Abstract
Fragile X syndrome is caused by loss of Fragile X Mental Retardation Protein (FMRP), an RNA binding protein that suppresses protein translation. Here, we identified Down Syndrome Cell Adhesion Molecule (Dscam) RNA, a molecule involved in neural development and implicated in Down syndrome, bound to FMRP. Elevated Dscam protein levels in Drosophila FMRP null animals and in animals with three copies of the Dscam gene both produced specific and similar synaptic targeting errors in a hard-wired neural circuit which impaired the animal’s sensory perception. Reducing Dscam levels in FMRP null animals reduced synaptic targeting errors and rescued behavioral responses. Our results demonstrate that excess Dscam protein may be a common molecular mechanism underlying altered neural wiring in major causes of intellectual disability.
Down syndrome and Fragile X syndrome are two of the most common causes of intellectual disability 1,2. A hallmark of both of these syndromes is elevated protein expression. In Down syndrome this is a consequence of having three copies of Chromosome 21 and the extraneous expression of the thousands of genes located there. In Fragile X syndrome, silencing of the Fragile X Mental Retardation gene leads to loss of its protein product, Fragile X Mental Retardation Protein (FMRP). FMRP binds RNA targets to suppress their protein translation; thus in Fragile X syndrome, loss of FMRP results in excessive protein synthesis of the RNAs that FMRP would normally suppress 2. Thousands of RNA targets of FMRP have been discovered using high-throughput RNA sequencing or microarray screens in an effort to identify key molecules involved in Fragile X syndrome and Autism Spectrum Disorders 3–5. However, it is not known whether unregulated expression of specific molecules common to Down syndrome, Fragile X syndrome, and Autism Spectrum Disorders might be responsible for their overlapping neural phenotypes.
One RNA target identified in these screens is Down Syndrome Cell Adhesion Molecule (Dscam) 4,5. In humans, Dscam is a large gene (~800 kilobases) located in the Down Syndrome Critical Region, a 4 Megabase region in Chromosome 21 implicated in many Down syndrome phenotypes 6–12. Dscam is an immunoglobulin cell-surface receptor and has conserved functions in neural development across invertebrates and vertebrates such as axon guidance, axonal and dendritic branching and targeting, and synapse maturation 13. Here, we identify Dscam RNA as a target for protein translation regulation by FMRP in Drosophila brains, and examine how overexpression of Drosophila Dscam protein through gene triplication or through loss of translational suppression by FMRP impairs synaptic targeting precision and neural circuit function.
Results
FMRP binds Dscam mRNA to suppress its translation
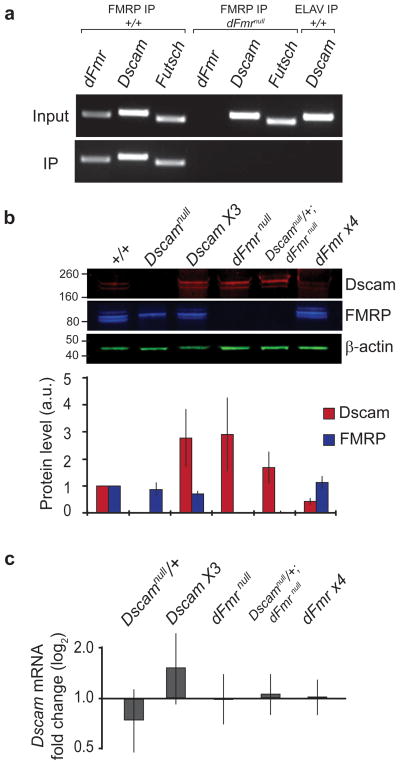
We identified Dscam RNA as a target of FMRP by immunoprecipitation of FMRP from Drosophila brains (Fig. 1a). This RNA-protein interaction was specific for FMRP, as Dscam RNA did not immunoprecipitate with a different neuronal RNA binding protein, ELAV, nor in FMRP null mutant brains (Fig. 1a). The Dscam mRNA and FMRP interaction is required for the suppression of Dscam protein translation, as Dscam protein levels were elevated in FMRP null mutants at amounts similar to animals with 3 copies of the Dscam gene (Fig. 1b). Conversely, animals with multiple copies of the Drosophila Fragile X Mental Retardation (dFmr) gene that overexpress FMRP had less Dscam protein expression than wildtype (Fig. 1b). Loss of FMRP resulted in large increases in Dscam protein levels, and conversely, even modest increases in FMRP levels decreased Dscam protein by approximately 60% (Fig. 1b), demonstrating a tight regulation of Dscam protein translation by FMRP. These results demonstrate that FMRP suppresses Dscam protein expression at the level of translation, as Dscam mRNA levels remained unchanged in FMRP null animals (Fig. 1c). To understand how this regulation of Dscam expression by FMRP is involved in neural wiring, we used the hard-wired mechanosensory neural circuit to quantitatively analyze axonal targeting decisions 14–16 (Fig. 2). A single mechanosensory neuron innervates a single bristle on the back of the fly, and because each bristle is uniquely identifiable, the same neuron among different animals can be identified based on the location of its corresponding bristle. In this study, we focused our analysis on the left and right posterior scutellar (pSc) neurons, and we verified FMRP expression within identified pSc neurons using immunohistochemistry in combination with fluorescent in situ hybridization for Dscam mRNA (Fig. 2a-d). The pSc neuron extends its axon into the central nervous system and synapses with specific interneurons, giving it a stereotyped and unique axonal arbour (Fig. 2e)14. To quantitate the variability of this synaptic targeting in wildtype animals, we measured the branch lengths and positions of the pSc axonal arbour in 74 wildtype animals and identified a prototypical “skeleton” comprised of 16 core branches occurring at >80% frequency for primary and secondary branches and >60% frequency for tertiary branches (Fig. 2f, g). Wildtype variability was then defined as branching phenotypes that occurred between 10% and 60% frequency, and targeting errors were defined as those occurring at <10% in wildtype (see Methods).
Figure 1. Fragile X Mental Retardation Protein (FMRP) suppresses Down syndrome cell adhesion molecule (Dscam) protein translation.
a, FMRP binds Dscam mRNA. FMRP-mRNA complexes were immunoprecipitated from Drosophila larval brains and specific targets were identified by RT-PCR. FMRP has been previously shown to bind its own mRNA and Futsch. No mRNAs were immunoprecipitated from Fragile X mutants (dFmrnull IP), and Dscam mRNA did not immunoprecipitate with another neuronal RNA-binding protein, ELAV.
b, Loss of FMRP in Fragile X mutants increases neuronal Dscam protein amounts. Representative fluorescent immunoblots of Dscam, FMRP, and actin in different genotypes. Protein samples for Dscamnull animals were prepared from embryos and showed restricted expression of FMRP isoforms. Dscam and FMRP protein intensities were normalized against actin (plotted in arbitrary units, a.u.), and the averages from 9 experiments are shown. Errors bars are standard error of the mean.
c, Quantitative real-time PCR analysis indicates that Dscam mRNA levels are not significantly altered in Fragile X mutants. Dscam mRNA for all experimental genotypes was measured as fold changes from wildtype levels. The averages from 6 experimental replicates are shown. Error bars are standard deviation of the mean.
Figure 2. The posterior scutellar (pSc) mechanosensory neuron is identifiable between animals based on the location of its corresponding bristle.
a-d, The pSc neuron expresses FMRP and Dscam. A cross section through a pSc bristle is shown in brightfield (a), and the corresponding FMRP immunofluorescence (green) within the pSc neuron (arrow) is shown in b. c, No detectable FMRP signal is observed in dFmrnull animals. d, Co-localization of FMRP and Dscam mRNA was observed in pSc neurons using fluorescence in situ hybridization for Dscam mRNA (magenta) combined with fluorescence immunohistochemistry for FMRP (green). Arrowheads point to Dscam mRNA puncta, arrow points to FMRP signal. Nuclei are stained in blue. Scale bars, 20μm.
e, A single mechanosensory neuron innervates a single bristle. The axonal projection into the central nervous system of the right posterior scutellar mechanosensory neuron is shown in red.
f, g, The stereotyped synaptic connectivity of the pSc neuron is used as a readout for synaptic targeting errors. f, The pSc axonal arbour has a complex and stereotyped branching pattern. Quantitative analysis of wildtype pSc axons revealed 16 core branches (yellow lines) and 2 variable branches occurring in 50% of animals (blue lines). g, Individual branches of the pSc axonal arbour can be identified between animals, and their lengths and variance can be quantified. Black lines represent the average lengths of each branch, red lines represent the standard deviations, and values are in μm.
Elevated Dscam levels produce axonal targeting errors
Quantitative analysis of dFmrnull animals revealed a significant increase in the total branch length of the pSc axonal arbour due to a significant increase in the number of ectopic branches in the mutants (3.4 branches, n = 99, p < 0.001) compared to wildtype (1.6 branches, n = 74) (Fig. 3a). These increases in axonal arbor sizes in Fragile X mutants were not due to non-specific overall growth, as the lengths of the branches that comprised the pSc “skeleton” were unaffected (Fig. 3b). Ectopic branches in the dFmrnull animals were highly specific and sprouted at identifiable locations from the prototypical pSc skeleton within the anterior, middle, and posterior regions of the central nervous system (Fig. 3). However, Fragile X mutants had many more targeting errors besides ectopic branches, and these errors were also stereotyped and included branch misrouting and midline crossing errors, and missing branches from the skeleton (Fig. 3). As expected from a total loss of FMRP regulation of many RNA targets, more than 85% of dFmrnull animals had targeting errors, with 59% also having multiple errors within their axonal arbours compared to only 2% of wildtype animals (p < 0.001). We confirmed that these errors were due to loss of FMRP within mechanosensory neurons by using a specific Gal4 driver (455-Gal4) to express dsRNA against dFmr only within the four neurons on the scutellum of the fly 16,17. Axonal targeting errors within these mosaic animals phenocopied the targeting errors observed in whole animal Fragile X mutants (Supplementary Fig. 1a).
Figure 3. Elevated Dscam protein levels produce specific axonal targeting errors.
a, Ectopic branch number and length are increased in dFmrnull and Dscam X3 animals.
b, The core pSc skeleton does not change in branch number or lengths among different genotypes.
c, d, Axonal branch targeting is impaired in Fragile X mutants. Compared to the stereotyped axonal branching pattern of wildtype pSc neurons (c), animals lacking FMRP (d) have specific targeting errors, such as misrouting and aberrant midline crossing branches (arrows). Dotted line marks the midline of the central nervous system. Scale bar, 50μm.
e, Dscam X3 animals have targeting errors (arrows) similar to those observed in Fragile X mutants.
f, Reducing Dscam levels in Fragile X mutants decreases targeting errors. Double mutant animals have a single null allele of Dscam and are homozygous null for dFmr.
g, The frequency and type of targeting errors phenocopied between dFmrnull and Dscam X3 animals can be rescued by reducing Dscam protein levels. Frequency of occurrence for ten error types that are significantly greater than wildtype for both Fragile X mutants and Dscam X3 is shown. Double mutant animals have a significant reduction in five axonal targeting errors (purple rectangles). Statistical significance comparisons to wildtype are indicated directly above the experimental genotypes’ bar; the double mutant comparison to dFmrnull animals are indicated above a connecting line. All error bars are standard error of the mean. * p < 0.05, ** p < 0.01, *** p < 0.001, and NS indicates not significant.
Dscam has previously been shown to have an essential function within the pSc neuron for axonal branch targeting, but not for the initial axon guidance into the central nervous system 15. We confirmed that loss of Dscam within pSc neurons rendered the axonal branches completely incapable of properly targeting in 100% of animals, with axonal branches extending in single directions before curving back onto the primary branch (Supplementary Fig. 1c). Thus, because Dscam is critical for pSc axonal arbour formation and its protein expression is regulated by FMRP, we sought to examine how axonal targeting is affected solely by increased Dscam protein levels rather than through loss of FMRP suppression. Therefore we analyzed the axonal arbours of flies with three copies of the Dscam gene (Dscam X3), reflecting the Down syndrome trisomy 21 case. We found that more than 65% of Dscam X3 flies had axonal targeting errors, and 30% had multiple errors within their arbours (n = 74, p < 0.001 compared to wildtype). Similar to Fragile X mutants, Dscam X3 animals also had a significant increase in the number of ectopic branches in their pSc axonal arbours (3.2 ectopic branches per animal) (Fig. 3). Analysis of the axonal targeting errors in Dscam X3 animals revealed that they were stereotyped and also similar to many of the errors observed in the Fragile X mutants (Fig. 3e). To measure the degree of overlap in targeting error phenotypes among different genotypes, we performed a blind analysis by shuffling the imaging data from the control and experimental groups (see Methods). Sixteen different error types were categorized among the data, occurring mostly within the dFmrnull genotype since Fragile X mutants had significantly higher occurrences of all error categories (Supplementary Fig. 2). Ten of these 16 errors were found to overlap between Dscam X3 and Fragile X mutants, and no targeting errors were observed in Dscam X3 animals that did not also occur in Fragile X mutants (Fig. 3g and Supplementary Fig. 2). These targeting errors were specific for Dscam and Fragile X mutants, as overexpression of other neuronal receptors did not result in these error types and did not produce stereotyped errors (Supplementary Fig. 3). Thus, overexpression of Dscam from having 3 copies of the gene can reproduce a large majority of axonal targeting phenotypes present in Fragile X mutant animals.
Reducing Dscam levels in Fragile X mutants decreases targeting errors
To determine how Dscam levels contribute to the axonal targeting defects in dFmrnull animals, we examined double mutant animals that are heterozygous null for Dscam and homozygous null for dFmr (Dscamnull/+; dFmrnull/dFmrnull). By removing one copy of the Dscam gene, this reduced the Dscam overexpression in dFmrnull animals by approximately 40% (Fig. 1b). Analysis of the axonal arbours of these Dscamnull/+; dFmrnull double mutant animals (n = 84) revealed significant reductions in the number of animals with errors (75%) compared to dFmrnull, and fewer of the double mutants (44%) had multiple errors within their pSc arbours compared to dFmrnull (p < 0.05). We also observed significant reductions in five out of the ten phenocopied axonal targeting errors compared to dFmrnull animals (p < 0.05) (Fig. 3g). However, this also led to a significant increase in one error phenotype from 4% in dFmrnull animals to 12% in Dscamnull/+; dFmrnull double mutants. Thus, the significant changes observed in these targeting errors represent the axonal targeting decisions that are most sensitive to FMRP regulation of Dscam, as the loss of one Dscam allele in the Fragile X mutants did not reduce the Dscam expression completely to wildtype levels, and we observed large variability in Dscam expression at the mRNA and protein levels in both the dFmrnull and the Dscamnull/+; dFmrnull double mutants (Fig. 1b, c). Thus, FMRP may also indirectly regulate Dscam transcription. FMRP also has multiple roles in mRNA splicing, processing, localization, and stabilization and this loss of regulation in Fragile X mutants likely results in large heterogeneities in protein expression throughout the nervous system 2,18–21.
Errors in synaptic targeting impair sensory perception
Do these axonal targeting errors in single neurons affect the mechanosensory circuit’s function? To measure the ability of a fly to perceive mechanical stimulation of its bristles, we developed a novel behavioural assay by applying a controlled amount of fluorescent dye to stimulate only the left and right posterior scutellar bristles (Fig. 4a). Stimulating these bristles evokes a cleaning reflex from the rear legs 22–25, and the fluorescent dye is transferred from the pSc bristles to the legs. Thus, we can combine this behavioral assay with the morphological and genetic analyses to examine how structural changes and axonal routing errors affect circuit function. We accomplished this by correlating behavioural responses with specific synaptic targeting patterns of the pSc neuron in individual animals (Fig. 4b). We examined the cleaning responses in mosaic animals that lack FMRP in only the scutellar neurons (455-Gal4; UAS-dsRNA-dFmr) and in Dscam X3 animals, and found that the altered synaptic connectivity of the pSc neurons in both of these mutants significantly reduced their cleaning responses compared to control flies (455-Gal4 control response rate was 26%, n = 121, compared to the 455-Gal4; UAS-dsRNA-dFmr response rate of 15%, n = 139, p < 0.01, and +/+ control response rate was 34%, n = 121, compared to Dscam X3 response rate of 20%, n = 120, p < 0.01) (Fig. 4c and Supplementary Fig. 4). Analysis of double mutant Fragile X mosaic animals lacking one copy of Dscam (Dscamnull/455-Gal4; UAS-dsRNA-dFmr) returned the response rate to that of control animals (23% response, n = 120, p < 0.05), indicating that reduction of Dscam protein levels can not only rescue synaptic targeting errors, but can restore touch perception in dFmr mutant animals.
Figure 4. Errors in synaptic targeting impair touch perception.
a, Mechanical stimulation of the pSc bristles using a controlled amount of fluorescent dye elicits a cleaning reflex from the rear legs. Transfer of the fluorescent dye from the back of the fly onto the rear legs is used to confirm a positive response.
b, Synaptic targeting of a single, identified neuron can be matched to the specific behavioral output for each animal. Representative images of the axonal arbours of previously stimulated pSc neurons are shown. Axonal arbours of mutant animals that either succeeded or failed to respond to bristle stimulation are compared to control responding animals. Arrows indicate targeting errors. Dotted line marks the midline. Scale bar, 50μm.
c, Synaptic targeting errors in the pSc neuron impair touch perception in the Fragile X mutant and Dscam X3 flies, and can be restored in the double mutant. The frequency of response is shown for mosaic animals with FMRP knocked down only in the scutellar neurons and for animals with 3 copies of Dscam, compared to their specific genetic controls (see Methods). The frequency of response to touch in mosaic double mutants, Dscamnull/455-Gal4; UAS-dsRNA-dFmr, was significantly higher (single asterisk) than mosaic Fragile X mutants, and was not significantly different from controls. n > 120 for each genotype. * p < 0.05, ** p < 0.01. Error bars are standard error of the mean.
FMRP binds multiple Dscam isoforms
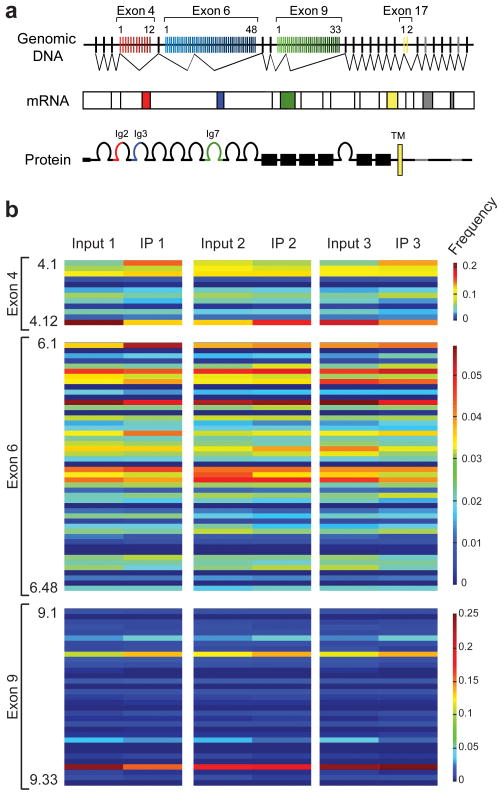
FMRP binds mRNAs in their untranslated regions through RNA secondary structures called “kissing complex RNA” (Supplementary Fig. 5) 5,26,27. Notably, alternative splicing of large exon arrays in the Drosophila Dscam gene can produce different immunoglobulin domains to create 19,008 different protein isoforms that differ only in their extracellular region (Fig. 5a) 28. Thus, to determine whether FMRP binds all of these multiple Dscam mRNA isoforms, we performed high-throughput pyrosequencing of Dscam bound to FMRP after immunoprecipitation. Pyrosequencing of Dscam enabled deep coverage of more than 1.2 million reads and long base pair read lengths 29. We confirmed that all possible Dscam isoforms expressed in the brain were also identified bound to FMRP (Fig. 5b), demonstrating that FMRP can suppress translation of tens of thousands of different Dscam protein forms. Comparisons of isoform distributions between Dscam in the input fraction and Dscam immunoprecipitated with FMRP showed that there was no significant bias in the isoforms that FMRP bound (Fig. 5b). These results demonstrate that FMRP regulation of Dscam is dependent on the splicing choices made in individual cells rather than through preferentially regulating specific mRNA isoforms. In addition, the specificity and quantitative overlap of the synaptic targeting errors between Dscam X3 and Fragile X mutants suggest that the effects of Dscam protein overexpression are most likely independent of Dscam isoform choice. Isoform-specific homophilic interactions of the Dscam receptor have been shown to induce dendritic branch repulsion 13, but nearly all of the targeting error phenotypes we observed in the pSc axons of Dscam X3 and Fragile X mutants consisted of ectopic branches, routing errors, and midline crossing errors, indicating an attraction function for the Dscam receptor. Thus, excessive Dscam protein levels in developing axonal branches most likely induces erroneous targeting decisions through inappropriate attraction to cells expressing Dscam ligands.
Figure 5. FMRP binds multiple Dscam isoforms.
a, Three large arrays of alternatively spliced exons in Drosophila Dscam (Exon 4, red; Exon 6, blue; Exon 9, green) encode for different extracellular immunoglobulin domains (Ig2, Ig3 and Ig7). Mutually exclusive splicing from each variable exon can produce 19,008 different extracellular domains. Exon 17 encodes for two alternate transmembrane domains (TM), and Exons 19 and 23 can be included or excluded in the intracellular domain.
b, High-throughput pyrosequencing of Dscam bound to FMRP identifies all possible Dscam isoforms. Dscam isoform distributions from a representative sequencing experiment of >1.2 million reads are shown as heatmaps for variable Exons 4, 6, and 9. Isoform distributions from the input and the FMRP IP from three separate experiments are shown. Dscam RNA isoforms immunoprecipitated with FMRP show no significant bias in representation compared to Dscam isoforms in the input fraction, indicating that FMRP binds all neuronal isoforms equally well.
Discussion
In this study, we found that an increase in Dscam protein levels due to either three copies of the Dscam gene or due to loss of translation suppression by FMRP impairs precise synaptic targeting and neural circuit function. Combining our behavioural analysis of mechanical stimulation of the pSc neuron with pSc axonal targeting patterns we confirmed that aberrant axonal targeting degrades sensory circuit function at levels appreciable enough to impact the animal’s perception. The restoration of the Dscamnull/+; dFmrnull double mutants’ cleaning response indicates that any other functions of Dscam independent of branch targeting that were impaired in the dFmrnull animals were also rescued. For example, Aplysia Dscam is required pre-and post-synaptically for synaptogenesis and synaptic plasticity induction, and Dscam signaling through trans-synaptic complexes leads to clustering of glutamate receptors 36.
Conversely, overexpression of FMRP may suppress many molecules involved in neural circuit function, such as synaptic transmission. Thus, when we overexpressed FMRP in the pSc neuron (455-Gal4; UAS-dFmr), this resulted in severe axon guidance and misrouting phenotypes, and also reduced the behavioural responses in these animals (Supplementary Fig. 6). Although the axon guidance and misrouting defects were suppressed when combined with Dscam overexpression (455-Gal4; UAS-dFmr/DscamBAC), the impaired behavioural response due to FMRP overexpression was not restored.
It is important to note that although the majority of the targeting errors that were phenocopied between Dscam X3 and dFmrnull mutants consisted of ectopic branches, four of the five targeting errors rescued in the double mutant Dscamnull/+; dFmrnull animals were branch misrouting and midline crossing problems (Fig. 3g). However, correlating specific synaptic targeting decisions with an individual animal’s behavioural output requires much larger data sets than our study given that we condensed all behavioural positive responses together into “yes response” rather than separating the positive responses into levels of cleaning efficiency in removing the fluorescent dye from the scutellum. The highly conserved “core” 16 skeletal branches observed in almost all pSc neurons are most likely the minimal aspects required for a basic cleaning response.
Although our results indicate that FMRP can bind all Dscam isoforms, pyrosequencing of Dscam mRNA isoforms in dFmrnull animals revealed specific differences in isoform splicing compared to wildtype (Supplementary Fig. 5). This may be due to loss of FMRP’s direct interaction with pre-mRNAs as an exonic splicing enhancer, or through unregulated expression of splicing proteins normally suppressed by FMRP 21,30. FMRP regulation of Dscam splicing may also be utilized in arthropod immune systems, as Dscam is expressed in insect and crustacean immune cells such as hemocytes 31,32, and FMRP is also expressed in hemocyte-derived S2 cells 33,34. In the arthropod immune system, specific Dscam receptor isoforms bind to different pathogens and become preferentially spliced and upregulated for pathogen clearance 31,32,35, but it remains unclear how the feedback to splicing and expression of Dscam isoforms occurs. FMRP might thus regulate Dscam isoform splicing in many different cell types for a wide range of functions.
Our study has found that neural circuit development and function are sensitive to increased in Dscam protein amounts. Previous studies using Dscam null heterozygous mice and mouse models of Down syndrome revealed that Dscam dosage is crucial for proper sorting of retinal ganglion cell axons and dendritic development 12,37, but thus far it has not been clear how Dscam overexpression might contribute to neurological impairments like Down syndrome. Dscam has also been associated with the congenital heart defects found in Down syndrome, which was identified using analysis of rare individuals with partial duplications of chromosome 21 (ref 10). Genetic interaction screens in Drosophila for congenital heart defect genes also identified Dscam, and overexpression of Dscam in the mouse produced physiological and morphological cardiac defects 38. Given its evolutionarily-conserved widespread functions throughout cardiac and neural development 13, and its conserved interaction with FMRP 4,5, Dscam expression levels are thus likely to be tightly regulated. Dysregulation of Dscam protein expression may therefore be a common molecular feature underlying a wide variety of neural developmental disorders such as in the dendritic spine pathologies found in Fragile X, Down, and Rett syndromes 39–41.
Methods
Drosophila Strains
The following dFmrnull fly stocks were used: dFmr3 (F. Bolduc, University of Alberta), dFmrΔ113, dFmrΔ50M, and Df(3R)Exel6265 (A.P. Haghighi, McGill University), and have been verified to lack FMRP (Fig. 1)42–45. Trans-heterozygous mutant flies were used in experiments (Supplementary Fig. 7a) and generated by mating dFmrΔ50/TM6b or dFmrΔ113/TM6b with dFmr3/TM6, Sb, Tb or Df(3R)Exel6265/TM6b. To overexpress FMRP, flies homozygous for an extra copy of the entire dFmr transcriptional unit were used, thus expressing four copies of dFmr 42. This dFmr genomic fragment (gdFmr) was confirmed to rescue FMRP protein expression and the pSc axonal targeting errors in the dFmrnull mutants (Supplementary Fig. 7b).
Site-specific insertions of a bacterial artificial chromosome (BAC) containing the entire genomic locus of Dscam were used to express an extra copy of the Dscam gene (H. Bellen, Howard Hughes Medical Institute, Baylor College of Medicine)46. Any dominant effects of the BACs were tested by analyzing the pSc axonal arbors in Dscam null flies expressing only the Dscam BACs (Dscamnull/Dscamnull; DscamBAC) (Supplementary Fig. 8), and lines 5-, 7-, 13-, 19-, 20- and 33-DscamBAC were used for experiments. Flies with three copies of Dscam were obtained by crossing DscamBAC homozygotes with w− flies. 5-DscamBAC/+ and 20-DscamBAC/+ are shown in Figure 3e.
Dscam21/CyO and Dscam23/CyO (W. Grueber, Columbia University) were used as Dscamnull mutants28. Dscamnull mutants are embryonic lethal, so Dscamnull early embryos were collected for negative controls in the immunoblotting experiments 15. Double mutant flies heterozygous for Dscam and homozygous null for dFmr were created by mating Dscam23/CyO; dFmr3/TM6b to dFmrΔ113/TM6b flies. Dscam23/+; dFmrΔ50M/dFmr3 and Dscam23/+; dFmrΔ113/dFmr3 are shown in Figure 3f.
For RNAi experiments, we used the following UAS-dsRNA-dFmr lines: RNAi lines (2-1), (1–7) and (1–10) (F. Bolduc, University of Alberta)43, and line 8933 from the Vienna Drosophila RNAi Center (Vienna, Austria). Fragile X mutants dFMRΔ113/dFmr3 and dFmrRNAi8933 are shown in Figure 3d. Gal4 expression within only the scutellar neurons was achieved using the 455-Gal4 line 16. To reduce Dscam levels in dFmr RNAi knockdowns, 455-Gal4/CyO; UAS-dsRNA-dFmr animals were crossed to Dscam23/CyO; UAS-dsRNA-dFmr.
Immunoprecipitation and RT-PCR
Immunoprecipitation experiments were performed in quintuplicate using adult fly brains and verified in sextuplicate from third instar wandering larval brains. FMRP-mRNA complexes were immunoprecipitated from wildtype or dFmrnull samples using mouse monoclonal anti-FMRP antibody 6A15 (Abcam, Cambridge, MA) coupled to protein G Dynabeads (Life Technologies, Carlsbad, CA). Eluted mRNAs were used as template for RT-PCR using the following gene-specific reverse transcription primers: dFmr CTCTCTCCACGCTGCTCATT, Dscam(Exon 11) TGATCATAATCACAGCCGAGAGG, and Futsch CTCGCTGGAAGTCTTTGTCC. PCR amplification was performed using the following forward and reverse primers (respectively for each gene): dFmr CGTGCCCGAGAGTATGAAAT, GTCTCAAAACCGATGTACGC; Dscam CAACGGAGATGTGGTTTCCT, GGTTATCTCGCTCCCAGACA; Futsch ATCACCGCAAGTTTTGAAGG, GCGAAGTCTTTTGGTGCTTC. All other mouse monoclonal antibodies used for immunoprecipitation were obtained from the Developmental Studies Hybridoma Bank. Immunoprecipitation of FMRP-mRNA complexes was also confirmed using another mouse monoclonal antibody 5B6 (developed by K.S. Broadie). Immunoprecipitation of ELAV-mRNA complexes using mouse monoclonal antibody 9F8A9 (developed by G.M. Rubin) and actin complexes using mouse monoclonal antibody JLA20 (developed by J.J-C. Lin) were used as negative controls (Supplementary Fig. 9) 47. ELAV has been shown to extend the 3′ untranslated region of the mRNA brain tumor (brat) 48, and this was used as a positive control of ELAV-mRNA complex precipitation (Supplementary Fig. 9).
Pyrosequencing
Immunoprecipitation was performed on adult fly brains using both the 5B6 and 6A15 monoclonal antibodies. Reverse transcription of mRNA extracted from input and immunoprecipitated samples was performed using Dscam-specific reverse primers for Exon 11 and Exon 7, CCGCCGATTCCTGGTCGTTTCTTAC. The cDNA was PCR amplified using 454 Lib-L unidirectional sequencing fusion primers containing the 454 adaptor sequence (Primer A/forward CCATCTCATCCCTGCGTGTCTCCGACTCAG; PrimerB/reverse CCTATCCCCTGTGTGCCTTGGCAGTCTCAG) and target-specific sequences for Exon 4 forward AAGCTGGTCTTCCCTCCATT and reverse CTCTCCAGAGGGCAATACCA, Exon 6 forward AGTGCCACAAAAGGACGATT and reverse GCTTGTTTACGGGTTGTTCC and Exon 9 forward CTACACTTGCGTTGCCAAGA and reverse TCAGCCTTGCATTCAACCTT. The PCR products were sequenced using the Roche GS-FLX Titanium sequencer. Samples were prepared in experimental triplicates and pyrosequencing experiments were verified in two sequencing runs. Sequences were analyzed using a custom written program in MatLab (MathWorks, Natick, MA) to identify isoforms, and positive identification of isoforms was established for ~70% of sequences. A frequency distribution of isoforms was generated for each exon, experimental replicate, and sample. A goodness-of-fit test based on the chi-square distribution was used to calculate statistical significance between frequency distributions of samples. For visual display of isoform frequency distributions, heatmaps were generated using MatLab (MathWorks).
Quantitative Real-Time PCR
Total RNA was extracted from adult fly heads. Reverse transcription was performed using a Dscam-specific reverse primer and a Ribosomal Protein 49 (Rp49)-specific reverse primer CATCAGATACTGTCCCTTGAAGC. Taqman Fast-Advanced Master Mix (Life Technologies) was used with the following primers and double quenched 5′-FAM/ZEN/IowaBlackFQ-3′ probes (Integrated DNA Technologies, Coralville, IA): Rp49 forward GCGCACCAAGCACTTCATC, Rp49 probe 5′-FAM-ATATGCTAAGCTGTCGCACAAATGGC-IBFQ-3′, Rp49 reverse GACGCACTCTGTTGTCGATACC, Dscam forward ACGATGTAGTTTACAATCAGACAA, Dscam probe 5′-FAM-ACCTGCGGGATGAGCTCGGATACA-IBFQ-3′, Dscam reverse GCCTCGCTTAATCCGGTCA. PCR amplification was detected using the Applied Biosystems StepOne Plus Real Time PCR System (Life Technologies) and cycle threshold (CT) values calculated using the StepOne software. Experiments were performed in six experimental replicates with three to six technical replicates. CT values were normalized to Rp49 control levels and technical replicates were averaged within each experimental replicate. Dscam mRNA levels from experimental genotypes were compared to wildtype levels from within the same experiment and reported as fold changes from wildtype.
Immunoblotting and Protein Quantification
Immunoblot protein quantification experiments were performed nine times using third instar wandering larval brains and replicated in duplicate in adult brains. Proteins were separated by electrophoresis on a NuPAGE Novex 12% Bis-Tris Gel (Life Technologies) and transferred to a polyvinylidene fluoride membrane. The membrane was incubated with the following antibody dilutions: 1:1000 anti-Dscam rabbit polyclonal (J. Clemens, Purdue University), 1:250 anti-dFmr 6A15 mouse monoclonal (Abcam), and anti-actin C4 mouse monoclonal (CedarLane, Burlington, ON). Secondary antibodies used were fluorescent anti-rabbit IRDye CW800 and anti-mouse IRDye CW800 (LI-COR, Lincoln, NE). Proteins were visualized using the Odyssey infrared imaging system (LI-COR). Protein bands were quantified by averaging the intensities of five randomly chosen 3×3 pixel regions, and Dscam and FMRP levels were normalized to actin.
Immunohistochemistry and Fluorescence In Situ Hybridization
Immunohistochemistry experiments on identified mechanosensory neurons were performed 15 times in wildtype, 12 times in dFmrnull, and 3 times for dFmr RNAi animals. Co-labeling of fluorescence in situ hybridization for Dscam mRNA with fluorescence immunohistochemistry for FMRP within identified pSc neurons was reproduced 8 times. Cryosections of the thorax along the rostral-caudal, dorsal-ventral axis were cut at 10 μm thickness from adult female flies. Custom fluorescent RNA probes against Dscam were designed to bind all isoforms within the constant mRNA sequences, and were conjugated to the Quasar670 dye (Biosearch Technologies, Novato, CA). Fluorescence immunohistochemistry with fluorescence in situ hybridization was performed as described 49. Mouse monoclonal antibody 5A11 for FMRP (developed by H. Siomi) at 1:100, or mouse monoclonal antibody 5B6 for FMRP at 1:100was added for overnight incubation. Secondary antibody goat anti-mouse AlexaFluor488 (Life Technologies) was applied during the wash steps, and a Hoechst dye was applied on the final wash to label nuclei.
Fluorescence microscopy was performed using an Olympus laser scanning confocal microscope FV1000. Images were acquired using a 60× oil objective, N.A. 1.4.Quantitative analysis of FMRP intensities was performed by measuring the average pixel intensity in the FMRP channel in a region of interest centered around the nucleus of the mechanosensory neuron. Efficiency of the UAS-dsRNA-dFmr was thus quantified (Supplementary Fig. 1) from three experiments and compared to FMRP intensities from wildtype neurons in three experiments.
Carbocyanine Dye Labeling and Imaging
Lipophilic dye labeling of single mechanosensory axons were conducted as previously described15,16. The left and right pSc neurons from two day old female flies were labeled with the fluorescent carbocyanine tracers DiI (D282) or DiD (D7757) (Life Technologies) dissolved in ethanol at 20 mg/mL and 40 mg/mL, respectively.
The thoracic ganglion was dissected out andslide-mounted with #1 thickness coverslips. Fluorescence and brightfield microscopy was performed using a Zeiss AxioScope A1, or an Olympus laser scanning confocal microscope FV1000. All images were acquired using a 40× objective, N.A. 1.0. Image analysis was performed on maximal intensity projections. Transmitted light images were acquired to measure the central nervous system (CNS) width and to verify there was no damage to the CNS or occlusions at the surface.
Image Analysis
Images were selected for analysis based on low background fluorescence and homogenous and strong labeling throughout a single pSc axon. Images were adjusted for contrast and brightness only. Axonal branch lengths and numbers were measured using a custom written program in MatLab (MathWorks). For qualitative analysis of pSc axon phenotypes, a prototypical skeleton of the wildtype pSc axonal arbor was first designated by identifying axonal branches that were invariant among 53 w flies. Primary and secondary branches were identified that occurred at greater than 80% frequency and tertiary branches that occurred at greater than 60% frequency. This wildtype pSc skeleton consisted of 16 primary, secondary and tertiary axonal branches ranging from 6μm for the smallest branch average to 130μm for the largest branch average. Branches were considered ectopic if they occurred in less than 10% of wildtype flies. Variable branches were thus defined as occurring at greater than 10% and less than 60% frequency, with an average frequency of 30% per wildtype fly. The midline was defined as a 10μm-wide region running along the anterior-posterior axis of the CNS. Any branch entering or crossing this region was considered a midline-crossing branch. The length of the primary axon entry point into the CNS (Fig. 1c, “branch 0”) is dependent on the number of images collected above the entry point as the axon travels within its fascicle, and so was not included in the branch length measurement calculations. Axon guidance errors of the primary axon entry point were quantified but not counted as axonal targeting errors and occurred at 1.1% and 1.4% in dFmrnulland Dscam X3 mutants, respectively, and did not occur in wildtype animals. Branch lengths among all genotypes were normally distributed from their means. One way ANOVA followed by Dunnett’s post-hoc pairwise comparison was used to determine statistical significance in branch lengths between wildtype and mutant genotypes. For statistical testing of discrete measurements, non-parametric Mann-Whitney U test was used to determine statistical significance in branch numbers between wildtype and mutant genotypes.
A total of 74 wildtype, 100 dFmrnull, 74 Dscam X3, 84 Dscamnull/+; dFmrnull double mutant animals were analyzed. Sample sizes were chosen based on previous studies15,16. Qualitative analysis of axonal targeting variability was performed blind to genotype by shuffling the axonal arbor data among all genotypes and 28 different targeting variability types were identified. Wildtype variability was identified (12 types), and 16 error types were found with a frequency of less than 10% in wildtype, and this was then used as a cutoff for the definition of a targeting error (Supplementary Fig. 2). Errors in all 16 categories were found in dFmrnull animals. Dscam X3 animals had errors in 15 categories, but five of these 15 error types were not significantly different from wildtype, thus the ten error types significantly higher in both dFmrnull and Dscam X3 mutants compared to wildtype were defined as the targeting error phenocopy. Targeting errors were considered rescued in the double mutant animals if the error frequency was significantly lower compared to dFmrnull. Statistical significance for each category between genotypes was determined by performing multiple comparisons using a two tailed t-test for proportions set at p < 0.05.
Behavioural Analysis
The scutellum specific Gal4 driver, 455-Gal4, was used to drive dFmr dsRNA only in the four scutellar mechanosensory neurons to ensure that the rest of the animal, in particular the post-synaptic neural circuitry, was left unperturbed by the gene manipulations. Dual color dye labeling of scutellar neurons and unaffected dorsocentral neurons were performed periodically to ensure specificity of the Gal4 driver16, and 455-Gal4>Dscam dsRNA flies, which lack all axonal branch targeting, were used as negative controls15,16. Experiments were performed on two day old female flies with the experimenter blind to genotype. All genotypes were assayed on the same day to control for seasonal growth effects, and at the same approximate time (early afternoon) to control for circadian effects. Flies were decapitated and left to recover for 1h in a humidified chamber. To ensure the integrity of the cleaning reflex circuit, flies were pre-selected by stimulating the notopleural bristles to elicit a cleaning response from the two front legs. The two pSc bristles of decapitated flies were stimulated by pressure injection of fluorescent dye (40 mg/ml DiD in ethanol or 2.5 mg/ml DiO in dimethylformamide). Success or failure to elicit a cleaning reflex was scored visually and then verified by the transfer of dye to the rear legs of the fly. The pSc bristles were then plucked from both responding and non-responding flies and the animals were prepared for subsequent dye filling and morphological analysis. A total of 121 w− controls, 121 455-Gal4/+ controls, 125 UAS-dsRNA-dFmr controls, 77 Dscam23/+ controls, 120 Dscam X3, 139 455-Gal4/+; UAS-dsRNA-dFmr mutants, and 120 double mutant (Dscam23/455-Gal4; UAS-dsRNA-dFmr) animals were analyzed using DiD stimulation (Supplementary Fig. 4). Sample sizes were chosen based on previous studies 22–25. All behavioral results were verified using DiO dissolved in dimethylformamide, a more viscous solvent, to stimulate the pSc bristles which produced greater response rates in all genotypes, and produced identical results among genotypes. Statistical significance in response rate between each genotype was determined using a two tailed t-test for proportions set at p < 0.05.
Supplementary Material
Acknowledgments
The authors thank Tsung-Jung Lin, Ibrahim Kays, and Viktoria Stoudenikina for assistance with experiments, Radu Suciu for assistance in pyrosequencing analysis, Alfredo Staffa and the Massively Parallel Sequencing Unit at Génome Québec for pyrosequencing assistance, and Bashar Douba for graphic arts assistance in the fly drawing. This work was supported by an Alfred P. Sloan Research Fellowship and a Canada Research Chair grant 950–212462 (to B.E.C.) and by funds from the Department of Medicine at McGill University and the Research Institute of the McGill University Health Centre.
Footnotes
Author Contributions
B.E.C. designed the experiments and supervised the project. V.C., A.D.H., F.E., and B.E.C. performed experiments and analyzed the data. V.C., F.E., and B.E.C. wrote the manuscript.
No competing financial interests to declare.
Supplementary Information is provided as 9 supplementary Fig.s.
References
- 1.Rachidi M, Lopes C. Mental retardation in Down syndrome: From gene dosage imbalance to molecular and cellular mechanisms. Neuroscience Research. 2007;59:349–369. doi: 10.1016/j.neures.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. S0896-6273(08)00847-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. S0092-8674(01)00568-2 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takashima S, Becker LE, Armstrong DL, Chan F. Abnormal neuronal development in the visual cortex of the human fetus and infant with down’s syndrome. A quantitative and qualitative Golgi study. Brain Res. 1981;225:1–21. doi: 10.1016/0006-8993(81)90314-0. 0006-8993(81)90314-0 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Antonarakis SE. 10 years of Genomics, chromosome 21, and Down syndrome. Genomics. 1998;51:1–16. doi: 10.1006/geno.1998.5335. S0888-7543(98)95335-6 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Yamakawa K, et al. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet. 1998;7:227–237. doi: 10.1093/hmg/7.2.227. [DOI] [PubMed] [Google Scholar]
- 9.Korenberg JR, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci U S A. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barlow GM, et al. Down syndrome congenital heart disease: a narrowed region and a candidate gene. Genetics in Medicine. 2001;3:91–101. doi: 10.1097/00125817-200103000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Hildmann T, et al. A contiguous 3-Mb sequence-ready map in the S3-MX region on 21q22.2 based on high-throughput nonisotopic library screenings. Genome Res. 1999;9:360–372. [PMC free article] [PubMed] [Google Scholar]
- 12.Alves-Sampaio A, Troca-Marin JA, Montesinos ML. NMDA-mediated regulation of DSCAM dendritic local translation is lost in a mouse model of Down’s syndrome. J Neurosci. 2010;30:13537–13548. doi: 10.1523/JNEUROSCI.3457-10.2010. 30/40/13537 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23:147–156. doi: 10.1101/gad.1752909. 23/2/147 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Ghysen A. The projection of sensory neurons in the central nervous system of Drosophila: choice of the appropriate pathway. Dev Biol. 1980;78:521–541. doi: 10.1016/0012-1606(80)90351-6. 0012-1606(80)90351-6 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Chen BE, et al. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. S0092-8674(06)00445-4 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Neufeld SQ, Hibbert AD, Chen BE. Opposing roles of PlexinA and PlexinB in axonal branch and varicosity formation. Molecular brain. 2011;4:15. doi: 10.1186/1756-6606-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinz U, Giebel B, Campos-Ortega JA. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. 0092-8674(94)90174-0 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 19.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nature reviews. Neuroscience. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 20.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Didiot MC, et al. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canal I, Acebes A, Ferrus A. Single neuron mosaics of the drosophila gigas mutant project beyond normal targets and modify behavior. J Neurosci. 1998;18:999–1008. doi: 10.1523/JNEUROSCI.18-03-00999.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corfas G, Dudai Y. Habituation and dishabituation of a cleaning reflex in normal and mutant Drosophila. J Neurosci. 1989;9:56–62. doi: 10.1523/JNEUROSCI.09-01-00056.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillis RW, et al. Isolation of mutations affecting neural circuitry required for grooming behavior in Drosophila melanogaster. Genetics. 1993;133:581–592. doi: 10.1093/genetics/133.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandervorst P, Ghysen A. Genetic control of sensory connections in Drosophila. Nature. 1980;286:65–67. doi: 10.1038/286065a0. [DOI] [PubMed] [Google Scholar]
- 26.Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet. 2009;18:3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darnell JC, et al. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 29.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guruharsha KG, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watson FL, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 32.Watthanasurorot A, Jiravanichpaisal P, Liu H, Soderhall I, Soderhall K. Bacteria-Induced Dscam Isoforms of the Crustacean, Pacifastacus leniusculus. PLoS pathogens. 2011;7:e1002062. doi: 10.1371/journal.ppat.1002062. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Monzo K, et al. Fragile X mental retardation protein controls trailer hitch expression and cleavage furrow formation in Drosophila embryos. Proc Natl Acad Sci U S A. 2006;103:18160–18165. doi: 10.1073/pnas.0606508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stetler A, et al. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum Mol Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 35.Dong Y, Taylor H, Dimopoulos G. AgDscam, a Hypervariable Immunoglobulin Domain-Containing Receptor of the Anopheles gambiae Innate Immune System. Plos Biol. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li HL, et al. Dscam mediates remodeling of glutamate receptors in Aplysia during de novo and learning-related synapse formation. Neuron. 2009;61:527–540. doi: 10.1016/j.neuron.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blank M, et al. The Down syndrome critical region regulates retinogeniculate refinement. J Neurosci. 2011;31:5764–5776. doi: 10.1523/JNEUROSCI.6015-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grossman TR, et al. Over-expression of DSCAM and COL6A2 cooperatively generates congenital heart defects. PLoS genetics. 2011;7:e1002344. doi: 10.1371/journal.pgen.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes, brain, and behavior. 2006;5(Suppl 2):48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 40.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 41.Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. 21/14/5139 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dockendorff TC, et al. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. S0896627302007249 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. nn.2175 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang YQ, et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. S0092-8674(01)00589-X [pii] [DOI] [PubMed] [Google Scholar]
- 45.Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. ng1312 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 47.Reeve SP, et al. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 48.Hilgers V, Lemke SB, Levine M. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 2012;26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.