally occurring tetraploid species of Oryza . Science Culture 23: 308–310. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oryza species and accessions used in this study.
(XLS)
Characteristics of 495 microsatellite markers producing clear amplified fragments of the expected sizes.
(XLS)