Abstract
Transcription factors comprise just over 7% of the human proteome and serve as the gatekeepers of cellular function, integrating external signal information into gene expression programs that reconfigure cellular physiology at the most basic levels. Surface-initiated, cell signaling pathways converge on transcription factors, decorating these proteins with an array of post-translational modifications (PTMs) that are often interdependent, being linked in time, space, and combinatorial function. These PTMs orchestrate every activity of a transcription factor over its entire lifespan—from subcellular localization to protein–protein interactions, sequence-specific DNA binding, transcriptional regulatory activity, and protein stability—and play key roles in the epigenetic regulation of gene expression. The multitude of PTMs of transcription factors also offers numerous potential points of intervention for development of therapeutic agents to treat a wide spectrum of diseases. We review PTMs most commonly targeting transcription factors, focusing on recent reports of sequential and linked PTMs of individual factors.
General overview of PTMs of transcription factors
Post-translational modifications regulate every aspect of transcription factor function and coordinate access of RNA polymerases to promoter templates. Site-specific, DNA-binding transcription factors (SSTFs) serve to nucleate repressor, activator, enhancer, or silencer complexes and associated enzymatic activities. To coordinate these activities, often with great spatial, temporal, and tissue-specific precision required of developmental and cell-cycle programs, the full range of cellular post-translational modifications (PTMs) of SSTFs may occur. In many cases, these PTMs occur as individual, isolated events and these modifications dictate some aspect of transcription factor function. In other cases, individual PTMs on proteins are sequentially linked—that is, one PTM may promote (or inhibit) the establishment of a second-site PTM within the same protein. These two PTMs are “linked” or “interconnected,” and as we describe below, this interconnectedness can be exploited therapeutically in the treatment of disease.
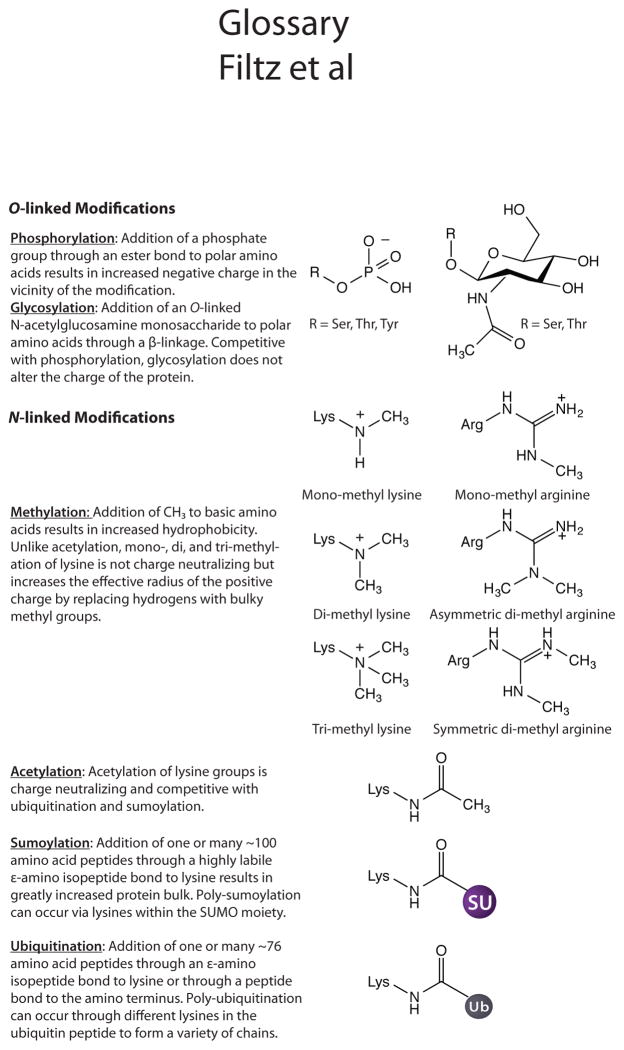
Among the more prominently studied PTMs of transcription factors are phosphorylation, sumoylation, ubiquitination, acetylation, glycosylation, and methylation (Figure 1). The analysis presented below (Figure 2) suggests that most of these PTMs occur on transcription factors at about the same rate as seen with other proteins, with the notable exceptions of ubiquitination, glycosylation, and sumoylation, which are found on transcription factors with moderately decreased, moderately increased and greatly increased frequencies, respectively (Figure 2). There is no obvious logic why ubiquitination would be somewhat lower or glycosylation somewhat higher among transcription factors. The many-fold increased incidence of sumoylation among transcription factors may reflect a real biological phenomenon. Alternatively, it is possible that this modification, which has historically been difficult to detect in native proteins, may be over-represented in transcription factor data sets due to a relative lack of information concerning this modification among non-nuclear proteins.
Figure 1. Types of PTMs.
Figure 2. Relative enrichment of PTMs in transcription factors.
Values are the relative ratio of PTMs identified in human transcription factors compared to those identified in non-transcription factor human proteins and are plotted to show the divergence from equality for the indicated PTM. Blue bars indicate the degree of over-enrichment in transcription factors and the red bar indicates the degree of under-ubiquitination. PTM data was drawn from the PhosphoSitePlus database [1]. Abbreviations: Ub, ubiquitination; Ac, acetylation; Me, methylation; PO4, phosphorylation; OG, O-GlcNAcylation, and SU, sumoylation.
PTMs may alter SSTF subcellular localization (transport into or out of the nucleus), stability, secondary structure and DNA binding affinity, or tertiary structure and association with co-regulatory factors. PTMs of SSTFs are of particular interest as a means of altering transcriptional regulatory activity of these proteins. Many excellent reviews have focused on the varied effects of transcription factor phosphorylation [2,3], sumoylation [4], ubiquitination [5], acetylation [6], and glycosylation [1,7]. In this review, we provide a few examples of small-mass modifications, including phosphorylation, acetylation, methylation, and glycosylation, and then focus on the larger modifications of sumoylation and ubiquitination, highlighting some examples of interconnected or sequentially-dependent modifications. Specific information about known PTMs of all proteins can be found at http://www.phosphosite.org [1–3] and information about sequentially linked PTMs in proteins can be accessed at the PTMcode website (http://ptmcode.embl.de) [4,8]. Both websites are actively curated and exceptionally informative.
Phosphorylation
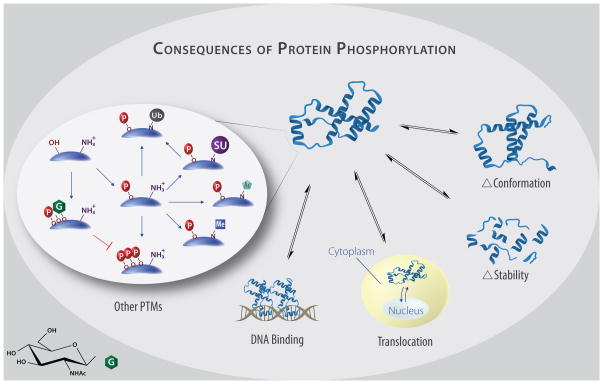
Phosphorylation is a gateway PTM; easily detected, phosphorylation is often the first PTM to be studied when looking at regulation of protein activity. Rapidly reversible phosphorylation, a ubiquitously utilized mechanism to transduce extracellular signals to the nucleus, may affect transcription factor stability, location, structure and/or protein–interaction network (Figure 3), all of which may impact target gene expression. Phosphorylation may also regulate the status of other PTMs in a time-dependent sequence that, for some transcriptional regulatory proteins may culminate in degradation. Straightforward examples of transcription factor regulation by single- or double-site phosphorylation leading to well-characterized, binary effects are described elsewhere [2,3,5].
Figure 3. Mechanisms by which PTMs may alter transcription factor activity.
To alter gene expression, phosphorylation may affect the secondary structure of a transcription factor (Δ Conformation) to reveal binding sites or alter affinity, increase or decrease protein degradation (Δ Stability), increase or decrease the nuclear occupancy and thus access to DNA (Localization), alter affinity for DNA regulatory regions (DNA Binding), or alter the modification of the factor by other PTMs including acetylation, sumoylation, methylation, O-GlcNAcylation, or ubiquitination.
Transcription factors may harbor multiple phosphorylation sites, serving as points of convergence of signaling pathways initiating at the plasma membrane. As such, transcription factors may function as coincidence detectors in which two or more pathways must be activated before gene transcription is altered. For example, TORC2 is normally a cytosolic protein in insulinoma cells that is activated to translocate to the nucleus by dephosphorylation, after which it becomes associated with the CREB transcriptional complex. TORC2 translocation requires both high glucose and incretin receptor activation to increase intracellular calcium and cAMP levels, respectively. Both calcium-induced activation of calcineurin and inhibition of the SIK2 serine/threonine kinase by cAMP are required to mediate TORC2 dephosphorylation at distinct sites to allow for translocation [2,3,6,9].
Multiple phosphorylation sites may also serve as tunable signal regulators with incremental phosphorylation leading to changes in amplitude of gene expression. The MSN2 transcription factor in yeast processes different stress responses by “tunable” accumulation in the nucleus. Phosphorylation of eight serines clustered within at least two regulatory domains of MSN2 leads to complex translocation kinetics and differentially tuned responses to stressors. Osmotic stress produces a single pulse of nuclear localization, glucose limitation induces sporadic pulses of nuclear localization, and oxidative stress produces sustained nuclear localization [4,10]. These differential responses are produced by dual regulation of both nuclear import and nuclear export rates by different phosphorylation states of MSN2.
A cooperative signaling response is another potential function of clusters of multiple phosphorylation sites in an SSTF. Although not a new story, the response of NFAT to dephosphorylation in T cells is the best-detailed example of multi-site phosphorylation/dephosphorylation providing a steep response curve for conformational change in response to signaling (reviewed by [5,11]). Dephosphorylation of NFAT by the calcium-sensitive phosphatase calcineurin allows for nuclear accumulation and transcriptional activity. Nuclear accumulation results from an alteration in the balance between nuclear import and export. Progressive dephosphorylation of 13 phosphorylation sites alters the likelihood that the NFAT nuclear localization signal subdomain is exposed and that the nuclear export signal is hidden due to a phosphorylation-dependent change in the folding energy of the protein [6,12,13]. This mechanism appears to provide a tightly defined, calcium concentration threshold for activation.
Although the above examples are all of multisite phosphorylation affecting nuclear localization in various ways, multisite phosphorylation of SSTFs may have other effects. A multisite phosphorylation gradient progressively impacts the stability and ubiquitination of ATF4 in cell cycle control, allowing for ATF4 to exert dose-dependent regulation of target genes in neurogenesis [7,14]. Dephosphorylation of RUNX1 by the tyrosine phosphatase SHP2 alters RUNX1 protein–protein interactions within the megakaryocyte nucleus, increasing association with the SWI/SNF chromatin remodeling complex, reducing association with other factors such as GATA1, and promoting development of megakaryocytes [2,3,15].
O-GlcNAcylation
Many transcription factors, in particular, but also other nuclear proteins, as well as cytosolic proteins are extensively modified by addition of β-D-N-acetylglucosamine (GlcNAc; Figure 3). The modification of serine and threonine by this monomeric GlcNAc moiety is distinguished from other forms of glycosylation by its relatively small size, subcellular location, and dynamic nature. GlcNAc addition and removal occurs in both the nucleus and cytoplasm, targeting the same motifs as phosphorylation, with which it competes. This reciprocal regulation is seen both on the nuclear factor substrates and on the enzymes affecting this regulation. Kinases are overrepresented among O-GlcNAcylation substrates [16] and the enzyme responsible for adding GlcNAc is itself phosphoactivated by a kinase that is regulated by O-GlcNAcylation. O-GlcNAcylation of CaMKIV on Ser189 limits phosphoactivation on a nearby Thr200 [7]. Similarly, O-GlcNAcylation of CK2 on Ser347 antagonizes phosphorylation at Thr344 and alters both stability and substrate specificity of the enzyme [17].
Much of the transcriptional proteome is modified by O-GlcNAcylation and the effects of this PTM on transcription factors and transcriptional regulatory proteins, including RNA polymerase II itself, may be either positive (via protein stabilization) or negative (via inhibition of transcriptional activation) [7,18,19].
O-GlcNAcylation occurs as a consequence of the activities of the single biosynthetic (O-GlcNAc transferase) and catabolic (O-GlcNAcase) enzymes in this pathway, and the availability of the high-energy donor substrate, UDP-GlcNAc [7]. Flux through the biosynthetic side of the O-GlcNAcylation pathway is dictated by nutrients (glucose), insulin, and cellular stress, and it has been proposed that alterations in the levels O-GlcNAcylation of transcription factors may underlie certain pathological aspects of metabolic diseases, such as diabetes, as well as neurodegenerative diseases and cancer [7].
Acetylation
Lysine residues are positively charged at physiological pH and acetylation of these residues, which generates an uncharged amide, may logically be expected to reduce the affinity of the SSTF for DNA. Accordingly, acetylation inhibits interaction of FOXO1 with the glucose-6-phosphatase promoter [20]. However, the data show an evolving picture that is substantially more complex as deacetylation of FOXO1 inhibits its interaction with the BIM promoter in transfected cells [21]. Acetylation of FOXO1 increases its nuclear localization in skeletal muscle, and this results in enhanced regulation of FOXO1 target genes [22]. These findings indicate the consequences of alterations in the acetylation status of FOXO1 are both promoter-specific and influenced by cell type, highlighting the complexity of this relatively simple modification.
Methylation
Arginine methylation may alter the transcriptional regulatory activity of SSTFs by altering the protein–interaction network of these factors. For example, methylation of RUNX1 by the arginine methyltransferase PRMT1 inhibits binding of this SSTF to the co-repressor SIN3A and promotes de-repression of RUNX1 target genes [23]. Similarly, methylation of C/EBPβ regulates its interaction with Mediator and SWI/SNF co-activator complexes, and alters C/EBPβ-mediated activation of myeloid and adipogenic target genes [24]. Reversible lysine modification of SSTFs, including NF-κB, STAT3, p53, and pRb, appears to be catalyzed on promoter templates by the same enzymes that place this modification on and remove it from the core histones [25] The effect of lysine methylation on the transcriptional regulatory activity of these proteins varies with the protein and the context within the protein [25].
Sumoylation
Reversible modification by small ubiquitin-like modifier (SUMO) proteins, SUMO1–4, can profoundly affect the activities, nuclear or sub-nuclear localization, and/or the protein–protein interaction network of SSTFs [4,26]. Protein sumoylation couples a glycine residue in the carboxyl terminus of the activated SUMO protein to the ε-amino group of an acceptor lysine in the target protein, resulting in a covalent, but highly labile, isopeptide bond. The majority of known SUMO-acceptor sites in target proteins conform to the sequence ψKx(D/E), where ψ corresponds to an aliphatic, hydrophobic amino acid and “x” can be any amino acid [4,27].
Importantly, the 10 kDa SUMO moiety is far larger than other common PTMs such as phosphorylation, acetylation, GlcNAc, and methylation, and larger also than ubiquitin. For example, mono-sumoylation of a 50 kDa transcription factor increases the size, and presumably the surface area, of that factor by 20%. The presence of multiple sites of mono-sumoylation and/or formation of SUMO chains at one or more these sites can easily double the mass and surface area of this transcription factor and alter the ability of the factor to participate in protein-protein interactions, which play a deterministic role in the activities of SSTFs.
Sumoylation of transcription factors is most often associated with enhanced transcriptional repressive activity [26], and several underlying mechanisms have been described or proposed. First, the presence of a large SUMO moiety may serve as a platform for interaction with proteins containing SUMO interaction motifs (SIMs; reviewed in [28]). Recruitment of the LSD1/CoREST1/HDAC complex to chromatin requires functional interaction of the CoREST1 SIM with SUMO2/3 [29], suggesting that sumoylation of a template-associated factor(s) nucleates assembly of a transcriptionally repressive complex. Second, sumoylation plays a key role in assembly and function of Polycomb group bodies [30], which serve as localized hubs of transcriptional repression [26]. Finally, sumoylation may directly enhance the enzymatic activity of DNA methylating enzymes, such as DNMT1 [31], which promote transcriptional repression.
Sumoylation stimulates the transcriptional activity of—or at least dampens transcriptional repression mediated by—a handful of transcription factors whose dysregulation has been linked to developmental defects and cancer, but the underlying mechanisms remain unknown. Sumoylation appears to stimulate transcriptional activation mediated by Ikaros [32], p53 [33], PAX6 [34], and BCL11B [35]. In the latter case, sumoylation of BCL11B results in recruitment of p300 to the BCL11B-NuRD complex and subsequent transcriptional activation of a BCL11B target gene [35]. Thus, sumoylation of BCL11B serves as a switch that converts BCL11B from a repressor to an activator of transcription, and this has relevance in the T-cell developmental program [35]. Finally, sumoylation of MBD1 inhibits its interaction with the histone-lysine methyltransferase SETDB, impairing repression mediated by the MBD complex [36].
Mono-sumoylated transcription factors may be further sumoylated to a state of poly-sumoylation, which may promote subsequent ubiquitination by E3 ligases harboring SUMO-Targeted Ubiquitin Ligase (STUbL) activity [37,38]. STUbL proteins, such as RNF4, harbor multiple SIMs that presumably dock with poly-sumoylated proteins via a multimerized SUMO–SIM interface [39,40]. STUbL proteins link sumoylation and ubiquitination pathways by catalyzing simultaneous hydrolysis of SUMO adducts and poly-ubiquitination of the target protein, preceding proteosomal degradation (see below).
Ubiquitination
There are many parallels between protein ubiquitination and sumoylation. Both modifications involve multiple processing steps that produce an active adduct, which is competent for transfer to target proteins, a reaction carried out by the cognate ligase in each pathway. Although consensus sequences surrounding the point of attachment diverge, SUMO and ubiquitin moieties share a common linkage to the lysine side chain of substrates [41]. Both SUMO and ubiquitin modify target proteins with a single copy at a single (mono-sumoylation or –ubiquitination) or multiple (multi-mono-sumoylation or –ubiquitination) lysine residues. SUMO and ubiquitin are both capable of chain extension at sites of modification, producing poly-sumoylated or –ubiquitinated target proteins [39,42,43]. Sumoylation and ubiquitination alter the functional properties of SSTFs in a myriad of overlapping ways [42].
Although the role of ubiquitination in protein degradation is well known, ubiquitination contributes to transcriptional processes via both proteolytic and non-proteolytic mechanisms. In general, mono-ubiquitination alters signaling mechanisms and/or activities of SSTFs [42,43]. For example, ubiquitination of FOXO4 drives nuclear translocation and stimulates transcriptional activation mediated by this factor [44]. In this case, ubiquitination may alter the interaction network of the target SSTFs, either by inhibiting basal protein-protein interactions or facilitating those involving ubiquitinated factors with proteins harboring ubiquitin binding domains [43]. In this way, ubiquitination likely facilitates nucleation of large protein complexes that are competent for transcriptional activation. Termination of the activity of ubiquitinated transcription factors may proceed by at least two pathways. First, the mono-ubiquitinated protein may continue to become more extensively ubiquitinated, producing a poly-ubiquitinated species, which is then subjected to proteosomal degradation. The kinetics of poly-ubiquitination would presumably dictate the active lifetime of the ubiquitinated transcription factor complex. Second, the ubiquitinated transcription factor may serve as a substrate for de-ubiquitinating enzymes, known as DUBs [45], the availability and catalytic activity of which determine the active lifetime of the ubiquitinated transcription factor complex.
The role of ubiquitination-induced proteolytic processing of transcription factors is equally complex. Poly-ubiquitination of SSTFs generally promotes degradation via the proteosomal system, and this dictates cellular levels and activities of SSTFs over time. However, the corollary is not always true: ubiquitination-stimulated proteolysis is necessary for transcriptional activation mediated by several types of SSTFs [46], and recruitment of the proteosomal machinery to the promoter template is integral to the transcriptional activation process [47]. This topic, “activation by destruction,” was recently reviewed by Geng and colleagues [42], who suggested that the transcriptional machinery marks particular SSTFs by phosphorylation when these factors are “spent.” They further suggested that phosphorylation of spent transcription factors both locks the proteins in an inactive state and recruits the proteosomal machinery to degrade the ubiquitinated transcription factor in situ. This proteolytic action of the ubiquitin-proteosomal system would then facilitate recruitment of non-marked transcription factor to the template for additional rounds of transcription.
Finally, ubiquitination does not necessarily doom transcription factors to the proteosomal system and degradation. For example, ubiquitination and subsequent, limited proteolytic processing of NF-κB is required for maturation and activation of this transcription factor (reviewed in [42]).
Interconnected PTMs
PTMs are often progressive with examples of alterations in phosphorylation leading to increased (or decreased) sumoylation, sumoylation leading to ubiquitination through the action of STUbL proteins, phosphorylation affecting acetylation, etc., in kinetically orchestrated sequences over the functional life of the protein. PTMs may also interact reciprocally as well as sequentially. Attempts have been made to quantitate the frequency of multiple, interconnected PTMs in the proteome using sequential isolation techniques and mass spectrometry [48,49]. In these studies, inhibiting the proteasome to reduce ubiquitination altered approximately 3% of all phosphorylations. Beltrao et al. used a computational evolutionary approach across the proteomes of 11 eukaryotic species to predict likely interconnected PTMs, and identified regulatory “hot spots” in protein sequences [50]. We provide a few examples of interconnected, transcriptionally relevant PTMs of SSTFs below.
Phosphorylation targets degradation
Absent in quiescent cells but essential for proliferation, c-MYC is regulated by a tight cycle of phosphorylation-driven control of activity and degradation. This cycle starts with ERK-mediated phosphorylation of Ser62, which is essential for transcriptional activity and required for subsequent GSK3-mediated phosphorylation of Thr58 (reviewed by [51]). These phosphorylations orchestrate, both individually and in combination, specific interactions that bring active c-MYC to target promoters while assuring rapid inactivation and degradation. Efficient association of PIN1, a peptidylprolyl isomerase that catalyzes production of trans-Pro63-MYC, requires Thr58 phosphorylation. This c-MYC conformation rapidly moves to target gene promoters [52] where it specifically recruits transcriptional coactivators and becomes multiply acetylated [53]. Phosphorylation on both sites is required for ubiquitination by AXIN2-scaffolded E3 ligase SCFFbw7, which targets c-MYC for proteosomal degradation. In addition to being more active, the trans-Pro63 conformation is also a substrate for trans-directed PP2A-mediated dephosphorylation of phospho-Ser62-MYC, resulting in inactivation of the protein. This normal cycle is disrupted in phosphonegative Thr58 mutants that are frequently observed in tumors and result in active phospho-Ser62-MYC accumulation [51].
Deacetylation promotes sumoylation
Competing for the same substrate target lysines, acetylation and sumoylation are competitively antagonist modifications. Regulation of protein sumoylation can occur through the action of deacetylases, which render the ε-amino groups accessible at sumoylation consensus sites. The tumor suppressor HIC1 is a SSTF required for mammalian development and silenced in many tumors. Sumoylation of HIC1 Lys314 is required for transcriptional repressor activity without affecting nuclear or subnuclear localization. Lys314 is also a substrate for acetylation by p300/CBP. Deacetylation of Lys314 by SIRT1 or HDAC4 increases sumoylation and thus the repressor activity of the protein [54].
Phosphorylation promotes sumoylation
Phosphorylation regulates the sumoylation and acetylation of some MEF2 proteins. The MEF2 transcription factors are required for myogenesis [55], and neuronal morphogenesis [56]. For MEF2A, MEF2C, and MEF2D, phosphorylation, acetylation and sumoylation are associated with regulation of transcriptional activity in several different tissues [57–59]. For each MEF2 factor, a phosphorylation-dependent sumoylation switch is present with the consensus motif of ψKxExxS/T. This motif is present in other transcription factors [60], including PPARγ2, HSF, and STAT1. Postsynaptic morphogenesis in cerebellar granule neurons is promoted by sumoylation and inhibited by acetylation of Lys403 of MEF2A. The phosphorylation status of Ser408 plays a key role in the phospho-SUMO switch, and ultimately the transcriptional regulatory activity of MEF2A. Ser408 of MEF2A is phosphorylated under basal or non-stimulated conditions and this promotes sumoylation at Lys403, which extinguishes the transcriptional activation activity of this factor. Dephosphorylation of phospho-Ser408, which appears to be catalyzed by the neuronal activity- and calcium-dependent phosphatase calcineurin, promotes a sumoylation to acetylation switch at Lys403, restoring the ability of MEF2A to activate expression of target genes [59].
Similarly to MEF2A, MEF2D sumoylation is dependent upon CDK5-mediated phosphorylation of Ser444 and sumoylation is opposed by calcineurin. Sumoylation of Lys439 of MEF2D inhibits its transcriptional regulatory activity and the ability to potentiate myogenesis. [61].
Phosphorylation-linked desumoylation
The transcriptional regulatory protein BCL11B provides a fascinating example of sequential, linked, and reversible PTMs with a clear transcriptional outcome. BCL11B exists as an ensemble of phosphorylated, sumoylated, and unmodified protein species under basal conditions in primary mouse thymocytes (Figure 4). The protein becomes rapidly phosphorylated by at least two MAP kinases, ERK1/2 and p38 [35], immediately following initiation of phorbol ester treatment. Within five minutes all three states of the BCL11B protein collapse into a species that is multiply phosphorylated co-incident with extensive desumoylation of the protein. The latter is due to the presence of a “phospho–deSUMO” switch within the BCL11B protein, the mechanistic basis of which is the phospho-BCL11B-dependent recruitment of SENP1 to the BCL11B complex and subsequent hydrolysis of SUMO-BCL11B [35]. Hydrolysis of SUMO-BCL11B is followed by a cycle of dephosphorylation and re-sumoylation, the latter of which is required for recruitment of the transcriptional co-activator p300 to SUMO-BCL11B complex and subsequent induction of expression of Id2, a gene that is repressed by BCL11B under basal conditions [35,62]. Finally, prolonged stimulation results in extensive poly-ubiquitination, perhaps via the action of an unidentified STUbL protein, and proteasomal degradation [35]. This pathway of intricate PTMs appears to serve as a molecular switch that converts the transcriptional repressor BCL11B into an activator of target gene expression, whereas the terminal step of poly-ubiquitination and proteosomal degradation likely serves as a mechanism of signal termination.
Figure 4. Sequential regulation of BCL11B by phosphorylation and sumoylation.
BCL11B, constitutively in the context of the NuRD repressor complex (NuRD), is subject to modification by kinases, phosphatases (PPTase), sumo-ligating enzymes (UBC9), sumo proteases (SENPx), and sumo-dependent ubiquitin-targeted ligases (StUBL) that alter its activity at the Id2 oncogene promoter over a 60 min time frame following mouse thymocyte stimulation. Termination of the stimulated signal involves ubiquitin-targeted degradation by the proteasome complex.
Phosphorylation-linked acetylation and ubiquitination
As described above, FOXO1, a negative regulator of insulin sensitivity [63], is a target for acetylation affecting interaction with target genes. Acetylation also promotes phosphorylation of FOXO1 by the insulin-dependent protein kinase B (PKB)/AKT, rendering a previously acetylated FOXO1 more sensitive to insulin signaling [20]. Progressive phosphorylation of FOXO1, first by PKB/AKT and then by CK1, reduces nuclear localization and DNA binding by FOXO1 [64–68], further reducing transcriptional regulatory activity [69]. Phosphorylation of FOXO1 and retention in the cytosol in insulin-sensitive, hepatic cells ultimately leads to its ubiquitination and proteasomal degradation [70].
Phosphorylation alters methylation
Prior to activated by extracellular signals C/EBPβ is maintained in a transcriptionally inactive state that is promoted by CARM1-mediated dimethylation of Arg3, which prevents interaction with SWI/SNF and Mediator complexes. MAP kinase-mediated phosphorylation of Thr235 induces a conformational changes that destabilizes CARM1–C/EBPβ interaction. Upon demethylation, C/EBPβ interacts productively with both SWI/SNF and Mediator complexes to induce transcription of target genes [24].
Interconnectedness of PTMs and possible clinical interventions
PTMs can be biomarkers of disease states and their utility in assessing and monitoring diseases of misregulation—cancer—is an emerging clinical priority. Our arsenal of clinically useful, PTM-directed drugs are few in general, and those that affect the PTM status of SSTFs are even more rare, likely because most compounds that affect PTMs lack specificity. However, the linkages between PTMs for any given disease state, once discerned, have the potential to identity novel drug targets. Several real and hypothetical drugs that impact transcription factor PTMs have been described and representative examples of these are summarized in Table 1. Of these, As2O3 is the one compound that most obviously targets linked PTMs, and does so on the oncogenic fusion protein PML-RARα [71]. The presence of PML-RARα is causative for acute promyelocytic leukemia, a subtype of acute myeloid leukemia, and the goal of chemotherapy is to eliminate this fusion protein from promyelocytes. As2O3 binds to the PML portion of the fusion protein and promotes oligomerization with subsequent sumoylation of the fusion protein. In this case, sumoylation of PML-RARα is linked to poly-ubiquitination via the action of the STUbL protein RNF4 [37,38]. Poly-ubiquitinated PML-RARα is then degraded via the proteasome, clearing the cell of this oncogenic protein and restoring the differentiative capacity of the affected promyelocytes [40].
Table 1.
Transcription factor PTMs as therapeutic targets
| PTM affected | Target Protein | Potential Use | Comments | Refs |
|---|---|---|---|---|
|
| ||||
| Phosphorylation | Tyrosine kinases | Treatment of cancer, rheumatoid arthritis, | Inhibition of tyrosine kinases by drugs, such as imatinib, ruxolitinib, and tofactinib, directly inhibits phosphorylation of STAT proteins, and indirectly inhibits phosphorylation of other transcription factors, including c-JUN, Rb1, Tp73, YAP1, and β-catenin. | [72] |
| Cyclin-dependent kinase 5 (CDK5) | Type 2 diabetes mellitus | Inhibition of CDK5-mediated phosphorylation of peroxisome proliferator-activated receptor γ (PPARγ) may be useful for T2DM. | [73] | |
| Peptidyl-prolyl cis–trans isomerase NIMA-interacting 1 (PIN1) | Treatment of cancer, including cancer of the breast | PIN1 isomerizes the pSer118–Pro119 bond of estrogen receptor α (ERα), increasing ligand-independent transcriptional activation mediated by ERα and inhibiting proteosomal-dependent degradation of the phospho-activated receptor. PIN1 expression is also elevated in some breast cancers that exhibit poor outcomes and numerous PIN1 inhibitors are in development. | [52,74–76] | |
| Calcineurin (protein phosphatase 3) | Immunosuppressant | Calcineurin activity is required to dephosphorylate and promote translocation of the transcription factor NFAT from the cytosol to the nucleus. Inhibition of calcineurin by FK506 (tacrolimus) prevents nuclear translocation of NFAT in T lymphocytes, leading to reduced expression of IL-2 and suppression of adaptive immunity. | [11] | |
| SMAD phosphatases | Spinal cord injury | Phosphorylated members of the SMAD family of transcription factors promote motor neuron axonal outgrowth. Inhibition of SMAD phosphatases may prolong the active lifetime of phospho-SMAD proteins. | [77,78] | |
|
| ||||
| Sumoylation | PML-RARα | Acute promyelocytic leukemia | Arsenic Trioxide (As2O3) promotes oligomerization and sumoylation of the PML portion of the PML- RARα fusion protein, with subsequent ubiquitination of the protein via SUMO-targeted ubiquitin ligases (STUbL proteins) and proteosomal degradation. | [71] |
| SENP proteins | Treatment of cancer | Inhibition of SENP proteins by betulinic acid and related compounds may prolong the lifetime of sumoylated transcription factors, such as SP1. | [79,80] | |
|
| ||||
| Ubiquitination | p53 | Treatment of cancer | Compounds such as Nutlin-3 bind to the MDM2 binding pocket of p53, inhibiting p53-MDM2 interaction and MDM2-mediated ubiquitination of p53, and prolonging the lifetime of activated p53. | [81] |
|
| ||||
| Methylation | Co-activator-associated arginine methyltransferase 1 (CARM1) | Treatment of cancer, potentially other disorders | Indole and/or pyazole inhibitors of the catalytic function of CARM1 may interrupt signaling by estrogen receptors in breast cancer, or other transcription factors in other hormone-dependent tumors. | [82,83] |
|
| ||||
| O-GlcNAcylation | O-GlcNAc transferase (OGT) | Treatment of cancer; particularly of the breast | Inhibition of OGT may sensitize breast cancer cells to tamoxifen therapy. | [84] |
| O-GlcNAc hydrolase (O-GlcNAcase) | As yet unknown | Inhibition of O-GlcNAcase by compounds, such as Thiamet-G, may be a means of altering the O- GlcNAc/phosphorylation reciprocal balance toward O-GlcNAc. This may serve to turn off transcription factors that require phosphorylation for activity. A similar observation has been made in the case of Tau kinases, which become hyperphosphorylated in Alzheimer’s disease and drive tau-mediated neurodegeneration. | [85,86] | |
|
| ||||
| Acetylation | Histone acetyltransferases (HATs) | Treatment of cancer, HIV | Garcinol, a natural compound, inhibits HAT activity of p300/CBP and may be useful to reprogram histone and transcription factor modifications that are characteristic of disease processes, such as cancer and HIV. | [87,88] |
Concluding remarks
Although the evidence is just beginning to accumulate on the frequency of multiply-modified proteins, it seems likely that evolving techniques, such as highly sensitive and quantitative mass spectrometry, will undercover many more examples. Multiple PTMs expand the possibilities for scalable transcriptional regulatory activity, and sequential, interdependent modifications allow for time-dependent control of gene expression in response to an initial stimulus with downstream effects that may play out over an extended time frame. Owing to the great importance of post-translational modification in dictating every aspect of transcription factor function and the key role that these proteins play in disease, it is perhaps not surprising that PTMs of transcription factors have become an attractive target for the development of therapeutic agents to treat a wide variety of diseases. Table 1 provides representative examples of current and future drug classes that target various transcription factor PTMs.
Highlights.
Transcription factors integrate extracellular signals
Transcription factor PTMs underlies signaling mechanism to nucleus
PTMs control every aspect of transcription factor function
Multiple signaling pathways converge at level of transcription factors
Sequential transcription factor PTMs allow for coincidence detection
Acknowledgments
The authors thank Mark Zabriskie for chemical structures, and Richard Hibbert for graphic design of the figures. This work was supported in part by the National Institutes of General Medical Sciences (grant GM096243 to T. M. F.) and Dental and Craniofacial Research (grant DE021879 to M. L.) of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucl Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitmarsh AJ, Davis RJ. Regulation of transcription factor function by phosphorylation. Cell Mol Life Sci. 2000;57:1172–1183. doi: 10.1007/PL00000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmberg CI, et al. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci. 2002;27:619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]
- 4.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 5.Conaway RC, et al. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ, Miska EA. Regulation of gene expression by transcription factor acetylation. Cell Mol Life Sci. 2000;57:1184–1192. doi: 10.1007/PL00000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart GW, et al. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minguez P, et al. PTMcode: a database of known and predicted functional associations between post-translational modifications in proteins. Nucl Acids Res. 2013;41:D306–11. doi: 10.1093/nar/gks1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Hao N, et al. Tunable signal processing through modular control of transcription factor translocation. Science. 2013;339:460–464. doi: 10.1126/science.1227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan PG, et al. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 12.Okamura H, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 13.Shen T, et al. The folding energy landscape and phosphorylation: modeling the conformational switch of the NFAT regulatory domain. FASEB J. 2005;19:1389–1395. doi: 10.1096/fj.04-3590hyp. [DOI] [PubMed] [Google Scholar]
- 14.Frank CL, et al. Control of activating transcription factor 4 (ATF4) persistence by multisite phosphorylation impacts cell cycle progression and neurogenesis. J Biol Chem. 2010;285:33324–33337. doi: 10.1074/jbc.M110.140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H, et al. A Src family kinase-Shp2 axis controls RUNX1 activity in megakaryocyte and T-lymphocyte differentiation. Genes Dev. 2012;26:1587–1601. doi: 10.1101/gad.192054.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trinidad JC, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarrant MK, et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat Chem Biol. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanover JA, et al. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 19.Ruan HB, et al. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab. 2013;24:301–309. doi: 10.1016/j.tem.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuzaki H, et al. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 22.Senf SM, et al. p300 Acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. Am J Physiol Cell Physiol. 2011;300:C1490–C1501. doi: 10.1152/ajpcell.00255.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowenz-Leutz E, et al. Crosstalk between C/EBPβ phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010;29:1105–1115. doi: 10.1038/emboj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark GR, et al. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res. 2011;21:375–380. doi: 10.1038/cr.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cubeñas-Potts C, Matunis MJ. SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YZ, et al. SUMOhydro: a novel method for the prediction of sumoylation sites based on hydrophobic properties. PLoS ONE. 2012;7:e39195. doi: 10.1371/journal.pone.0039195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perry JJP, et al. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Ouyang J, et al. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luis NM, et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell. 2011;9:233–246. doi: 10.1016/j.stem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Lee B, Muller MT. SUMOylation enhances DNA methyltransferase 1 activity. Biochem J. 2009;421:449–461. doi: 10.1042/BJ20090142. [DOI] [PubMed] [Google Scholar]
- 32.Gómez-del Arco P, et al. Ikaros SUMOylation: switching out of repression. Mol Cell Biol. 2005;25:2688–2697. doi: 10.1128/MCB.25.7.2688-2697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez MS, et al. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan Q, et al. Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proc Natl Acad Sci U S A. 2010;107:21034–21039. doi: 10.1073/pnas.1007866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang LJ, et al. Coordinated regulation of transcription factor Bcl11b activity in thymocytes by the mitogen-activated protein kinase (MAPK) pathways and protein sumoylation. J Biol Chem. 2012;287:26971–26988. doi: 10.1074/jbc.M112.344176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyst MJ, et al. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J. 2006;25:5317–5328. doi: 10.1038/sj.emboj.7601404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 38.Lallemand-Breitenbach V, et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 39.Ulrich HD. The fast-growing business of SUMO chains. Mol Cell. 2008;32:301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 40.de Thé H, et al. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198:11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jadhav T, Wooten MW. Defining an Embedded Code for Protein Ubiquitination. J Proteomics Bioinform. 2009;2:316. doi: 10.4172/jpb.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geng F, et al. Ubiquitin and proteasomes in transcription. Annu Rev Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 44.van der Horst A, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 45.Clague MJ, et al. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 46.Reid G, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 47.Métivier R, et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 48.Swaney DL, et al. Global analysis of phosphorylation and ubiquitylation crosstalk in protein degradation. Nat Meth. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Meth. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beltrao P, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold HK, Sears RC. A tumor suppressor role for PP2A-B56α through negative regulation of c-Myc and other key oncoproteins. Cancer Metastasis Rev. 2008;27:147–158. doi: 10.1007/s10555-008-9128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farrell AS, et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol. 2013;33:2930–2949. doi: 10.1128/MCB.01455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez-Arévalo Lobo VJ, et al. Dual regulation of Myc by Abl. Oncogene. 2013 doi: 10.1038/onc.2012.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stankovic-Valentin N, et al. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved ψKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 56.Heidenreich KA, Linseman DA. Myocyte enhancer factor-2 transcription factors in neuronal differentiation and survival. Mol Neurobiol. 2004;29:155–166. doi: 10.1385/MN:29:2:155. [DOI] [PubMed] [Google Scholar]
- 57.Ma K, et al. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol Cell Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao X, et al. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shalizi A, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 60.Yang XJ, Grégoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 61.Grégoire S, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- 62.Kastner P, et al. Bcl11b represses a mature T-cell gene expression program in immature CD4+ CD8+ thymocytes. Eur J Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakae J, et al. Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 64.Nakae J, et al. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 65.Brunet A, et al. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 66.Biggs WH, et al. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, et al. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277:45276–45284. doi: 10.1074/jbc.M208063200. [DOI] [PubMed] [Google Scholar]
- 68.Zhao X, et al. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem J. 2004;378:839–849. doi: 10.1042/BJ20031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rena G, et al. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 2002;21:2263–2271. doi: 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsuzaki H, et al. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang XW, et al. Arsenic trioxide controls the fate of the PML-RARα oncoprotein by directly binding PML. Science. 2010;328:240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 72.Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi JH, et al. Antidiabetic actions of a non-agonist PPARγ ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rajbhandari P, et al. Regulation of estrogen receptor α N-terminus conformation and function by peptidyl prolyl isomerase Pin1. Mol Cell Biol. 2012;32:445–457. doi: 10.1128/MCB.06073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rajbhandari P, et al. Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene. 2013 doi: 10.1038/onc.2013.78. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore JD, Potter A. Pin1 inhibitors: Pitfalls, progress and cellular pharmacology. Bioorg Med Chem Lett. 2013;23:4283–4291. doi: 10.1016/j.bmcl.2013.05.088. [DOI] [PubMed] [Google Scholar]
- 77.Bruce DL, Sapkota GP. Phosphatases in SMAD regulation. FEBS Lett. 2012;586:1897–1905. doi: 10.1016/j.febslet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Kelly CE, et al. Rnf165/Ark2C enhances BMP-Smad signaling to mediate motor axon extension. PLoS Biol. 2013;11:e1001538. doi: 10.1371/journal.pbio.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan Y, et al. Betulinic acid-induced programmed cell death in human melanoma cells involves mitogen-activated protein kinase activation. Clin Cancer Res. 2003;9:2866–2875. [PubMed] [Google Scholar]
- 80.Hsu TI, et al. Betulinic acid decreases specificity protein 1 (Sp1) level via increasing the sumoylation of sp1 to inhibit lung cancer growth. Mol Pharmacol. 2012;82:1115–1128. doi: 10.1124/mol.112.078485. [DOI] [PubMed] [Google Scholar]
- 81.Shangary S, Wang S. Small-molecule inhibitors of the MDM2–p53 protein–protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sack JS, et al. Structural basis for CARM1 inhibition by indole and pyrazole inhibitors. Biochem J. 2011;436:331–339. doi: 10.1042/BJ20102161. [DOI] [PubMed] [Google Scholar]
- 83.Kawabe YI, et al. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11:333–345. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanwal S, et al. O-GlcNAcylation-inducing treatments inhibit estrogen receptor α expression and confer resistance to 4-OH-tamoxifen in human breast cancer-derived MCF-7 cells. PLoS ONE. 2013;8:e69150. doi: 10.1371/journal.pone.0069150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuzwa SA, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 86.Yu Y, et al. Differential effects of an O-GlcNAcase inhibitor on tau phosphorylation. PLoS ONE. 2012;7:e35277. doi: 10.1371/journal.pone.0035277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sarli V, Giannis A. Selective inhibition of CBP/p300 HAT. Chem Biol. 2007;14:605–606. doi: 10.1016/j.chembiol.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 88.Collins HM, et al. Differential effects of garcinol and curcumin on histone and p53 modifications in tumour cells. BMC Cancer. 2013;13:37. doi: 10.1186/1471-2407-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]