Abstract
In Lafora disease (LD), deficiency of either EPM2A or NHLRC1, the genes encoding the phosphatase laforin and E3 ligase respectively, causes massive accumulation of less-branched glycogen inclusions, known as Lafora bodies, also called polyglucosan bodies (PBs), in several types of cells including neurons. The biochemical mechanism underlying the PB accumulation, however, remains undefined. We recently demonstrated that laforin is a phosphatase of muscle glycogen synthase (GS1) in PBs, and that laforin recruits malin, together reducing PBs. We show here that accomplishment of PB degradation requires a protein assembly consisting of at least four key enzymes: laforin and malin in a complex, and the glycogenolytic enzymes, glycogen debranching enzyme 1 (AGL1) and brain isoform glycogen phosphorylase (GPBB). Once GS1-synthesized polyglucosan accumulates into PBs, laforin recruits malin to the PBs where laforin dephosphorylates, and malin degrades the GS1 in concert with GPBB and AGL1, resulting in a breakdown of polyglucosan. Without fountional laforin-malin complex assembled on PBs, GPBB and AGL1 together are unable to efficiently breakdown polyglucosan. All these events take place on PBs and in cytoplasm. Deficiency of each of the four enzymes causes PB accumulation in the cytoplasm of affected cells. Demonstration of the molecular mechanisms underlying PB degradation lays a substantial biochemical foundation that may lead to understanding how PB metabolizes and why mutations of either EPM2A or NHLRC1 in humans cause LD. Mutations in AGL1 or GPBB may cause diseases related to PB accumulation.
Keywords: Laforin, Malin, AGL1, GPBB, Polyglucosan body, Polyglucosan, Lafora disease
Introduction
Lafora disease (LD) is an early-onset fatal inherited neurological disorder characterized by three hallmarks: progressive myoclonic epilepsy (PME), severe broad types of neurological deterioration [1-2], and massive accumulation of poorly branched starch-like glycogen inclusions called Lafora bodies (LBs), or polyglucosan bodies (PBs), in several types of cells including neurons [3,2,4]. The disease-causative genes have been identified to be either the phosphatase EPM2A encoding laforin [5] or the E3 ubiquitin ligase NHLRC1 encoding malin [6]; both are mutated deficiently in most cases of LD found so far [7-8]. Mutations in other gene loci have been predicted due to instances of LD without mutations in either EPM2A or NHLRC1 [9]. These genetic studies have established a fundamental base to investigate the molecular pathogenic mechanisms underlying LD development.
Biological functional studies demonstrated that laforin is associated with the inactivation of muscle glycogen synthase (GS1) by activating kinases of glycogen synthase kinase 3 beta (GSK3β) [10-12] and AMP-activated protein kinase (AMPK) [13-14]. Laforin also inhibited glycogenesis by reducing glucose uptake and insulin sensitivity [15-16]; nevertheless, one report showed that laforin did not affect whole body glucose metabolism and insulin sensitivity [17]. Forming a complex with malin, laforin participated in the degradation of both the protein targeting glycogen (PTG)[18-19], an activator of glycogen synthase, and the glycogen generated by overexpressed PTG in neuronal cells [20]. Besides the involvement in glycogen metabolism, laforin and malin are associated with maintaining the autophagosomal-lysosomal function [21-23]. These biochemical studies emphasize an indirectly causative mechanism underlying PB accumulation. We recently demonstrated that laforin directly acts on PB metabolism through a mechanism by which laforin recruits malin to PBs where laforin dephosphorylates GS1 and together they work on PB degradation [24-25].
The rate-limiting enzymes in glycogenesis, GS1, and in glycogenolysis, the glycogen debranching enzyme AGL1 and brain isoenzyme of glycogen phosphorylases GPBB, are all ubiquitously expressed and widely distributed in mammalian cells, including astrocytes and neurons [26-29]. GS1 and GPBB are regulated by reversible phosphorylations and by allosteric effectors such as glucose-6-phosphate (G6P, a primary activator of GS1) and AMP (a primary activator of GPBB) [30-32]. Glycogen breakdown requires both GPBB and AGL1, an enzyme possessing the activities of both amylo-α1,6-glucosidase and 4-α-glucanotransferase. AGL1 removes α1,6 branch to allow glycogen to be broken down continually when glycogen phosphorylase halts four glucose residuals away from the branch during glycogenolysis [33]. Deficient mutations of AGL1 in humans cause Cori's disease (glycogen storage disease type III), an autosomal recessive disorder [34-35]. Symptoms of Cori's disease are associated with abnormal glycogen deposition in liver, skeletal muscle and heart [36]. Deficient mutations in muscle and liver isoenzymes of glycogen phosphorylases cause glycogen storage diseases of type V (in muscle) and VI (in liver) [37-38]. While mutations in the ubiquitously expressed GPBB have not been reported, we predicate its deficiency might be involved in PB accumulation.
Our results here suggest that there are two glycogen metabolic systems in mammalian cells, one for conventional normal glycogen metabolism and the other for abnormal glycogen (polyglucosan) metabolism. The polyglucosan degradation is accomplished by a protein and/or enzyme assembly, which consists of at least four key enzymes, laforin, malin, GPBB and AGL1, and proteasomal components. Knockdown of each of the four enzymes causes an inefficient degradation of PB by the assembly. Neurons are extremely vulnerable to the damage imposed by the accumulated PBs so that they succumb to the consequences early on. Further definition of the assembly will elucidate the pathogenesis of neurological disorders related to PB accumulation other than LD.
Materials and Methods
Mice and Cells
Epm2a KO mice, originally from a 129Sv strain, have been backcrossed for more than 10 generations onto a C57BL/6 background [39]. The human embryonic kidney 293FT cell line (HEK293) and the mouse neuroblastoma cell line Neuro2a (N2A) were from Invitrogen and ATCC, respectively. HEK293 cells were cultured in DMEM medium supplemented with 4.5g glucose, 2 mM glutamine, 2% penicillin, and 10% fetal bovine serum (FBS). N2A cells were cultured in MEM medium supplemented with 2 mM glutamine, 2% penicillin, and 10% FBS.
Antibodies and Reagents
In the present studies we used rabbit polyclonal antibodies to muscle GS1 (CG1183, Cell Application), phosphor-GS1 (Ser641/645) (NSB1092, Novus), AGL1 (AP2402b, Abgent), GFAP (Z0334, DAKO), GAPDH (SC-257, Santa Cruz), Lysine 48-linked polyubiquitin (clone Apu2, Millipore) and Myc and Flag tags (Sigma); and mouse monoclonal antibodies to Flag tag (M2, Sigma), GPBB (G8170-11, US Biological), Myc and V5 tags (Invitrogen) and -actin (Sigma). Enzymes and substrates of G6P dehydrogenase (G6378), phosphoglucomutase (P3397), amyloglucosidase (A7420), α-amylase (A6814), glycogen (G0885), NADP+ (N5755), glucose-1-phosphate (G1P) (G6750) were purchased from Sigma. The Periodic acid-Schiff (PAS) staining kit (Richard-Allan Scientific) and Glucose (GO) assay kit (GAGO20, Sigma) were purchased from the indicated vendors.
Plasmids and RT-PCR
The coding regions of mouse GS1 (Gys1), mouse laforin and human malin were cloned as previously described [24]. The coding region cDNA of GPBB from C57BL/6 mouse bone marrow was cloned into a pcDNA vector with Flag tag at the C-terminus of the target. The GS1 catalytically inactive double mutant E510A/E518A (GS1-EE2A) [40] and the sites1&2 glycogen-binding multiple mutant W129A/W160A/E164A/D451A/R461A/F466A (GS1-B-Mt) [41] were created by site-directed mutagenesis using Gys1 as a template. Gys1 was truncated at its N- or C-terminus or both, named ΔN10, Δ639, or ΔN10-639, respectively, as previously described [25]. The cDNA expression plasmid of Flag-tagged mouse Agl1 was a generous gift from Alan R. Saltiel [42]. The gene silencing sequence of Agl1 was 5-gaccctcttcagacttgaaca-3. The silencing sequences of Epm2a, Nhlrc1, and Pygb were described before [25]. The silencing sequence of Agl1 was cloned into a shRNA lentiviral vector [24].
Transfection and Western Blot
Generally, for a single plasmid transfection, HEK293 and N2A cells were transiently transfected with a total of 0.25 μg plasmid and 0.75 μl Lipofectamine 2000 per well in a 24-well plate for 24 hrs in Opti-MEM medium containing 10% FBS; for a multiple plasmid transfection and to obtain a high transfection efficiency over 70% for each plasmid in HEK293 cells, the total DNA amount of plasmids did not exceed 0.75μg per well in a 24-well plate. Endotoxin-free purification of plasmids is strongly recommended for the high transfection efficiency. The transfected cells were lysed with 1% Triton X-100 lysis buffer containing 20mM Tris-HCl (pH7.4), 150mM NaCl, 40mM NaF, 2 mM DTT, and a protease and phosphatase inhibitor cocktail (Sigma). Lysate supernatants were resolved on a reducing, heat-denaturing 10% SDS-PAGE.
Immunofluorescence and PAS Staining
Mouse brains were perfused with 0.9% saline followed by a freshly prepared 4% PFA in PBS buffer. Frozen brain tissues were cut into 12 μm thick sections. Prepared brain sections and cold methanol-fixed cells were permeabilized with 0.3% Triton X-100 and stained with primary and then secondary antibodies, as previously described [25]. PAS staining was performed on permeabilized cells after pretreatment with 0.5% periodic acid for 1 hr at 37°C. In α-amylase-resistant staining, the fixed cells were pretreated with 0.2U/ml α-amylase in PBS buffer at 37°C for 30 min before staining with PAS reagent.
PB Isolation, Purification and Digestion
PBs were isolated, as described before [24]. PBs were digested completely in PBS pH7.4 containing 1U/ml of amyloglucosidase and 5U/ml of α-amylase at 37°C overnight. After centrifugation at 18000g for 30 min, the glucose and protein concentrations in the supernatant were determined by the glucose GO kit and the Quick Start Bradford (BioRed) assay, respectively. The absorbance was measured at 540 nm for glucose and 595 nm for protein.
PB Degradation in vitro by Laforin-malin Complex
Equal amounts of GS1-ΔN10's PBs, isolated by a serial centrifugation method described before [24] were mixed with corresponding controls or laforin-malin complexes, which were isolated from cotransfected HEK293 cells, as previously described [24], in a buffer containing 50mM Hepes, pH 5.5-6.0, 50mM NaCl, 5 mM EGTA, 5mM mercaptoethanol and 2 mM AMP. The reaction was carried out at 37°C for 1 hr (for G1P determination) or overnight (for better disruption). The supernatant, after centrifugation at 18,000g for 30 min, was mixed with 10U/ml phosphoglucomutase dissolved in a buffer containing 0.1M Tris-HCl, pH8.0 and 5 mM MgCl2. After the phosphoglucomutase converted G1P to G6P, the G6P content was determined spectrophotometrically, as previously described [43]. The absorbance was measured at 340 nm and G6P content was calculated using the standard molar absorption of NADPH.
Results
Proteasome participates in PB metabolism
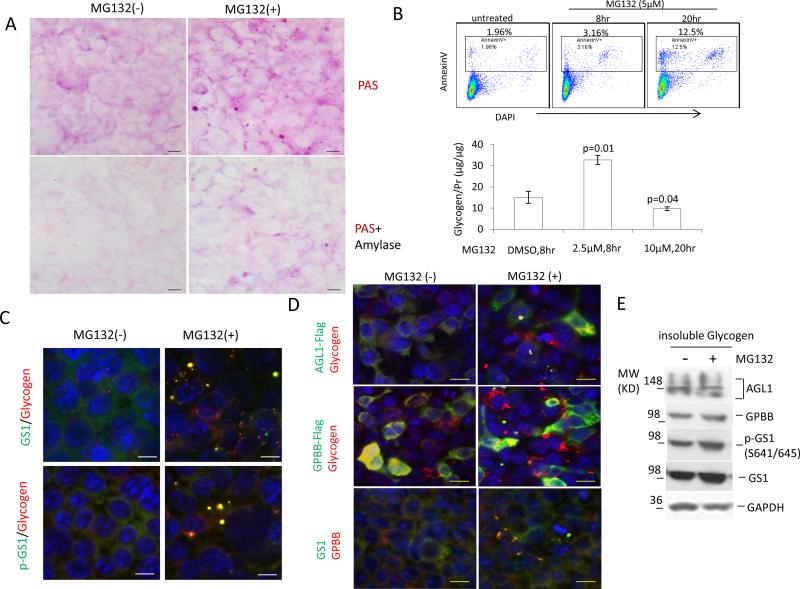
PB accumulation in LD with malin deficiency and in mice with targeted deletion of the E3 ligase suggests that proteasomes are associated with PB metabolism. To demonstrate the biochemical process, we treated HEK293 cells with a commonly used proteasomal inhibitor, MG132, at a low dose (2.5 to 5 μM) over 4 to 8 hrs. These conditions did not significantly induce cell death or apoptosis (seen in the upper panel of Fig.1b). Surprisingly, this proteasomal inhibition increased the α-amylase-resistant, PAS-positive glycogen granules or bodies in the cytoplasm of the cells and elevated the insoluble glycogen content (Fig. 1a,b), indicating that abnormal glycogen bodies, PBs, were induced or not degraded. These PBs contained the GS1 which was phosphorylated at the GSK3 target sites Ser641/645, and the glycogen breakdown enzymes, AGL1 and GPBB (Fig.1c,d). Western blotting on the induced PB deposit confirmed the presence of these components (Fig. 1e).
Fig. 1.
Proteasomal inhibition increases glycogen-positive bodies that intertwine with proteins GS1, AGL1 and GPBB. a Proteasomal inhibition by MG132 generates PBs. HEK293 cells grown on cover-glass were treated with or without 2.5μM MG132 in the culture medium for 8 hrs before methanol fixation and PAS staining. The staining was carried out after the fixed cells were pretreated with or without 0.2U/ml α-amylase for 15 min. b Short-term, low-dose treatment with MG132 increases insoluble glycogen without apoptotic induction. After treatment of HEK293 with 2.5 μM MG132 for 8 hrs or 10 μM for 20 hrs, insoluble glycogen was isolated, quantified and expressed as a value divided by protein content in the amyloglucosidase/amylase-digested solution. Vehicle DMSO treatment of the cells for 8 or 20 hrs gave a similar glycogen content and the 8-hr treatment shown as control. The cells after treated with 5 μM MG132 for 8 or 10 hrs were subjected to apoptotic analysis by flow cytometry after being stained with Annexin V (positive for apoptosis) and nuclear dye DAPI (positive for death). c,d MG132-induced PBs contain GS1, p-GS1, AGL1 and GPBB. Untransfected or transfected HEK293 cells with AGL1-Flag or GPBB-Flag were or were not treated with 2.5μM MG132 for 8 hrs before being fixed for immunofluorescent staining with antibodies to endogenous GS1, pS641/645 GS1, GPBB or glycogen, or to exogenous Flag-tagged proteins. Only merged fluorescent photos are shown. Scale bar, 10μm. e Western-blot detected these components in the isolated PBs from HEK293 cells, treated as in c.
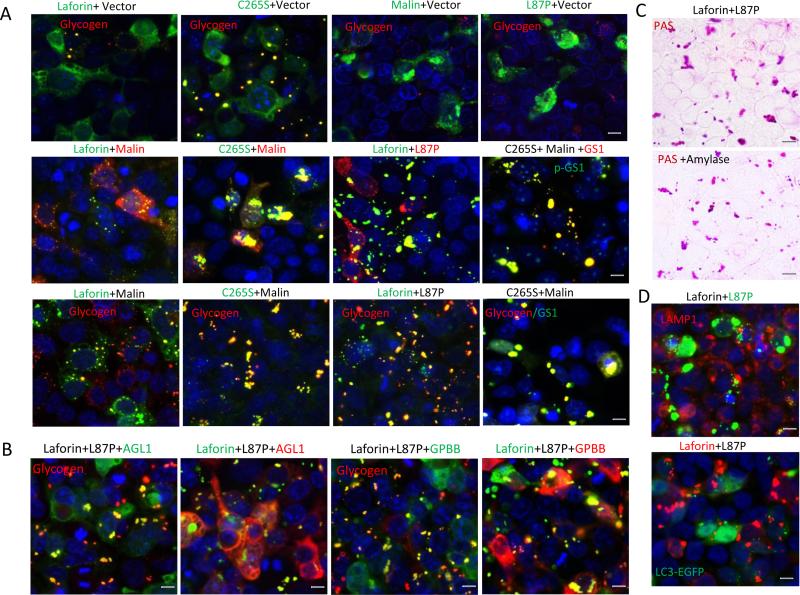
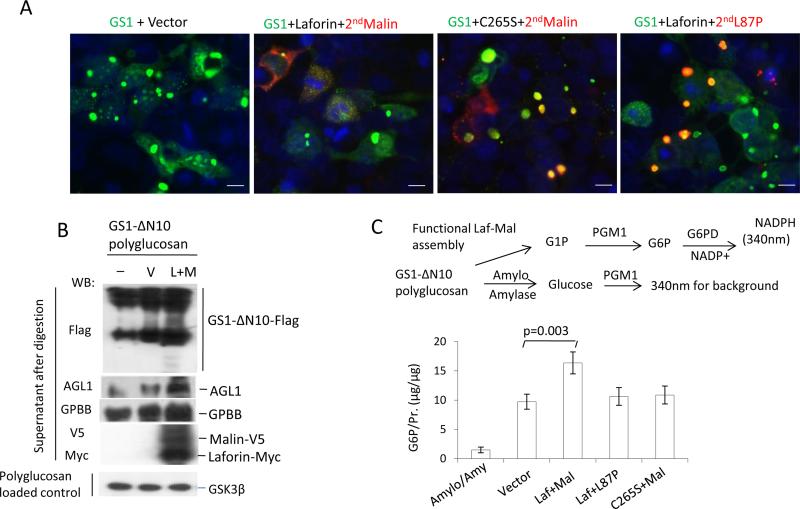
Instead of inhibiting proteasomes by MG132 to induce PBs, we were able to generate PBs by creating a dysfunctional laforin-malin complex in HEK293 cells through transient transfection of plasmids of wild type (WT) laforin and a ligase-inactive mutant L87P of malin or a phosphatase-inactive mutant C265S of laforin and WT malin. Single transfection of laforin plasmid formed much smaller and fewer glycogen-positive granules or bodies than that of its mutant C265S (Fig. 2a, top row). Single transfection of malin or its mutant L87P generated protein aggregates but did not generate glycogen-positive granules in HEK293 cells expressing low levels of laforin, because laforin was required for the stabilization and glycogen-recruiting ability of malin [24]. However, double transfection of laforin and L87P or C265S and malin to create a dysfunctional laforin-malin complex in HEK293 cells led to massive PBs accumulated in the cytoplasm (Fig. 2a). Like the PBs generated by MG132, the PBs generated by the two dysfunctional complex assemblies also contained GS1, pS641/645-GS1 (p-GS1), AGL1 and GPBB (Fig. 2a,b), and resisted amylase hydrolysis (Fig. 2c). These accumulated cytoplasmic PBs were not located in lysosomes and autophagy vesicles because they did not co-localize with the lysosomal marker LAMP1 and the autophagosomal marker LC3 (Fig. 2d). Taken together, these results show that PB metabolism takes place in cytoplasm and requires the activities of both laforin and malin, and their functional complex assembly.
Fig. 2.
Functional laforin-malin complex prevents PB accumulation. a,b Dysfunctional laforin-malin complex accumulates PBs that are stained positively for GS1, p-GS1, AGL1, GPBB and glycogen. HEK293 cells were transfected with combined plasmids for 36 hr as indicated above the photos. After transfection, the cells were fixed by methanol and doubly stained with antibodies to tags (above the photos) or endogenous glycogen, GS1 or pS641/645 GS1 (in the photos). c Laforin-L87P complex-generated PBs are resistant to α-amylase hydrolysis. Transfected HEK293 cells, as in a, were pretreated with or without 0.2U/ml α-amylase for 30 min before being stained with PAS reagents. d Laforin-L87P complex-generated PBs were located in the cytoplasm but not in lysosomes and autophagosomes. The transfected HEK293 cells were fixed by methanol or by 4% paraformaldehyde (for the LC3-EGFP transfection) and doubly stained, either with antibodies to tags or LAMP1; the LC3-EGFP was not stained with antibody. Scale bar, 10μm.
Functional laforin-malin complex degrades PB-binding GS1 to prevent synthesized PB accumulation
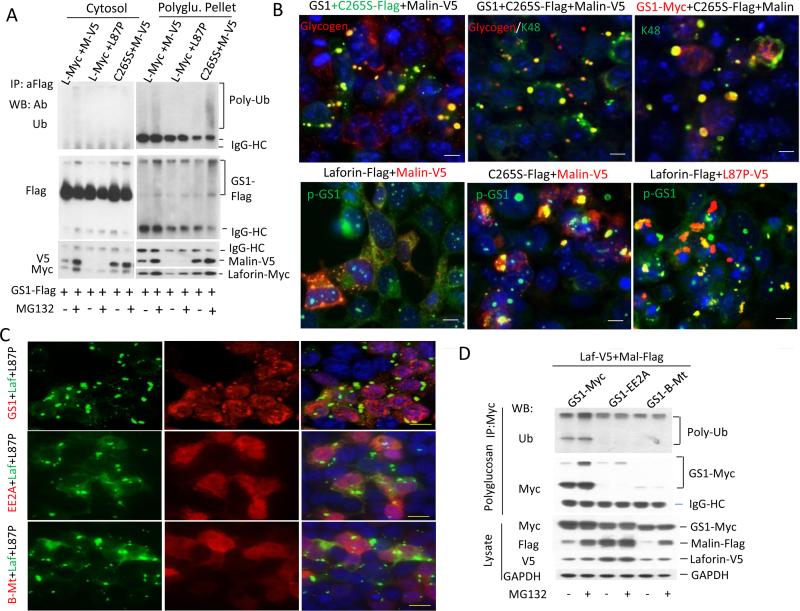
Proteasomal inhibition and the blockage of functional laforin-malin complex result in deposits of markedly accumulated GS1 in PBs, indicating that this complex targets GS1 for proteasome-mediated degradation in order to suppress the GS1-led polyglucosan synthesis. Therefore, the malin-mediated GS1 degradation takes place on PBs because laforin preferably recruits malin to PBs over soluble glycogen [24,44]. To prove this assumption, we triply transfected GS1-Flag, laforin-Myc and malin-V5 into HEK293 cells and separated the transfected GS1 into two fractions, the cytosolic soluble portion and the insoluble PB deposit. Consistent with the GS1 accumulation in the PB deposit by proteasomal inhibition, the ubiquitination of GS1 nearly occurred in the PB fraction but not in the cytosolic portion (Fig. 3a). The GS1 ubiquitination depended on malin E3 activity because the L87P-laforin complex failed in the GS1 ubiquitination (Fig. 3a). Smaller amounts of laforin and L87P were detected due to the difficult digestion and solubilization of PBs generated by the L87P-laforin complex (Fig.3a). Laforin phosphatase activity was not required for GS1 ubiquitination; rather, it was required for its degradation by malin since the C265S-malin complex caused the accumulation of K48-linked ubiquitinated GS1 in PBs (Fig. 3b last photo, top row). This result further demonstrated our recent finding that laforin is a phosphatase of GS1 on PBs [25], and that it dephosphorylates GS1 to facilitate its degradation by malin. The dephosphorylation and degradation of GS1 could take place simultaneously, since blocking its ubiquitination by mutant L87P causes phosphorylated GS1 accumulation in PBs (Fig. 3b last photo, bottom row). In other words, the dephosphorylation and ubiquitination of GS1 are both required for GS1 degradation by the laforin-malin complex so that a deficiency of either laforin or malin causes GS1 accumulation in LBs [45-49]. Moreover, the GS1 degradation continued after it bound to glycogen and began to synthesize PBs because the laforin-L87P complex did not trap the GS1 that lost its glycogen-binding ability (mutant GS1-B-Mt) [41] and its catalytic activity (mutant GS1-EE2A) [40], in the PBs (Fig. 3c), and because the laforin-malin complex did not ubiquitinate and degrade the two GS1 mutants as well (Fig. 3d). Taken together, these results show that once polyglucosan synthesizes and accumulates into bodies, laforin dephosphorylates and malin ubiquitinates the GS1 on PBs for its degradation by proteasome.
Fig. 3.
Laforin-malin complex degrades GS1 in PB pool. a Functional laforin-malin complex degrades GS1 in PB deposit. HEK293 cells were transfected for 36 hrs with the indicated tagged plasmids, and were lysed by 0.55% NP40 lysis buffer to separate cytosol and polyglucosan pellet by serial centrifugation. Ubiquitination was detected in the two fractions after the pellet was digested completely with amyloglucosidase and amylase. b Both laforin and malin are required for the degradations of GS1 and PBs. The transfected HEK293 cells, as in a, were fixed and doubly stained with antibodies to tags or endogenous p-GS1 or K48-linked polyubiquitin. c,d Laforin-malin complex did not degrade the GS1, which was incapable of binding to and synthesizing glycogen. HEK293 cells were cotransfected with laforin-L87P or laforin-malin complex, together with GS1, its catalytically inactive mutant GS1-EE2A, or its glycogen-binding mutant GS1-B-Mt, for 36 hrs before being fixed for immunostaining (c) or PB isolation (d). IP-Western blotted on PBs after completely digested with amyloglucosidase and amylase. Scale bars in b, 10μm; in c, 20μm
Laforin-malin complex acts in concert with AGL1 and GPBB to degrade PBs
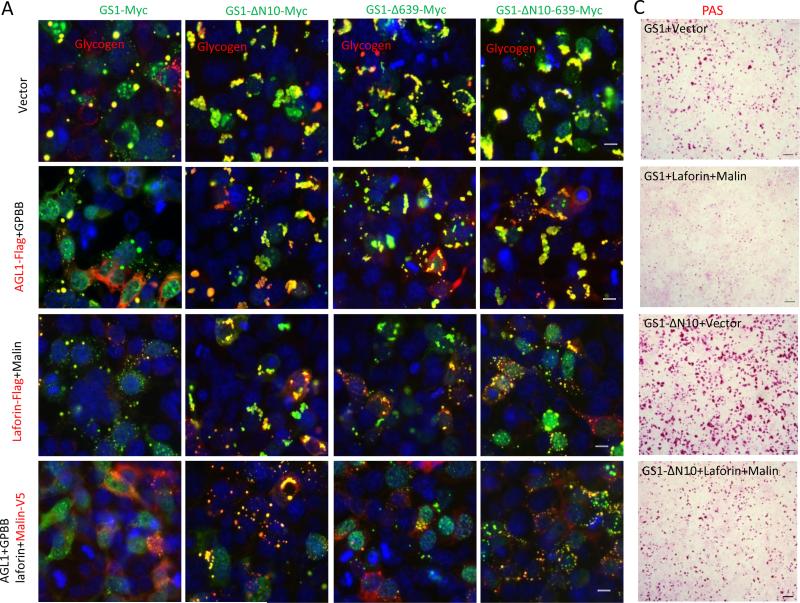
Recruitment of the four key enzymes, laforin, malin, AGL1 and GPBB, to PBs indicates that there is a cooperative process for GS1 degradation and polyglucosan breakdown. To demonstrate the process, we transfected GS1 with each of the four enzymes, or in combinations, into HEK293 cells. Overexpressed AGL1, together with overexpressed GPBB, moderately reduced the PB size and number, likely due to the presence of their highly expressed endogenous proteins. Overexpressed laforin together with overexpressed malin greatly removed the GS1-synthesized PBs (Fig. 4a), indicating that HEK293 cells express low levels of endogenous laforin and malin. This led to the laforin-malin complex becoming more efficient than the AGL1-GPBB combination for the removal of large amounts of PBs synthesized by more active truncated forms of GS1, which were made by deleting the glycogenesis-inhibiting phosphorylation regions in the N terminus (GS1-ΔN10), C-terminus (GS1-Δ639) or both (GS1-ΔN10-639) (Fig. 4a). None of the four enzymes alone affected the size and number of the more active GS1-synthesized PBs (Fig. 4b). The GS1 synthesizes polyglucosans, which always form granules, bodies, or big chunks, synthesized especially by its more active forms (Fig. 4a top row).
Fig. 4.
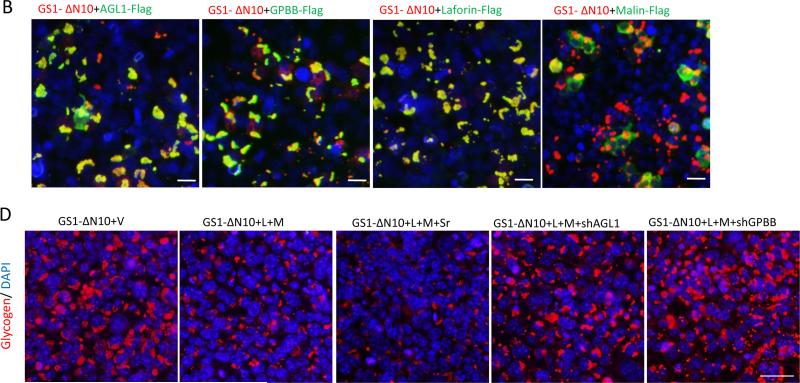
Functional laforin-malin complex, in concert with AGL1 and GPBB, degrades PBs in vivo. a,b Laforin-malin complex efficiently removes PBs synthesized by GS1 or its active truncated forms in vivo. HEK293 cells were transfected alone or in combinations with the indicated plasmids for 36 hrs before being fixed for double staining by using antibodies to tags (out of the photos) or glycogen (in the photos). c PAS staining shows PB removal by the laforin–malin complex in the cotransfected HEK293 cells as indicated. d AGL1 and GPBB are required for PB removal by laforin-malin complex. HEK293 cells were cotransfected with plasmids of GS1-ΔN10, Laforin (L), Malin (M), shAGL1, shGPBB, empty vector (V) or Scrambled sh control (Sr) as indicated for 36 hrs, and then fixed with methanol for PB staining by using antibody to glycogen. Scale bars: a,b, 10 μm; c,d, 50 μm .
The laforin-malin complex was able to remove PBs, synthesized by WT, and these truncated forms of GS1 through a comparable in vivo efficiency that did not depend on the locations of GS1-phosphorylating regions or polyglucosan sizes (Fig. 4 a,c). This indicates that it is not the phosphates on GS1 or glycogen [50] that make a great difference in the PB removal by laforin-malin complex. The laforin-malin complex removed PBs depending on the expressions of AGL1 and GPBB because knockdown of AGL1 or GPBB dramatically reduced PB removal by the complex (Fig. 4c). Generally, these results show that the laforin-malin complex, in concert with AGL1 and GPBB, degrades PBs.
To demonstrate that the PB-binding GS1 degradation is accomplished by PB breakdown, we performed a sequential transfection of GS1, laforin and malin, allowing the GS1-premade PBs to disappear from the cytoplasm by subsequent expression of functional laforin-malin complex, not dysfunctional laforin-L87P or C265S-malin complex (Fig. 5a). To see this in vivo process occurring in vitro, we combined the purified GS1-ΔN10's PBs with the isolated functional laforin-malin complex, which contains endogenous AGL1 and GPBB, in a glycogen hydrolysis buffer. As shown in Fig. 5b, the laforin-malin complex was able to disrupt the GS1-ΔN10's PBs, judging from the formation of a gel ladder of GS1-ΔN10, whereas the negative control that contains PBs and the buffer alone did not show the GS1-ΔN10 ladder. In the digested supernatant, only the laforin-malin complex, not the C265S-malin or laforin-L87P complexes, significantly increased the G1P formation (Fig. 5c). The trace G1P from the amyloglucosidase- and amylase-digested PBs demonstrated that the G1P from laforinmalin complex-digested PBs specifically comes from the breakdown of polyglucosan.
Fig. 5.
Functional laforin-malin complex, together with AGL1 and GPBB, degrades PBs in vitro. a Sequential transfection shows PB removal by the functional laforin-malin complex. HEK293 cells were transfected first with GS1 along with laforin or C265S for 24 hrs to allow PB formation, then transfected a second time with malin or its mutant L87P for another 24 hrs. After the sequential transfection, the cells were fixed for double staining to detect PBs. b,c Functional laforin-malin complex disrupts PBs in vitro. The isolated PBs of GS1-ΔN10 were digested with the isolated functional or dysfunctional laforin-malin complex, which were all isolated from transfected HEK293 cells, in glycogen hydrolysis buffer for 24 hrs (b) or 1 hr (c) at 37°C. After completing the digestion and following a 18,000g, 30-min centrifugation, the supernatant of PBs was subjected to Western-blot (b) or G1P determination (c). The parallel, isolated components from empty vector-transfected HEK293 cells served as the control for the laforin-malin complex. GSK3β acts as loading control of GS1-ΔN10 polyglucosan. The G6P was determined after conversion of G1P to G6P by phosphoglucomutase (PGM). G6P content in the digested supernatant of GS1-ΔN10 polyglucosan with amyloglucosidase and α-amylase served as a negative control for G1P (c). Scale bars, 10μm.
Functional laforin-malin assembly is required for PB degradation in neurons
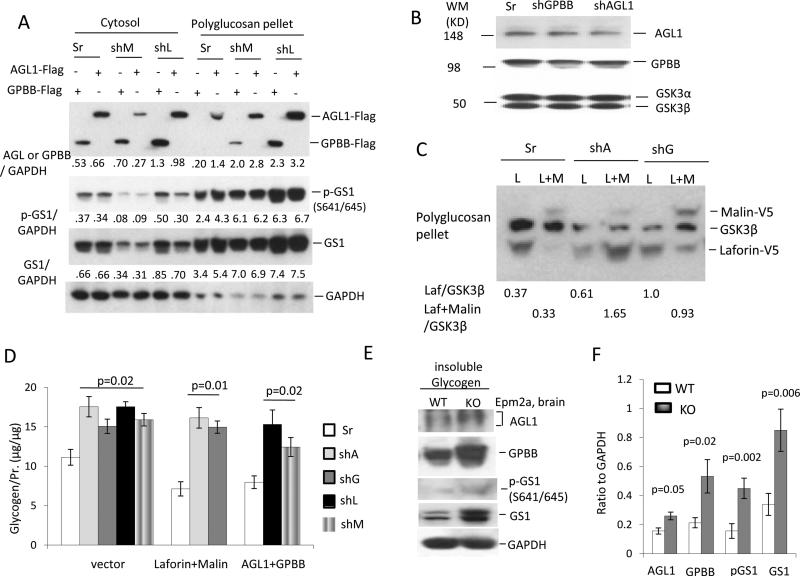
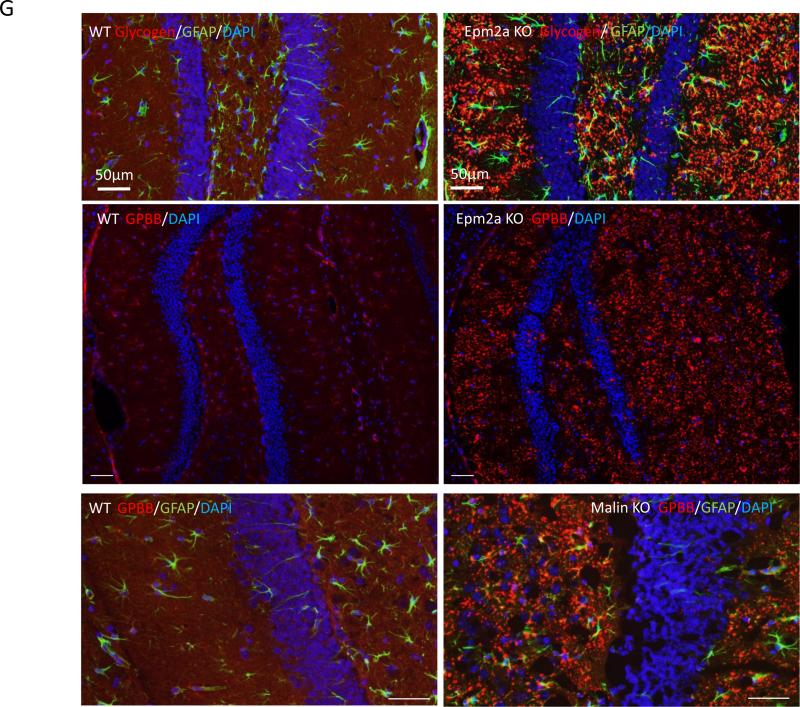
Next we examined whether each of the four key enzymes is essential for PB degradation in neuronal cells. We transfected small hairpin (sh) RNA of Epm2a, Nhlrc1, Agl1 or Pygb into N2A cells and obtained relatively stable shRNA-expressing cells after drug selection to kill the untransfected cells. AGL1 and GPBB expression in the insoluble PB portion of shEpm2a (shL) or shMalin (shM) was higher in cells expressing the scrambled shRNA (Sr). With the accumulation of AGL1 and GPBB, greater amounts of GS1 and p-GS1 also accumulated in the PB deposit versus the Sr control, although endogenous Agl and Gygb were knocked down by 50% (Fig. 6a,b). Comparably, laforin and malin expression in Agl1 or Pygb knocked-down N2A cells was also increased in the insoluble polyglucosan deposits (Fig. 6c). The accumulation of laforin and malin in the shAgl1 or shPygb cells significantly reduced the PB degradation; similarly, the AGL1 and GPBB accumulation in the shL or shM cells significantly decreased the breakdown of PBs (Fig. 6d). The shL and shM N2A cells used here are the same ones used before [24]. These results demonstrate that deficiency of each of the four enzymes causes the accumulation and degradation blockage of PBs in neuronal cells. Consistent with the above observations, AGL1, GPBB, GS1 and p-GS1 accumulated more in the isolated PBs of brain neurons from Epm2a KO mice versus those of WT mice (Fig. 6e,f). More importantly, immunohistochemistry revealed that GPBB was increased in hippocampal LBs of either laforin or malin KO mice; whereas it was evenly distributed in the hippocampi of WT mice (Fig. 6g). These results demonstrate that each of the four enzymes is essential for the PB degradation in neurons.
Fig. 6.
The laforin-malin complex in concert with AGL1 and GPBB degrades PBs in neuronal cells. a AGL1 and GPBB are accumulated in the PB deposits isolated from N2A cells that stably express the silencer of small hairpin (sh) RNA of Epm2a (shL) or Nhlrc1 (shM). AGL1 or GPBB was transfected into shL or shM or scrambled control (Sr) cells for 36 hrs before PB isolation. Western-blot detected the distribution of AGL1, GPBB, GS1 and p-GS1 in the cytosolic portions and PB deposits. The densitometry quantitation relative to GAPDH was shown in the bottom of each of the western blotting. b Knockdown efficiency of shRNA of Agl (shA) or Pygb (shG). N2A cells were lentivirally infected with shA or shG silencer for 48 hrs, and then selected with 6.5 μg/ml blasticidin for one week (about 70% cells positive for the silencer marker EGFP) before being lysed for detection of endogenous Agl1 and Gpbb. c,d Knockdown of Agl1 or Gygb increases the accumulation of laforin and malin in PB deposits and reduced PB degradation by laforin-malin complex. N2A cells stably expressing shA or shG were transiently transfected with plasmids of malin, laforin or both for 36 hrs. The accumulation of laforin and malin in shA or shG N2A cells was detected by Western-blot (c). Quantitation of glycogen content in PB deposits was determined after complete digestion (d). e,f AGL1 and GPBB are accumulated in neurons of Epm2a KO mice. LBs isolated from brains of 5-month-old Epm2a KO mice were digested and subjected to analysis of AGL1 and GPBB by western-blot (e). Insoluble glycogen isolated from age-matched WT mice acted as a control. Quantitation of the accumulation of indicated components in the insoluble glycogen pools of the two strains is shown (f). g GPBB accumulation in LBs of Epm2a KO and malin KO mice. Hippocampal sections of 5-month-old mice of the two strains were stained with antibodies to GPBB or glycogen or the astrocyte marker GFAP. Scale bars, 50 μm.
Discussion
Our results show that, in mammalian cells, there is a laforin-malin complex assembly on PBs, once they have been synthesized and accumulated, to ensure their efficient degradation in the cytoplasm. The functional assembly requires at least four key enzymes (laforin, malin, AGL1 and GPBB) and proteasomal components. These proteins may assemble like a ‘polyglucosan metabolisome’ to degrade polyglucosan granules or bodies into glucose for energy supply to cells on demand. Like normal glycogen metabolism, polyglucosan metabolism occurs in the cytoplasm of cells and, compared to normal glycogen, it supplies energy glucose more often when cellular metabolic stress has been imposed on the cells because polyglucosan formation (followed by degradation) is induced by stress [25]. Unlike normal glycogen, which is soluble and fairly evenly distributed in the cytoplasm of most cells, and has a metabolism that is highly regulated by multi-faceted factors, polyglucosan is insoluble and distributed in the form of particles or granules in the cytoplasm of diverse types of cells, such as neurons, with an extremely low normal glycogen metabolism. This lack of high regulation of polyglucosan metabolism could be beneficial for cells with a rapid turnover of polyglucosan, especially when cells encounter stress for survival. The obvious differences between polyglucosan and normal glycogen – their solubility, morphological structure, glycogen-branching extent, interior protein and glycogen [50-51] phosphorylations and fibril contaminants – predicate that polyglucosan metabolism needs a regulatory system that sustains the homeostasis and avoids the abnormal accumulation of polyglucosans.
PB formation is led by GS1, its activity being controlled primarily by its allosteric activator G6P [32]. The G6P binds to and drives phosphorylated GS1 to synthesize PBs while interwining with both G6P and GS1 [51-52]. Once PBs form, the laforin-malin complex assembles on the PBs for a simultaneous degradation of both GS1 and PBs in concert with polyglucosan breakdown by the enzymes AGL1 and GPBB. Laforin dephosphorylates the GS1 to facilitate its degradation by malin-mediated proteasome. And laforin may restrain the allosteric activation of GS1 by G6P through dephosphorylating G6P in an event that takes place on PBs. The dephosphorylation of G6P by laforin may lead to complete removal of PBs simultaneously by laforin-malin complex assembly. This assumption would explain why laforin-malin complex degrades PBs in vivo in an efficiency that does not primarily depend on the phosphates in GS1 or glycogen at physical conditions.
PB degradation by laforin-malin assembly in cytoplasm does not require the assistance of autophaghosomes and lysosomes because the nonmembrane-bound PBs cannot be engulfed by these digesting vesicles [53]. That is why almost all PBs are not located in autophaghosomes and lysosomes and why a few LBs from Epm2a KO or malin KO mice were found in these digesting systems [21-22]. Although the undegradable, cytoplasmic, abnormal glycogen, which is piling up with protein aggregates, may elicit a secondary response that allows autophagosomes-lysosomes to digest these inclusions, the event was impaired in the Epm2a or malin KO mice [23,22,21]. Our finding that PB degradation requires laforin, malin, GPBB and AGL1 lays the foundation for studying the regulation between the PB degradation machinery and the autophagomal-lysosomal system. Meanwhile, our data show that knockdown of each of the four enzymes causes an increase or sequestration of the others in the insoluble PB deposits. Whether the accumulation is caused by a secondary compensatory response related to autophagosomes-lysosomes or by the deficient PB degradation, remains to be determined.
Under stress conditions, polyglucosan formation is associated with the phosphorylation of GS1 at Ser641/645 sites by GSK3 [25]. How GSK3 regulates polyglucosan metabolism remains unknown. Endoplasmic reticulum stress increases GS1 phosphorylation in PB deposits by GSK3, eliciting neuronal degeneration [25]. This indicates that the increased phosphorylation of GS1 by GSK3 could not be beneficial for stress-induced PB degradation, and thus, for neuronal survival under stress conditions. Because laforin, malin and GSK3 work around a GS1-containing polyglucosan pool, defining their exact interactions will stimulate the development of drugs to treat or prevent the progression of patients with LD or other neurological disorders related to PB accumulation. We are the first to raise the concept of the presence and function of PB degradation machinery in mammalian cells. Our findings will stimulate further studies on the components, assembling process and regulation of the PB degradation machinery and on the effect of PB accumulation on normal glycogen metabolism and glucose metabolic abnormalities.
Acknowledgements
This work is supported by grants from the National Institutes of Health (1R21NS062391 and 5R21CA164469) and start-up package funds from Children's National Medical Center. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We gratefully acknowledge Berge B. Minassian and Peixiang Wang, who supplied the brain tissues of the malin KO mice.
Abbreviations
- G6P
Glucose-6-phosphate
- G1P
Glucose-1-phosphate
- GDE1/AGL1
Glycogen debranching enzyme 1
- GSK3
Glycogen synthase kinase 3
- GPBB
Glycogen phosphorylase brain isoenzyme
- GS1
Muscle glycogen synthase
- LB
Lafora body
- LD
Lafora disease
- PB
Polyglucosan body
- N2A
Neuro-2a
- PAS
Periodic acid Schiff
Footnotes
Author contributions
Y.W. performed tissue isolation and animal experiments and conducted the data analysis and experimental design. K.M. performed the biochemical assays. L.Z prepared brain sections. O.B. provided reagents. P.Z. and Yang L. provided advice for experimental design and manuscript preparation. Yan L. performed the molecular cloning, Western blot, immunocytostaining and PAS staining, conducted data analysis, developed the experimental design, and supervised the project. Yan L. and Y.W. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Minassian BA. Progressive myoclonus epilepsy with polyglucosan bodies: Lafora disease. Adv Neurol. 2002;89:199–210. [PubMed] [Google Scholar]
- 2.Sakai M, Austin J, Witmer F, Trueb L. Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 1970;20(2):160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 3.Gambetti P, Di Mauro S, Hirt L, Blume RP. Myoclonic epilepsy with lafora bodies. Some ultrastructural, histochemical, and biochemical aspects. Arch Neurol. 1971;25(6):483–493. doi: 10.1001/archneur.1971.00490060017002. [DOI] [PubMed] [Google Scholar]
- 4.Cavanagh JB. Corpora-amylacea and the family of polyglucosan diseases. Brain Res Brain Res Rev. 1999;29(2-3):265–295. doi: 10.1016/s0165-0173(99)00003-x. [DOI] [PubMed] [Google Scholar]
- 5.Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281(41):30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentry MS, Worby CA, Dixon JE. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci U S A. 2005;102(24):8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead OC, 3rd, Lopes-Cendes I, Tsui LC, Delgado-Escueta AV, Rouleau GA, Scherer SW. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20(2):171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 8.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, Bohlega S, Andermann E, Rouleau GA, Delgado-Escueta AV, Minassian BA, Scherer SW. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet. 2003;35(2):125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 9.Chan EM, Omer S, Ahmed M, Bridges LR, Bennett C, Scherer SW, Minassian BA. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology. 2004;63(3):565–567. doi: 10.1212/01.wnl.0000133215.65836.03. [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Wang L, Chen C, Liu Y, Zhou P, Wang Y, Wang X, Turnbull J, Minassian BA, Liu Y, Zheng P. Laforin negatively regulates cell cycle progression through glycogen synthase kinase 3beta-dependent mechanisms. Mol Cell Biol. 2008;28(23):7236–7244. doi: 10.1128/MCB.01334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet. 2005;14(18):2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Liu Y, Wu C, Zhang H, Zheng X, Zheng Z, Geiger TL, Nuovo GJ, Liu Y, Zheng P. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer cell. 2006;10(3):179–190. doi: 10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, Aguado C, Knecht E, Serratosa J, Guinovart JJ, Sanz P, Rodriguez de Cordoba S. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet. 2008;17(5):667–678. doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 14.Moreno D, Towler MC, Hardie DG, Knecht E, Sanz P. The laforin-malin complex, involved in Lafora disease, promotes the incorporation of K63-linked ubiquitin chains into AMP-activated protein kinase beta subunits. Mol Biol Cell. 2010;21(15):2578–2588. doi: 10.1091/mbc.E10-03-0227. doi:E10-03-0227 [pii] 10.1091/mbc.E10-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh PK, Singh S, Ganesh S. The laforin-malin complex negatively regulates glycogen synthesis by modulating cellular glucose uptake via glucose transporters. Mol Cell Biol. 2012;32(3):652–663. doi: 10.1128/MCB.06353-11. doi:MCB.06353-11 [pii] 10.1128/MCB.06353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vernia S, Heredia M, Criado O, Rodriguez de Cordoba S, Garcia-Roves PM, Cansell C, Denis R, Luquet S, Foufelle F, Ferre P, Sanz P. Laforin, a dual specificity phosphatase involved in Lafora disease, regulates insulin response and whole-body energy balance in mice. Hum Mol Genet. 2011;20(13):2571–2584. doi: 10.1093/hmg/ddr157. doi:ddr157 [pii] 10.1093/hmg/ddr157. [DOI] [PubMed] [Google Scholar]
- 17.DePaoli-Roach AA, Segvich DM, Meyer CM, Rahimi Y, Worby CA, Gentry MS, Roach PJ. Laforin and malin knockout mice have normal glucose disposal and insulin sensitivity. Hum Mol Genet. 2012;21(7):1604–1610. doi: 10.1093/hmg/ddr598. doi:ddr598 [pii] 10.1093/hmg/ddr598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worby CA, Gentry MS, Dixon JE. Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG). J Biol Chem. 2008;283(7):4069–4076. doi: 10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernia S, Solaz-Fuster MC, Gimeno-Alcaniz JV, Rubio T, Garcia-Haro L, Foretz M, de Cordoba SR, Sanz P. AMP-activated protein kinase phosphorylates R5/PTG, the glycogen targeting subunit of the R5/PTG-protein phosphatase 1 holoenzyme, and accelerates its down-regulation by the laforin-malin complex. J Biol Chem. 2009;284(13):8247–8255. doi: 10.1074/jbc.M808492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, Garcia-Rocha M, Soriano E, Rodriguez de Cordoba S, Guinovart JJ. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat Neurosci. 2007;10(11):1407–1413. doi: 10.1038/nn1998. doi:nn1998 [pii] 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 21.Puri R, Suzuki T, Yamakawa K, Ganesh S. Dysfunctions in endosomal lysosomal and autophagy pathways underlie neuropathology in a mouse model for Lafora disease. Hum Mol Genet. 2012;21(1):175–184. doi: 10.1093/hmg/ddr452. doi:ddr452 [pii] 10.1093/hmg/ddr452. [DOI] [PubMed] [Google Scholar]
- 22.Criado O, Aguado C, Gayarre J, Duran-Trio L, Garcia-Cabrero AM, Vernia S, San Millan B, Heredia M, Roma-Mateo C, Mouron S, Juana-Lopez L, Dominguez M, Navarro C, Serratosa JM, Sanchez M, Sanz P, Bovolenta P, Knecht E, Rodriguez de Cordoba S. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum Mol Genet. 2012;21(7):1521–1533. doi: 10.1093/hmg/ddr590. doi:ddr590 [pii] 10.1093/hmg/ddr590. [DOI] [PubMed] [Google Scholar]
- 23.Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E, Rubinsztein DC. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet. 2010;19(14):2867–2876. doi: 10.1093/hmg/ddq190. doi:ddq190 [pii] 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng L, Wang Y, Baba O, Zheng P, Liu Y. Laforin is required for the functional activation of malin in endoplasmic reticulum stress resistance in neuronal cells. FEBS J. 2012;279(14):2467–2478. doi: 10.1111/j.1742-4658.2012.08627.x. doi:10.1111/j.1742-4658.2012.08627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Ma K, Wang P, Baba O, Zhang H, Parent JM, Zheng P, Liu Y, Minassian BA. Laforin prevents stress-induced polyglucosan body formation and lafora disease progression in neurons. Mol Neurobiol. 2013;48(1):49–61. doi: 10.1007/s12035-013-8438-2. doi:10.1007/s12035-013-8438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellegri G, Rossier C, Magistretti PJ, Martin JL. Cloning, localization and induction of mouse brain glycogen synthase. Brain Res Mol Brain Res. 1996;38(2):191–199. doi: 10.1016/0169-328x(95)00305-c. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffer-Guglielmi B, Fleckenstein B, Jung G, Hamprecht B. Immunocytochemical localization of glycogen phosphorylase isozymes in rat nervous tissues by using isozyme-specific antibodies. Journal of neurochemistry. 2003;85(1):73–81. doi: 10.1046/j.1471-4159.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- 28.Narahara E, Makino Y, Omichi K. Glycogen debranching enzyme in bovine brain. J Biochem. 2001;130(3):465–470. doi: 10.1093/oxfordjournals.jbchem.a003007. [DOI] [PubMed] [Google Scholar]
- 29.Herszberg B, Mata X, Giulotto E, Decaunes P, Piras FM, Chowdhary BP, Chaffaux S, Guerin G. Characterization of the equine glycogen debranching enzyme gene (AGL): Genomic and cDNA structure, localization, polymorphism and expression. Gene. 2007;404(1-2):1–9. doi: 10.1016/j.gene.2007.07.034. doi:S0378-1119(07)00409-X [pii] 10.1016/j.gene.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 30.Crerar MM, Karlsson O, Fletterick RJ, Hwang PK. Chimeric muscle and brain glycogen phosphorylases define protein domains governing isozyme-specific responses to allosteric activation. J Biol Chem. 1995;270(23):13748–13756. doi: 10.1074/jbc.270.23.13748. [DOI] [PubMed] [Google Scholar]
- 31.Johnson LN. Glycogen phosphorylase: control by phosphorylation and allosteric effectors. FASEB J. 1992;6(6):2274–2282. doi: 10.1096/fasebj.6.6.1544539. [DOI] [PubMed] [Google Scholar]
- 32.Bouskila M, Hunter RW, Ibrahim AF, Delattre L, Peggie M, van Diepen JA, Voshol PJ, Jensen J, Sakamoto K. Allosteric regulation of glycogen synthase controls glycogen synthesis in muscle. Cell Metab. 2010;12(5):456–466. doi: 10.1016/j.cmet.2010.10.006. doi:S1550-4131(10)00358-X [pii] 10.1016/j.cmet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Villar-Palasi C, Larner J. Glycogen metabolism and glycolytic enzymes. Annu Rev Biochem. 1970;39:639–672. doi: 10.1146/annurev.bi.39.070170.003231. doi:10.1146/annurev.bi.39.070170.003231. [DOI] [PubMed] [Google Scholar]
- 34.Horinishi A, Okubo M, Tang NL, Hui J, To KF, Mabuchi T, Okada T, Mabuchi H, Murase T. Mutational and haplotype analysis of AGL in patients with glycogen storage disease type III. J Hum Genet. 2002;47(2):55–59. doi: 10.1007/s100380200000. doi:10.1007/s100380200000. [DOI] [PubMed] [Google Scholar]
- 35.Cheng A, Zhang M, Okubo M, Omichi K, Saltiel AR. Distinct mutations in the glycogen debranching enzyme found in glycogen storage disease type III lead to impairment in diverse cellular functions. Hum Mol Genet. 2009;18(11):2045–2052. doi: 10.1093/hmg/ddp128. doi:ddp128 [pii] 10.1093/hmg/ddp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfsdorf JI, Weinstein DA. Glycogen storage diseases. Rev Endocr Metab Disord. 2003;4(1):95–102. doi: 10.1023/a:1021831621210. doi:5109165 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Wolfsdorf JI, Holm IA, Weinstein DA. Glycogen storage diseases. Phenotypic, genetic, and biochemical characteristics, and therapy. Endocrinol Metab Clin North Am. 1999;28(4):801–823. doi: 10.1016/s0889-8529(05)70103-1. [DOI] [PubMed] [Google Scholar]
- 38.Dimaur S, Andreu AL, Bruno C, Hadjigeorgiou GM. Myophosphorylase deficiency (glycogenosis type V; McArdle disease). Curr Mol Med. 2002;2(2):189–196. doi: 10.2174/1566524024605770. [DOI] [PubMed] [Google Scholar]
- 39.Ganesh S, Delgado-Escueta AV, Sakamoto T, Avila MR, Machado-Salas J, Hoshii Y, Akagi T, Gomi H, Suzuki T, Amano K, Agarwala KL, Hasegawa Y, Bai DS, Ishihara T, Hashikawa T, Itohara S, Cornford EM, Niki H, Yamakawa K. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum Mol Genet. 2002;11(11):1251–1262. doi: 10.1093/hmg/11.11.1251. [DOI] [PubMed] [Google Scholar]
- 40.Cid E, Gomis RR, Geremia RA, Guinovart JJ, Ferrer JC. Identification of two essential glutamic acid residues in glycogen synthase. J Biol Chem. 2000;275(43):33614–33621. doi: 10.1074/jbc.M005358200. doi:10.1074/jbc.M005358200 M005358200 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Baskaran S, Chikwana VM, Contreras CJ, Davis KD, Wilson WA, DePaoli-Roach AA, Roach PJ, Hurley TD. Multiple glycogen-binding sites in eukaryotic glycogen synthase are required for high catalytic efficiency toward glycogen. J Biol Chem. 2011;286(39):33999–34006. doi: 10.1074/jbc.M111.264531. doi:M111.264531 [pii] 10.1074/jbc.M111.264531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori's disease. Genes Dev. 2007;21(19):2399–2409. doi: 10.1101/gad.1553207. doi:21/19/2399 [pii] 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cervenansky C, Arias A. Glucose-6-phosphate dehydrogenase deficiency in pleiotropic carbohydrate-negative mutant strains of Rhizobium meliloti. J Bacteriol. 1984;160(3):1027–1030. doi: 10.1128/jb.160.3.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan EM, Ackerley CA, Lohi H, Ianzano L, Cortez MA, Shannon P, Scherer SW, Minassian BA. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum Mol Genet. 2004;13(11):1117–1129. doi: 10.1093/hmg/ddh130. [DOI] [PubMed] [Google Scholar]
- 45.Valles-Ortega J, Duran J, Garcia-Rocha M, Bosch C, Saez I, Pujadas L, Serafin A, Canas X, Soriano E, Delgado-Garcia JM, Gruart A, Guinovart JJ. Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO Mol Med. 2011;3(11):667–681. doi: 10.1002/emmm.201100174. doi:10.1002/emmm.201100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J Biol Chem. 2008;283(49):33816–33825. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbull J, Wang P, Girard JM, Ruggieri A, Wang TJ, Draginov AG, Kameka AP, Pencea N, Zhao X, Ackerley CA, Minassian BA. Glycogen hyperphosphorylation underlies lafora body formation. Ann Neurol. 2010;68(6):925–933. doi: 10.1002/ana.22156. doi:10.1002/ana.22156. [DOI] [PubMed] [Google Scholar]
- 48.Tiberia E, Turnbull J, Wang T, Ruggieri A, Zhao XC, Pencea N, Israelian J, Wang Y, Ackerley CA, Wang P, Liu Y, Minassian BA. Increased Laforin and Laforin Binding to Glycogen Underlie Lafora Body Formation in Malin-deficient Lafora Disease. J Biol Chem. 2012;287(30):25650–25659. doi: 10.1074/jbc.M111.331611. doi:M111.331611 [pii] 10.1074/jbc.M111.331611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to lafora bodies and the accumulation of insoluble laforin. J Biol Chem. 2010;285(33):25372–25381. doi: 10.1074/jbc.M110.148668. doi:M110.148668 [pii] 10.1074/jbc.M110.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tagliabracci VS, Heiss C, Karthik C, Contreras CJ, Glushka J, Ishihara M, Azadi P, Hurley TD, DePaoli-Roach AA, Roach PJ. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13(3):274–282. doi: 10.1016/j.cmet.2011.01.017. doi:S1550-4131(11)00042-8 [pii] 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nitschke F, Wang P, Schmieder P, Girard JM, Awrey DE, Wang T, Israelian J, Zhao X, Turnbull J, Heydenreich M, Kleinpeter E, Steup M, Minassian BA. Hyperphosphorylation of glucosyl C6 carbons and altered structure of glycogen in the neurodegenerative epilepsy Lafora disease. Cell Metab. 2013;17(5):756–767. doi: 10.1016/j.cmet.2013.04.006. doi:S1550-4131(13)00148-4 [pii] 10.1016/j.cmet.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Baskaran S, Roach PJ, DePaoli-Roach AA, Hurley TD. Structural basis for glucose-6-phosphate activation of glycogen synthase. Proc Natl Acad Sci U S A. 2010;107(41):17563–17568. doi: 10.1073/pnas.1006340107. doi:1006340107 [pii] 10.1073/pnas.1006340107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kakhlon O, Glickstein H, Feinstein N, Liu Y, Baba O, Terashima T, Akman HO, Dimauro S, Lossos A. Polyglucosan neurotoxicity caused by glycogen branching enzyme deficiency can be reversed by inhibition of glycogen synthase. J Neurochem. 2013 doi: 10.1111/jnc.12277. doi:10.1111/jnc.12277. [DOI] [PubMed] [Google Scholar]