Abstract
The phage T4 gene 45 protein (gp45), Escherichia coli beta and the eukaryotic proliferating cell nuclear antigen (PCNA) function in replication as processivity factors of their corresponding DNA polymerases. The T4 gp45 also functions as the transcriptional activator that connects expression of viral late genes to DNA replication. DNA tracking is an essential component of the replication and transcription regulatory functions of T4 gp45. The ability of gp45, beta and PCNA to track along DNA has been analyzed by photocrosslinking. Each of these proteins must be loaded onto DNA by a species-specific assembly factor. For gp45 and beta, the density of traffic along DNA is determined by a dynamic balance between continuous protein loading and unloading, and is also dependent on interaction with the conjugate single-stranded DNA binding protein.
Full text
PDF
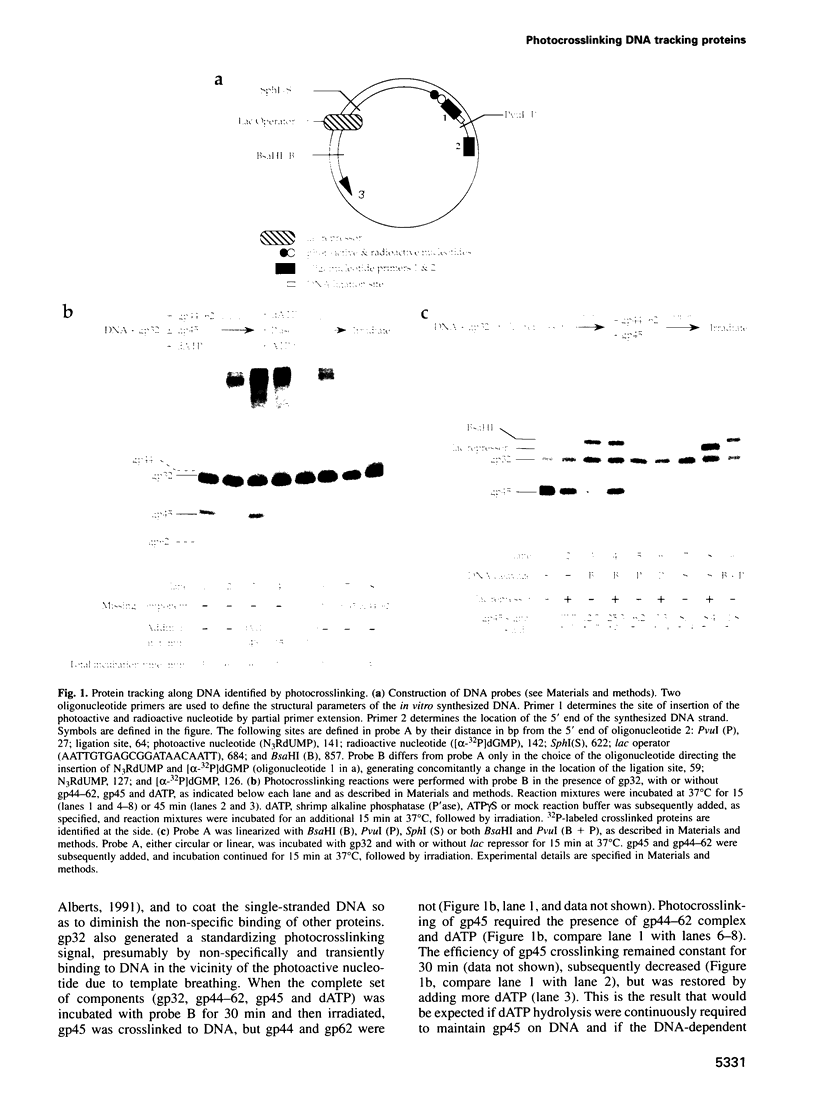


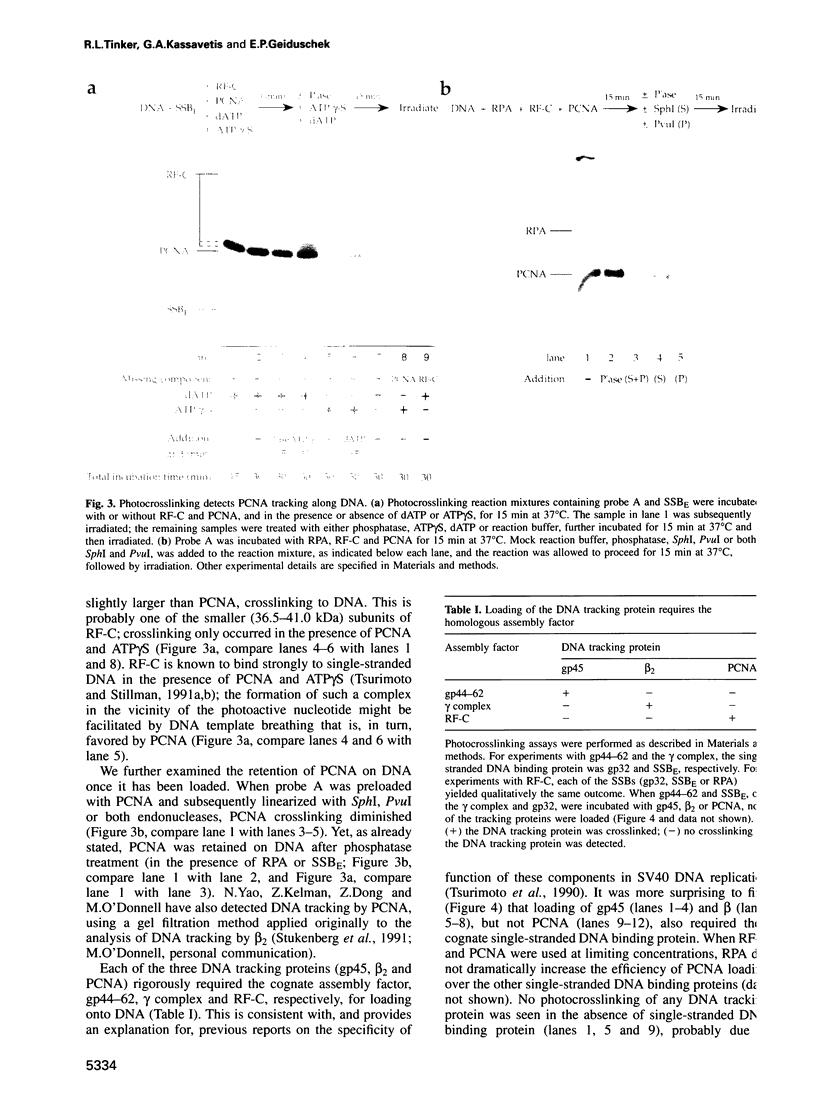



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartholomew B., Kassavetis G. A., Braun B. R., Geiduschek E. P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990 Jul;9(7):2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis G. A., Geiduschek E. P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991 Oct;11(10):5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G. A., Burgers P. M. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990 Jan 25;18(2):261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M., Yoder B. L. ATP-independent loading of the proliferating cell nuclear antigen requires DNA ends. J Biol Chem. 1993 Sep 25;268(27):19923–19926. [PubMed] [Google Scholar]
- Capson T. L., Benkovic S. J., Nossal N. G. Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell. 1991 Apr 19;65(2):249–258. doi: 10.1016/0092-8674(91)90159-v. [DOI] [PubMed] [Google Scholar]
- Coppo A., Manzi A., Pulitzer J. F. Host mutant (tabD)-induced inhibition of bacteriophage T4 late transcription. II. Genetic characterization of mutants. J Mol Biol. 1975 Aug 25;96(4):601–624. doi: 10.1016/0022-2836(75)90141-2. [DOI] [PubMed] [Google Scholar]
- Formosa T., Burke R. L., Alberts B. M. Affinity purification of bacteriophage T4 proteins essential for DNA replication and genetic recombination. Proc Natl Acad Sci U S A. 1983 May;80(9):2442–2446. doi: 10.1073/pnas.80.9.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradkin L. G., Kornberg A. Prereplicative complexes of components of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1992 May 25;267(15):10318–10322. [PubMed] [Google Scholar]
- Gogol E. P., Young M. C., Kubasek W. L., Jarvis T. C., von Hippel P. H. Cryoelectron microscopic visualization of functional subassemblies of the bacteriophage T4 DNA replication complex. J Mol Biol. 1992 Mar 20;224(2):395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- Grossman L., Thiagalingam S. Nucleotide excision repair, a tracking mechanism in search of damage. J Biol Chem. 1993 Aug 15;268(23):16871–16874. [PubMed] [Google Scholar]
- Henricksen L. A., Umbricht C. B., Wold M. S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994 Apr 15;269(15):11121–11132. [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Barry J., Alberts B. M., Geiduschek E. P. Enhancement of bacteriophage T4 late transcription by components of the T4 DNA replication apparatus. Science. 1989 Sep 1;245(4921):952–958. doi: 10.1126/science.2672335. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Williams K. P., Kassavetis G. A., Geiduschek E. P. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science. 1990 May 4;248(4955):573–578. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- Huang C. C., Hearst J. E., Alberts B. M. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J Biol Chem. 1981 Apr 25;256(8):4087–4094. [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., Hockensmith J. W., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12717–12729. [PubMed] [Google Scholar]
- Johanson K. O., Haynes T. E., McHenry C. S. Chemical characterization and purification of the beta subunit of the DNA polymerase III holoenzyme from an overproducing strain. J Biol Chem. 1986 Sep 5;261(25):11460–11465. [PubMed] [Google Scholar]
- Kenny M. K., Lee S. H., Hurwitz J. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerases alpha and delta. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., O'Donnell M. Sliding clamps of DNA polymerases. J Mol Biol. 1993 Dec 20;234(4):915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- Mace D. C., Alberts B. M. The complex of T4 bacteriophage gene 44 and 62 replication proteins forms an ATPase that is stimulated by DNA and by T4 gene 45 protein. J Mol Biol. 1984 Aug 5;177(2):279–293. doi: 10.1016/0022-2836(84)90457-1. [DOI] [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the gamma subunit essential for processive synthesis. J Biol Chem. 1988 May 15;263(14):6555–6560. [PubMed] [Google Scholar]
- Matsuoka S., Yamaguchi M., Matsukage A. D-type cyclin-binding regions of proliferating cell nuclear antigen. J Biol Chem. 1994 Apr 15;269(15):11030–11036. [PubMed] [Google Scholar]
- Morris C. F., Hama-Inaba H., Mace D., Sinha N. K., Alberts B. Purification of the gene 43, 44, 45, and 62 proteins of the bacteriophage T4 DNA replication apparatus. J Biol Chem. 1979 Jul 25;254(14):6787–6796. [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. The T4 DNA polymerase accessory proteins form an ATP-dependent complex on a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20024–20033. [PubMed] [Google Scholar]
- O'Donnell M., Studwell P. S. Total reconstitution of DNA polymerase III holoenzyme reveals dual accessory protein clamps. J Biol Chem. 1990 Jan 15;265(2):1179–1187. [PubMed] [Google Scholar]
- Pan Z. Q., Hurwitz J. Reconstitution of cyclin-dependent cdc2 and cdk2 kinase activities in vitro. J Biol Chem. 1993 Sep 25;268(27):20433–20442. [PubMed] [Google Scholar]
- Piperno J. R., Kallen R. G., Alberta B. M. Analysis of a T4 DNA replication protein complex. Studies of the DNA recognition site for T4 gene 44/62 and 45 protein-catalyzed ATP hydrolysis. J Biol Chem. 1978 Jul 25;253(14):5180–5185. [PubMed] [Google Scholar]
- Prelich G., Kostura M., Marshak D. R., Mathews M. B., Stillman B. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature. 1987 Apr 2;326(6112):471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shivji K. K., Kenny M. K., Wood R. D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992 Apr 17;69(2):367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- Sigal N., Delius H., Kornberg T., Gefter M. L., Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A., Tsaneva I. R., West S. C., Benson C. J., Yu X., Egelman E. H. The Escherichia coli RuvB branch migration protein forms double hexameric rings around DNA. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studwell P. S., O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990 Jan 15;265(2):1171–1178. [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Tinker R. L., Williams K. P., Kassavetis G. A., Geiduschek E. P. Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell. 1994 Apr 22;77(2):225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989 Feb;9(2):609–619. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991 Jan 25;266(3):1950–1960. [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem. 1991 Jan 25;266(3):1961–1968. [PubMed] [Google Scholar]
- Waga S., Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994 May 19;369(6477):207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- West S. C. The processing of recombination intermediates: mechanistic insights from studies of bacterial proteins. Cell. 1994 Jan 14;76(1):9–15. doi: 10.1016/0092-8674(94)90168-6. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Zhang H., Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992 Oct 30;71(3):505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]






