Abstract
Background
Use of a levonorgestrel-releasing intrauterine system (LNG-IUS) in humans may alter vaginal microbial populations and susceptibility to pathogens. This study evaluated the time-dependent effects of an LNG-IUS on the vaginal microbiome of the baboon, a useful animal model for reproductive studies.
Methods
LNG-IUS were inserted into three reproductively mature, female baboons. The animals were evaluated for six months by physical examination and Gram-stained cytology. The vaginal microbiota was characterized at each timepoint by culture-independent analysis of the16S rRNA-encoding gene.
Results
Each baboon harbored a diverse vaginal microbiome. Inter-individual variation exceeded intra-individual variation. Diversity declined over time in one baboon and showed mild fluctuations in the other two. There were no significant community differences from early to late post LNG-IUS placement.
Conclusions
The baboon vaginal microbiome is unique to each individual and is polymicrobial. In this pilot study, the vaginal microbiome remained stable from early to late post LNG-IUS placement.
Keywords: Levonorgestrel, nonhuman primate, pyrosequencing
Introduction
The vaginal microbiome is an important source of defense against sexually transmitted infections (STIs) in women, in part by maintaining a low pH and serving as a physical barrier to colonization [4, 10]. A healthy vaginal microbiome in women has most commonly been defined as a predominance of Lactobacillus species [12]. However, recent findings using culture-independent pyrosequencing techniques have demonstrated that the range of typical vaginal microbiota in healthy women may be greater than previously thought [34, 46].
Fluctuations in the structure and composition of the vaginal microbiome can be influenced by endogenous and exogenous factors such as age, menstrual cycle, sexual behavior, and method of birth control [10, 17, 46]. While some women have highly stable microbial populations, in others there can be rapid community fluctuations [13]. Whether and how these fluctuations impact susceptibility to pathogen invasion remains unknown. Bacterial vaginosis (BV) is a condition characterized by lasting alteration of the vaginal microbiome to a polymicrobial state, accompanied by a decrease in relative abundance of Lactobacillus species. This condition has been epidemiologically associated with increased risk for acquisition of genital pathogens such as Chlamydia trachomatis, and subsequent development of pelvic inflammatory disease (PID) [4, 31, 43]. The magnitude of change in the microbial community and the ability of the community to re-stabilize may be important factors governing susceptibility to infection [17].
Natural and induced hormonal changes have been hypothesized to impact the vaginal microbiome [6, 28, 44]. The levonorgestrel intrauterine system (LNG-IUS) is a hormone-releasing contraceptive method that provides continuous release of low-dose synthetic progestin. Previous studies have not demonstrated a significant impact on the vaginal microbiome following insertion of this device [7, 21, 30]. However, most of these studies evaluated only a single post-insertion timepoint to determine microbial changes. Only a single study evaluated microbial populations at multiple timepoints post-IUS insertion – this study found increased incidence of candidiasis in long-term users (4-7 years) but based this conclusion on cytology only and found no other microbiological alterations [21].
Additional studies have similarly used vaginal or cervical smears to evaluate the microbiome, techniques with limited sensitivity and specificity [18, 42]. Increasingly, culture-free, next-generation sequencing techniques based on analysis of 16S rRNA genes are being utilized for microbial profiling due to their speed, accuracy, and ability to identify species that are difficult or impossible to cultivate by traditional methods [11, 12, 20]. Longitudinal microbial profiling of LNG-IUS related changes by next-generation sequencing would be a useful technique to understand the potential impact of this device on susceptibility to disease.
While monitoring techniques can be performed in women, the identification of animal models that could subsequently be utilized in challenge studies would be an important means of hypothesis-testing for microbial profiles thought to be associated with disease susceptibility. The baboon is an excellent model for reproductive studies due to the similar size and shape of the reproductive tract to women, the straight cervical canal that facilitates cervical examination, cyclic perivulvar skin swelling that enables external monitoring of reproductive cycling, and the lack of significant zoonotic disease risk in this species [3, 15, 19]. Our group has already demonstrated that a LNG-IUS can be successfully placed and maintained in baboons [2]. The aim of this study was to determine if the vaginal microbiome changes over time following placement of a LNG-IUS, as assessed by culture-independent 16S rRNA gene pyrosequencing in a prospective baboon model.
Methods
Humane Care Guidelines
This research was reviewed and approved by the Texas Biomedical Research Institute (TBRI) Institutional Animal Care and Use Committee, where the research was conducted. An exemption for off-site animal work was received from the University of Michigan Committee for the Care and Use of Animals.
Study Population and IUS Placement
The study population and LNG-IUS placement method utilized in this study have been previously described [2]. Briefly, a LNG-IUS device (Mirena®, Bayer Healthcare Pharmaceuticals, Wayne, NJ, USA) was inserted in three reproductively mature female olive baboons (Papio anubis) housed in a large research colony (Texas Biomedical Research Institute, San Antonio, TX, USA). The three baboons were uniquely identified at TBRI as 16594, 16999, and 26502, and are hereafter referred to as baboon 1, baboon 2, and baboon 3, respectively. Insertion of the device was analogous to that in women without alteration to the device or introducer. After placement, the strings were trimmed to the level of the cervical os to prevent premature removal by the baboons, a variation from the normal procedure in women in which the string is left in place. Animals were sedated for complete physical examinations, and transabdominal ultrasounds were performed once weekly for four weeks, and then monthly for five months [2]. Devices were removed at 24 weeks (six months) post-insertion.
Sample Collection
Vaginal samples were taken at baseline and at each subsequent physical examination point, for a total of ten samples per animal, representing six months of LNG-IUS placement. During each sedated physical exam, two Dacron vaginal swabs (Fischer Scientific, 1490723, Pittsburgh, PA, USA) were taken from the distal half of the vagina. One was used to create a Gram-stained vaginal cytology smear, and then stored in liquid Amies media. The other was immediately stored in liquid Amies media. Swabs were immediately frozen (−80°C) after collection. Gram-stained smears were evaluated for the presence of Gram-positive and Gram-negative organisms using the Nugent scoring system [33]. In brief, this system utilizes the prevalence of pathogenic bacterial morphotypes and absence of Lactobacilli to assign a numerical score characterizing the vaginal microbial community. Scores range from one to ten, and, in women, scores greater than or equal to seven reportedly correspond to bacterial vaginosis, a dysbiotic state. In addition to sample collection, reproductive cycle stage was recorded at baseline and at each physical examination. Cycle stage was estimated by a numerical score corresponding to the degree of observable tumescence of the perivulvar skin. The score was converted to a cycle stage according to previously published criteria [41]. Vaginal pH was measured by swabbing the vaginal wall with a Dacron-tipped swab, which was then rolled on a pH indicator strip.
DNA Extraction
Total DNA was extracted from baboon vaginal swab tips using a Mo Bio PowerSoil®–htp 96 Well Soil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA) on the epMotion 5075 automated pipetting system (Eppendorf AG, Hamburg, Germany).
Amplification and Sequencing of 16S rRNA Genes
The generation and analysis of 16S rRNA-encoding gene amplicons was performed according to the protocol adopted by the NIH Human Microbiome Project (http://www.hmpdacc.org/doc/16S_Sequencing_SOP_4.2.2.pdf) for the V3-V5 hypervariable regions. Samples were amplified using Primer A, containing an “A” adapter oligonucleotide sequence + barcode + primer 926R (5’-CCATCTCATCCCTGCGTGTCTCCGACTCAGXXXXXCCGTCAATTCMTTTRAGT-3’) and Primer B, containing a “B” adapter oligonucleotide sequence + primer 357F (5’- CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCCTACGGGAGGCAGCAG-3’). The bold portions are 926R and 357F, respectively, and XXXXX in primer A indicates the location of a unique, 5-10 nucleotide barcode sequence. Primary PCR was carried out using 2 μL of DNA, 0.2 μM of each primer (A and B), 0.15 μL AccuPrime Taq DNA Polymerase High Fidelity and 1X AccuPrime PCR Buffer II (Invitrogen, 12346-094) under the following thermal cycler conditions: 95° C for 2 minutes followed by 30 cycles of 95° C (20 sec), 50° C (30 sec), 72° C (5 min) and a final hold temperature of 4° C. A 2 uL sample of each primary PCR product was verified to consist of the expected ~660 bp band fragment on a 2% agarose E-gel containing SYBR Safe (Invitrogen, G7208-02). Subsequently each sample was purified using AMPure beads according to manufacturer instructions, with the exception of the use of 0.6× for the bead volume rather than the recommended 1.8× in order to optimize output (Beckman Coulter Genomics, A63881). This was followed by quantification in duplicate with a Quant-iT PicoGreen® dsDNA Assay Kit (Invitrogen, P11496). A pooled library was created, normalizing for number of molecules, and subjected to a final round of purification using AMPure beads. Quality control of the pooled library was monitored by first observing a single band on an Agilent Bioanalyzer using a High Sensitivity DNA Kit (# 5067-4627) and then by determining molecules/μL using qPCR (KAPA Biosystems, KK4851). Pyrosequencing was performed on the finished library on the Roche 454 GS Junior Titanium platform according to the manufacturer's instructions (Roche 454 Life Sciences, Branford, CT, USA).
Sequence Processing
Bacterial 16S rRNA gene sequences were processed using the software program mothur [36, 38-40]. A total of 100,661 sequences were obtained. Sequences with less than 200 bases, with ambiguous bases, with homopolymers longer than eight bases, or with missing or erroneous barcodes were removed. Sequences were aligned to a SILVA alignment database using the Needleman-Wunsch and NAST algorithms, and sequences that did not share a defined alignment space were trimmed [36, 38]. Chimeras were removed using the uchime algorithm [8]. The Ribosomal Database Project (RDP) naïve Bayesian classifier within mothur was used to assign 16S rRNA gene sequences to a taxonomy outline to the genus level, using a confidence cutoff of 80%, and sequences classified as “Chloroplast”, “Mitochondria”, or “unknown” were removed. This resulted in a total of 79,997 reads, averaging 255 bases in length, with an average of 2512 sequences per sample. For analysis, the number of sequences in each sample was normalized to 1129 by subsampling. Relative abundances were calculated and represented as bar graphs and pie charts. Sequences were grouped into operational taxonomic units (OTUs) based on at least 97% sequence similarity [36, 39].
Data Transformation and Statistical Analysis
Data transformation and statistical analysis was performed using algorithms within mothur. To enable data analysis across timepoints and individuals, a distance matrix was first generated by calculating the θYC dissimilarity coefficients across all timepoints and all individuals. The θYC dissimilarity coefficient (1 - θYC similarity) is a function of OTU proportions from both shared and non-shared OTUs between two communities [45]. Statistical analyses for each type of data comparison were applied to the θYC distance matrix as described below.
Inter-individual microbiome variation between baboons at all timepoints was evaluated as follows. All timepoints for a given baboon were combined to form a single group that represented the overall microbiome of that baboon. Inter-individual comparison was by analysis of molecular variance (AMOVA). Data clustering between timepoints was evaluated by AMOVA, with significance defined as p < 0.05 [1, 9, 27]. Bonferroni correction for multiple comparisons is included in the mothur algorithm for AMOVA comparisons. Principal Coordinates Analysis (PCoA) was applied for data visualization, using algorithms within mothur. Diversity scores at each timepoint were calculated within mothur using the inverse Simpson (1/D) and nonparametric Shannon diversity indices. These calculations were applied to a shared file containing relative abundance data for each OTU at each timepoint. This same shared file was used to determine the number of OTUs shared across all three baboons, and the number of OTUs unique to each baboon. OTUs present at a relative abundance of at least 1% in all three baboons were considered shared across all three. OTUs present at a relative abundance of at least 1% in only one baboon were considered unique to that baboon.
Microbiome changes between baseline (day 0) and post-LNG-IUS placement were evaluated for all baboons grouped and for each baboon individually. For the grouped comparison, the day 0 timepoints for all three baboons were combined. Each subsequent timepoint was also combined across all 3 baboons. AMOVA was used to compare the day 0 group to each subsequent timepoint group. For intra-individual comparisons, the microbiome at day 0 for an individual baboon was compared to each subsequent timepoint for that individual using a weighted UniFrac algorithm (which considers relative abundance) within mothur [24, 25, 37]. Bonferroni correction for multiple comparisons was applied to the significance scores.
Microbiome alterations between early (one month post-placement) and late LNGIUS timepoints were evaluated both inter-individually and intra-individually. For inter-individual comparisons, data from timepoints day 8 through 29 (first month after IUS placement) were grouped across all three baboons. This group was then compared by AMOVA to a group consisting of the microbiome from all three baboons at remaining timepoints (days 57 through 174). For intra-individual comparisons, the early microbiome timepoints (days 8-29) for each individual baboon were combined and compared to a group consisting of the microbiome at remaining timepoints (days 57-174). These timepoint groupings were chosen based on clinical practice and device manufacturer guidelines suggesting increased risk of infection in the first 20 days of LNG-IUS placement and reports of increased occurrence of abnormal vaginal discharge confined to the early time period of IUD placement [30].
Results
Reproductive Cycle Phase and Vaginal pH
Baboons 1 and 2 were in stage zero (luteal phase) during device placement and baboon 3 was in stage three (follicular phase), as defined by degree of perivulvar skin tumescence [41]. As previously described, all baboons converted to the external appearance of luteal phase (no tumescence) within one month of device placement and remained in that state for the six-month duration of the study [2]. No adverse clinical signs were noted, and all baboons retained the LNG-IUS devices for the duration of the study [2].
All three baboons had vaginal pH values > 4.5 at day 0, and the pH remained relatively alkaline for the remainder of the study. There was no consistent trend in pH changes across baboons, although baboons 1 and 3 had pH values of 6.5 for the majority of the study, while baboon 2 had a less alkaline pH of 5.5 at most timepoints (Table 1).
Table 1.
Vaginal pH pre-IUS placement (Day 0) and post-IUS placement.
| Baboon 1 | Baboon 2 | Baboon 3 | |
|---|---|---|---|
| Day 0 | 5 | 6 | 5 |
| Day 8 | 6.5 | 6.5 | 6.5 |
| Day 15 | 6.5 | 6.5 | 6.5 |
| Day 22 | 7 | 5.5 | 6.5 |
| Day 29 | 6.5 | 5.5 | 6.5 |
| Day 57 | 7.5 | 6 | 6.5 |
| Day 85 | 6.5 | 7 | 5.5 |
| Day 113 | 6.5 | 5.5 | 6.5 |
| Day 141 | 6.5 | 5.5 | 6 |
| Day 174 | 6.5 | 5.5 | 6.5 |
Gram Stain
Gram-stained slides made from vaginal swabs taken at each timepoint were scored according to Nugent scoring criteria [33]. For baboons 1 and 3, smears consistently demonstrated very low numbers of Lactobacillus morphotypes, moderate to high numbers of Gardnerella or Bacteroides morphotypes, and high numbers of curved Gram-negative bacilli at all timepoints, resulting in Nugent scores greater than or equal to seven. Smears from baboon 2 contained higher numbers of Lactobacillus morphotypes, although Gardnerella/Bacteroides morphotypes and curved Gram-negative bacilli remained at similar levels as for the other two baboons. Thus, three timepoints for baboon 2 (day 15, day 57 and day 113) had Nugent scores below seven, although the remaining scores were greater than or equal to seven as for the other baboons. While, overall, Nugent scores did not demonstrate a trend in any baboon following placement of the IUS, there was an increase in Nugent score at day eight for all baboons, which subsequently declined to equal or below original values for baboons 2 and 3, but remained elevated for baboon 1 (Table 2).
Table 2.
Nugent scores pre-IUS placement (Day 0) and post-IUS placement.
| Baboon 1 | Baboon 2 | Baboon 3 | |
|---|---|---|---|
| Day 0 | 7 | 7 | 9 |
| Day 8 | 10 | 10 | 10 |
| Day 15 | 10 | 6 | 10 |
| Day 22 | 10 | 8 | 10 |
| Day 29 | 9 | 8 | 10 |
| Day 57 | 10 | 6 | 10 |
| Day 85 | 10 | 7 | 10 |
| Day 113 | 10 | 5 | 8 |
| Day 141 | 9 | 8 | 10 |
| Day 174 | 10 | 7 | 10 |
Vaginal microbial communities: descriptive evaluation of community membership and structure
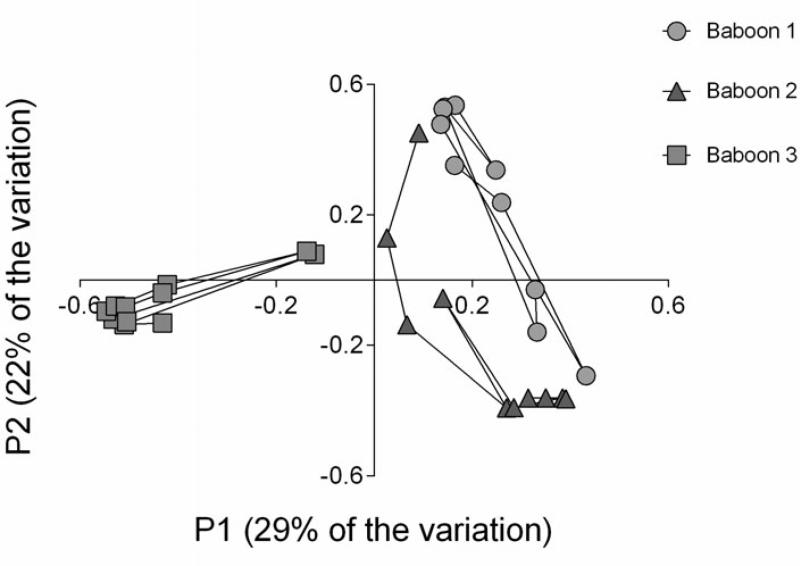
The vaginal bacterial communities of the three baboons at baseline before IUS insertion and during six months with an IUS were compared using 16S rRNA gene sequence analysis. Pyrosequencing results of 16S rRNA were compared across all timepoints and all individuals for descriptive evaluation of overall community membership (ie. which organisms are present) and community structure (ie. the relative abundance of these organisms). Overall there were significant inter-individual differences in vaginal bacterial communites based on θYC dissimilarity (AMOVA, p<0.001) and illustrated by PCoA (Figure 1). Taking into account both the operational taxonomic units (OTUs) present and their relative abundances, θYC dissimilarity is a measure of community structure difference. With respect to membership, 164 OTUs were shared across all baboons. Baboons 1, 2, and 3 had 21, 14, and 16 unique OTUs, respectively. Thus, the majority of OTUs were shared across all baboons.
Fig. 1.
Principal Coordinates Analysis (PCoA) of bacterial 16S rRNA genes reveal clustered vaginal microbiome communities structured by individual baboon. Each point corresponds to the vaginal bacterial community for a single timepoint from baboon 1 (blue circles), baboon 2 (red triangles) or baboon 3 (green squares). The percentage of variation explained by the plotted principal coordinates is indicated on the axes. The microbial communities for each individual were significantly different from that of other individuals as determined by AMOVA (P < 0.001). Data fit best on three axes (R2 = 0.79), two of which are shown.
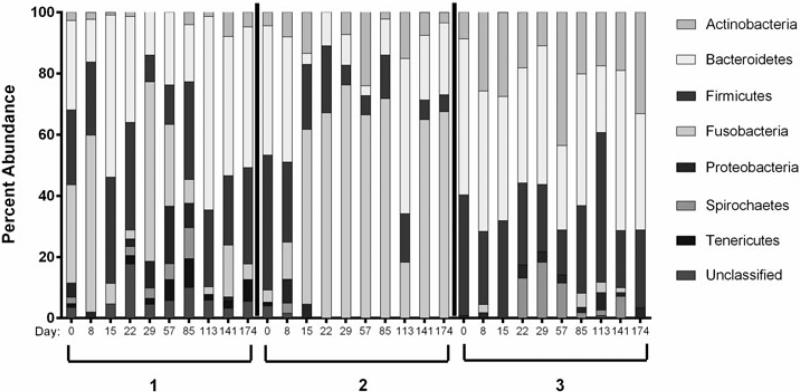
There were significant inter-individual differences in community structure, as evidenced when comparing relative abundances across baboons and across timepoints (Figure 2). The most common phyla across all baboons at all timepoints were Bacteroidetes, Firmicutes, Fusobacteria, and Actinobacteria. Baboons 1 and 2 had a higher proportion of Fusobacteria in comparison to baboon 3, in which this phylum appeared infrequently and in low proportion. Baboon 3 had similar relative abundances of Bacteroidetes and Firmicutes but higher levels of Actinobacteria and lower Fusobacteria in comparison to the other baboons. For the most part, the genera within these phyla consisted of anaerobes, including Sneathia within the phylum Fusobacteria, Porphyromonas and Prevotella within the phylum Bacteroidetes, and Peptostreptotoccus within the phylum Firmicutes.
Fig. 2.
Phylum-level relative abundance for all three baboons at day 0 before IUS placement, and for 6 months following placement. Days postplacement are indicated on the x axis.
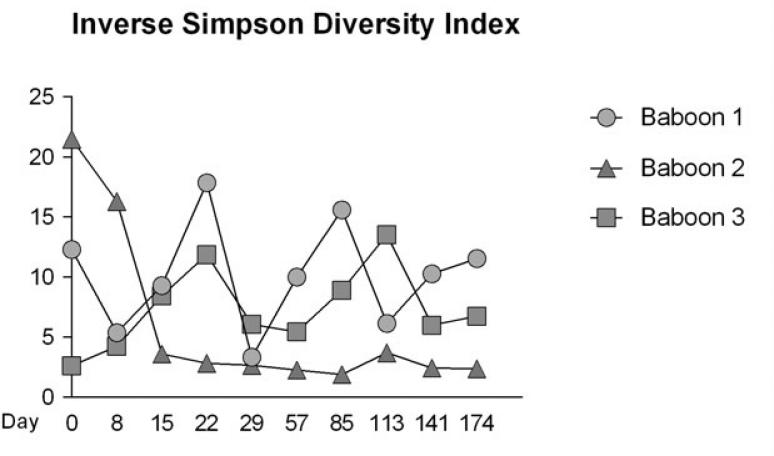
Diversity scores over time for each baboon, as measured by the inverse Simpson diversity index, were highest for baboons 1 and 3, in comparison to baboon 2 (Figure 3). Baboon 2 showed a decrease in diversity up to day 15, at which point the diversity index stabilized at a low value. For this baboon, day 15 corresponded to Fusobacteria becoming the dominant phylum (Figure 2). Diversity was also evaluated using the nonparametric Shannon index, and results were similar to those found for inverse Simpson (data not shown).
Fig. 3.
Plot of inverse Simpson diversity index at each timepoint for each baboon. Mean diversity index and standard deviation for each baboon are as follows: baboon 1 mean 10.18 _ 4.49; baboon 2 mean 5.97 _ 0.16; baboon 3 mean 7.15 _ 3.29. Baboons 1 and 3 had higher mean diversity and higher standard deviations, but did not show increasing or decreasing trends in diversity. Baboon 2 showed a decrease in diversity from day 0 to day 15, at which point the diversity index stabilized at a low value. Nonparametric Shannon diversity indices were also calculated for each timepoint, and showed similar trends (data not shown).
Vaginal microbial communities: pre- and post-LNG-IUS placement
Microbial community differences between the pre-LNG-IUS placement timepoint (day 0) and other timepoints were evaluated for all baboons together and for each individual baboon. When day 0 timepoints representing the baseline microbiome were grouped across baboons and compared to cross-baboon groups representing each post-placement timepoint, no significant community differences were found at any timepoint (AMOVA, p >0.05). When making the same comparison of day 0 vs other timepoints for each baboon individually, there were significant differences only for Baboon 2 between days 0 and 85. (weighted UniFrac, p<0.001).
Vaginal microbial communities: early vs late LNG-IUS placement
Since current clinical practice is based on the assertion that LNG-IUS pose the greatest risk of infection in the first 20 days post-placement, we also compared microbial communities between early (within the first month post-placement) and late (the last five months post-placement) timepoints. There were no significant differences between the communities present in the first month and those in the last five months following device placement when considering all baboons grouped together (AMOVA, p>0.05) or when considered separately (AMOVA, p > 0.05). Baboon 2 approached significance (p = 0.057), perhaps corresponding to the higher proportion of Fusobacteria at timepoints of day 15 and later for this animal (Figure 2).
Vaginal microbial communities: specific phyla and further taxonomical evaluation
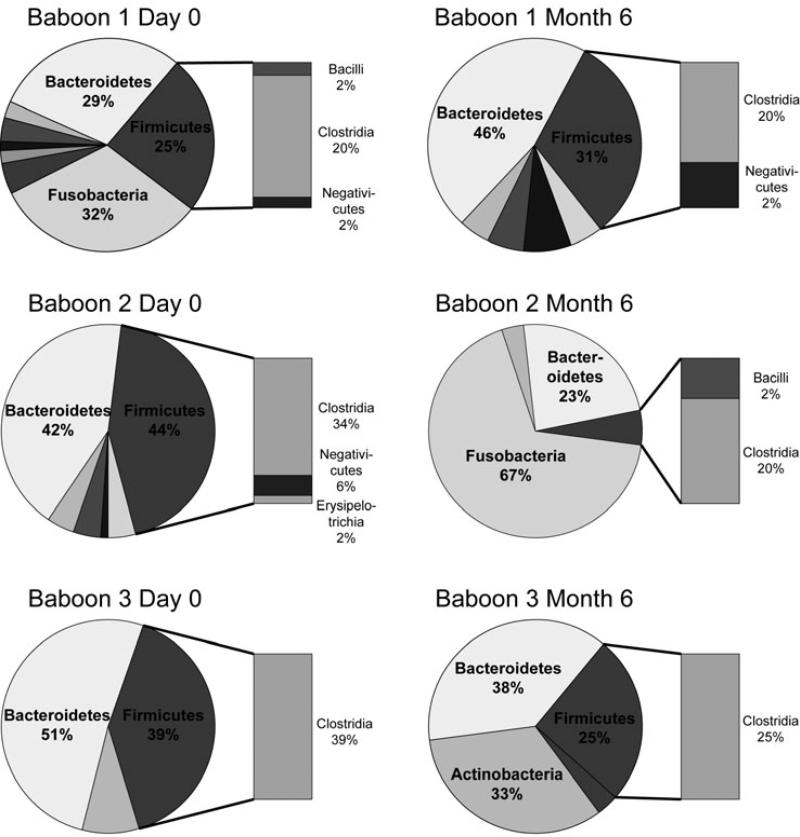
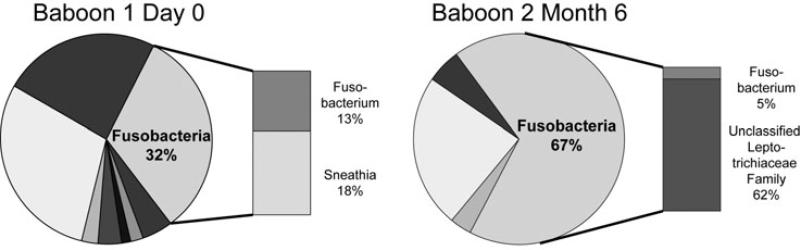
We further evaluated selected phyla known to be of particular relevance to the human condition of bacterial vaginosis. These included Firmicutes, which includes the Lactobacilli, and Fusobacteria, since increased abundance of Fusobacteria has been associated with bacterial vaginosis in humans [23]. In the baboons, Firmicutes was composed primarily of Clostridia at the class level, with relatively low abundances of Bacilli and the Gram-negative anaerobes Negativicutes (Figure 4). At the genus level, the relative abundance of sequences within Firmicutes classified as Lactobacillus (class Bacilli) was below one percent for baboons 1 and 3. Baboon 2 was the only animal that ever had Lactobacilli present above the 1% level, with relative abundances of this genus present at 4%, 6%, and 9% at days 57, 85, and 113, respectively. Members of the phylum Fusobacteria were present in higher relative abundance at certain timepoints (days 0, 8, 29, 57) in baboon 1 and at timepoints after day 15 in baboon 2 (Figure 2). These consisted predominantly of Sneathia (in the Leptotrichiaceae family) in baboon 1 and unclassified members of the Leptotrichiaceae family in baboon 2 (Figure 5).
Fig. 4.
Phylum level relative abundance for the three baboons at timepoints pre-IUS placement and at 6 months post-placement. Class level is represented for Firmicutes in the bar graph, and shows that the Firmicutes in the baboon consisted predominantly of Clostridia, as opposed to Lactobacilli (included in the class Bacilli). The graphs shown are representative of Firmicutes class composition for the majority of timepoints.
Fig. 5.
Genus level percent abundance for the phylum Fusobacteria. Genera consisted primarily of Sneathia (a member of the family Leptotrichiaceae) for baboon 1 and unclassified members of the Leptotrichiaceae family in baboon 2. The graphs shown are representative of Fusobacteria genus composition at days 0, 8, 29, and 57 for baboon 1 and timepoints after day 15 for baboon 2.
Discussion
This study was intended as a preliminary comparison of the effects of LNG-IUS placement on the vaginal microbiome in a baboon model. Previous evaluations of the effects of LNG-IUS on the microbiome are sparse and equivocal, suggesting either an increased incidence of bacterial vaginosis or aerobic vaginitis, as assessed by cytology [7], or increased stability of the vaginal microbiome over time. Increased community stability is proposed as an effect both of continuous release of low-dose (20 μg/day) progestin [14], and by the inhibition of endometrial proliferation and decrease or cessation of menstrual flow [32]. The menstrual phase of the reproductive cycle has been associated with a temporarily increased fluctuation in diversity of the vaginal microbiome in women [13]. In our study, the persistence of a luteal state, consistent with the anticipated effects of levonorgestrel release, was confirmed by detumescence of perineal/perivulvar sex skin and amenorrhea in all three animals although hormone levels were not directly measured.
The principal goal of this study was to determine if the vaginal microbiome changes over time following placement of a levonorgestrel-releasing IUS in a baboon model. Because only one baseline timepoint was available, we compared early (first month post-placement) to late timepoints in LNG-IUS placement as well as pre (day 0) and post placement timepoints. We found significant inter-individual differences but no consistent shift in microbial membership (organisms present) or structure (relative abundances) that could be associated with the presence of the LNG-IUS. The only time-dependent significant difference present was between day 0 and 85 for baboon 2. This animal also was the only one to approach significance for early vs late LNG-IUS placement, perhaps due to an increase in Fusobacteria. Because of the high level of inter-individual variation observed at each timepoint in this study, these alterations likely represent variation over time not specifically related to the LNG-IUS. Monitoring of additional pre-LNG-IUS timepoints in comparison to post-placement will be necessary to determine if the fluctuations seen have any relationship to the LNG-IUS.
This study confirmed that the baboon vaginal microbiome has greater diversity and decreased Lactobacilli in comparison to the majority of women. In women, the vaginal microbiome is a relatively low diversity niche in comparison to other body sites [22]. In baboons 1 and 3, vaginal microbial diversity remained relatively high across the pre and post-LNG-IUS timepoints and in the early vs. late period of LNG-IUS placement, with greater fluctuations than in baboon 2. If greater stability (not just lower diversity) correlates with greater disease resistance, we would expect baboon 2 to be more disease resistant than baboons 1 and 3. The dominant phylum in baboon 2 was Fusobacterium, which would be indicative of a dysbiotic state in women. However, if low diversity and community stability is more important than absolute membership with respect to disease susceptibility, then baboon 2 may be more disease resistant than an animal with greater diversity, despite the presence of Fusobacterium. These are testable hypotheses for future studies. For the most part, the vaginal microbiome for the three baboons in this study consisted of anaerobes, including some that have been associated with bacterial vaginosis in women. This is consistent with previous single timepoint studies in baboons [35]. The diagnosis of a bacterial vaginosis-like state in a baboon would be difficult since part of the diagnostic criteria in women involves self-reported discomfort. Nevertheless, the lack of discharge, odor, or clinical signs of discomfort in the animals suggests that disease was not present and that the diverse vaginal microbial communities present regardless of LNG-IUS placement represent the normal state for this species. Although Nugent scores were high in the baboons throughout the study, this scoring system is heavily influenced by the presence (or absence) of Lactobacillus as in women. Thus, high scores (corresponding to low Lactobacilli) cannot be considered diagnostic of a disease state in baboons, a species for which a low Lactobacillus state appears to be normal. In women, recent evidence suggests that approximately 25% of healthy, asymptomatic women have a bacterial vaginosis-like state at any one time, characterized by a higher microbial diversity and lack of Lactobacilli [13, 34]. The susceptibility of a diverse vaginal microbial community, as found in the baboon, to perturbation or to pathogen challenge may have relevance for this subset of women. Structurally different communities could still provide similar functionality. For example, some of the bacteria identified in our baboon samples (e.g. Leptotrichia and Sneathia) are lactic acid producers and may play the functional role of Lactobacillus in regulating pH [16, 26, 29]. Whether a diverse, polymicrobial state influences susceptibility to bacterial vaginosis or sexually transmitted diseases is still unknown.
There were several limitations to this study. First, the small sample size limits generalization of our conclusions, although the study does include longitudinal analysis, which has not been previously performed for this species. Secondly, because only one pre-LNG-IUS timepoint was available for each baboon, we cannot compare our LNG-IUS findings to natural time-dependent fluctuations or lack thereof in the baboon vaginal microbiome. Future longitudinal comparisons pre- and post-LNG-IUS placement will be helpful. Study designs in which each baboon serves as its own (pre-LNG-IUS) control may be more useful than comparison between control and LNG-IUS groups, since this study suggests that inter-individual variation prevents one baboon from serving as a consistent control for the microbiome of another baboon. The lack of consistent fluctuations in community membership or structure following LNG-IUS placement does not necessarily indicate that this device has no effect on the microbiome or on disease susceptibility. If the LNG-IUS promotes microbial community stability this may contribute to resistance to pathogen challenge. Further study using animals stratified by community stability over time, or animals specifically treated to disturb the community stability, might be helpful in evaluating this hypothesis. Finally, trimming of the IUS strings in our animals represents a variation from the technique used in women, in which the string is left in place. To date there has only been one investigation into the impact of IUD strings in women, and this focused on Candida species, so comparable data from women is lacking [5]. In this study the strings were trimmed to prevent premature removal by the animals so we may not be able to precisely mimic this aspect of IUD use in women, although it is worth exploring in future studies.
In summary, the vaginal microbiome did not measurably change from early to late post LNG-IUS placement in the baboons used in this study, although this stability could not be specifically ascribed to the LNG-IUS. The vaginal microbial community of the baboons in this study had low numbers of Lactobacilli but high microbial diversity, which decreased to a stable, low value in one animal and fluctuated about a higher mean in two animals. Inter-individual variation consistently exceeded intra-individual variation. This system may represent a useful model for prospective investigation of reproductive disease susceptibility in the context of a highly polymicrobial state, particularly in the evaluation of the roles of stability and increased diversity in pathogen susceptibility. Incorporation of functional parameters (i.e. lactic acid production) and inclusion of larger sample sizes and increased pre-manipulation timepoints will be helpful in ascertaining factors influencing community structure changes and whether these changes have functional consequences.
Acknowledgments
Assistance with the in vivo portion of this study was provided by Pat Frost and the veterinary and husbandry staff at the Texas Biomedical Research Institute Southwest National Primate Research Center, which is supported by NIH grant P51 OD011133 from the Office of Research Infrastructure Programs, National Institutes of Health. We thank Patrick Schloss and members of his laboratory at the University of Michigan for consultation on microbial sequence data analysis.
This work was funded by NIH/NICHD WRHR K12 HD065257-01.
References
- 1.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. [Google Scholar]
- 2.Bell JD, Bergin IL, Natavio MF, Jibrel F, Zochowski MK, Weadock WJ, Swanson SD, Aronoff DM, Patton DL. Feasibility of LNG-IUS in a baboon model. Contraception. 2012 doi: 10.1016/j.contraception.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell JD, Bergin IL, Schmidt K, Zochowski MK, Aronoff DM, Patton DL. Nonhuman primate models used to study pelvic inflammatory disease caused by Chlamydia trachomatis. Infect Dis Obstet Gynecol. 2011;2011:675360. doi: 10.1155/2011/675360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brotman RM. Vaginal microbiome and sexually transmitted infections: an epidemiologic perspective. J Clin Invest. 2011;121:4610–4617. doi: 10.1172/JCI57172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caliskan S, Keceli Ozcan S, Cinar S, Corakci A, Caliskan E. [In vitro biofilm formation and relationship with antifungal resistance of Candida spp. isolated from vaginal and intrauterine device string samples of women with vaginal complaints]. Mikrobiyol Bul. 2011;45:697–706. [PubMed] [Google Scholar]
- 6.Cauci S, Driussi S, De Santo D, Penacchioni P, Iannicelli T, Lanzafame P, De Seta F, Quadrifoglio F, de Aloysio D, Guaschino S. Prevalence of bacterial vaginosis and vaginal flora changes in peri- and postmenopausal women. J Clin Microbiol. 2002;40:2147–2152. doi: 10.1128/JCM.40.6.2147-2152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donders GG, Berger J, Heuninckx H, Bellen G, Cornelis A. Vaginal flora changes on Pap smears after insertion of levonorgestrel-releasing intrauterine device. Contraception. 2011;83:352–356. doi: 10.1016/j.contraception.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farage MA, Miller KW, Sobel JD. Dynamics of the Vaginal Ecosystem - Hormonal Influences. Infectious Diseases: Research and Treatment. 2010;3:1–15. [Google Scholar]
- 11.Fredricks DN. Molecular methods to describe the spectrum and dynamics of the vaginal microbiota. Anaerobe. 2011;17:191–195. doi: 10.1016/j.anaerobe.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 13.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttinger A, Critchley HO. Endometrial effects of intrauterine levonorgestrel. Contraception. 2007;75:S93–98. doi: 10.1016/j.contraception.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Hafez ESE, Jaszczak S. Comparative Anatomy and Histology of the Cervix Uteri in Non-Human Primates. Primates. 1972;13:297–316. [Google Scholar]
- 16.Harwich MD, Jr., Serrano MG, Fettweis JM, Alves JM, Reimers MA, Buck GA, Jefferson KK. Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics. 2012;13(Suppl 8):S4. doi: 10.1186/1471-2164-13-S8-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Translational research : the journal of laboratory and clinical medicine. 2012;160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaya D, Demirezen S, Hascelik G, Gulmez Kivanc D, Beksac MS. Comparison of PCR, culturing and Pap smear microscopy for accurate diagnosis of genital actinomyces. J Med Microbiol. 2013 doi: 10.1099/jmm.0.053165-0. [DOI] [PubMed] [Google Scholar]
- 19.Kyama CM, Mihalyi A, Chai D, Simsa P, Mwenda JM, D'Hooghe TM. Baboon model for the study of endometriosis. Womens Health (Lond Engl) 2007;3:637–646. doi: 10.2217/17455057.3.5.637. [DOI] [PubMed] [Google Scholar]
- 20.Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP, Romero R. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG. 2011;118:533–549. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessard T, Simoes JA, Discacciati MG, Hidalgo M, Bahamondes L. Cytological evaluation and investigation of the vaginal flora of long-term users of the levonorgestrel-releasing intrauterine system (LNG-IUS). Contraception. 2008;77:30–33. doi: 10.1016/j.contraception.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Li K, Bihan M, Yooseph S, Methe BA. Analyses of the microbial diversity across the human microbiome. PLoS ONE. 2012;7:e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, Li L, Nelson KE, Xia Y, Xiang C. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics. 2010;11:488. doi: 10.1186/1471-2164-11-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marconi C, Donders GG, Parada CM, Giraldo PC, da Silva MG. Do Atopobium vaginae, Megasphaera sp. and Leptotrichia sp. change the local innate immune response and sialidase activity in bacterial vaginosis? Sex Transm Infect. 2013;89:167–173. doi: 10.1136/sextrans-2012-050616. [DOI] [PubMed] [Google Scholar]
- 27.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller L, Patton DL, Meier A, Thwin SS, Hooton TM, Eschenbach DA. Depomedroxyprogesterone-induced hypoestrogenism and changes in vaginal flora and epithelium. Obstet Gynecol. 2000;96:431–439. doi: 10.1016/s0029-7844(00)00906-6. [DOI] [PubMed] [Google Scholar]
- 29.Mirmonsef P, Gilbert D, Zariffard MR, Hamaker BR, Kaur A, Landay AL, Spear GT. The effects of commensal bacteria on innate immune responses in the female genital tract. Am J Reprod Immunol. 2011;65:190–195. doi: 10.1111/j.1600-0897.2010.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neale R, Knight I, Keane F. Do users of the intrauterine system (Mirena) have different genital symptoms and vaginal flora than users of the intrauterine contraceptive device? Int J STD AIDS. 2009;20:423–424. doi: 10.1258/ijsa.2008.008391. [DOI] [PubMed] [Google Scholar]
- 31.Ness RB, Kip KE, Hillier SL, Soper DE, Stamm CA, Sweet RL, Rice P, Richter HE. A cluster analysis of bacterial vaginosis-associated microflora and pelvic inflammatory disease. Am J Epidemiol. 2005;162:585–590. doi: 10.1093/aje/kwi243. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson CG. Comparative quantitation of menstrual blood loss with a d-norgestrel-releasing iud and a Nova-T-copper device. Contraception. 1977;15:379–387. doi: 10.1016/0010-7824(77)90001-4. [DOI] [PubMed] [Google Scholar]
- 33.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivera AJ, Frank JA, Stumpf R, Salyers AA, Wilson BA, Olsen GJ, Leigh S. Differences between the normal vaginal bacterial community of baboons and that of humans. Am J Primatol. 2011;73:119–126. doi: 10.1002/ajp.20851. [DOI] [PubMed] [Google Scholar]
- 36.Schloss PD. 454 SOP [Internet] University of Michigan Medical School, Department of Microbiology & Immunology; Ann Arbor (MI): 2009. [2013 June 19]. [updated 2013 June 3]. Available from: http://www.mothur.org/wiki/Schloss_SOP. [Google Scholar]
- 37.Schloss PD. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2008;2:265–275. doi: 10.1038/ismej.2008.5. [DOI] [PubMed] [Google Scholar]
- 38.Schloss PD. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS ONE. 2009;4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.VandeBerg JL, Williams-Blangero S, Tardif SD. The Baboon in Biomedical Research. Springer; 2009. pp. 89–110. [Google Scholar]
- 42.Verbruggen BS, Boon ME, Melkerl P, van Haaften M, Heintz AP. Microscopic diagnosis of dysbacteriosis in stained vaginal smears in clinical practice. Diagn Cytopathol. 2006;34:686–691. doi: 10.1002/dc.20530. [DOI] [PubMed] [Google Scholar]
- 43.Wiesenfeld HC, Hillier SL, Krohn MA, Landers DV, Sweet RL. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis. 2003;36:663–668. doi: 10.1086/367658. [DOI] [PubMed] [Google Scholar]
- 44.Wira CR, Fahey JV, Ghosh M, Patel MV, Hickey DK, Ochiel DO. Sex hormone regulation of innate immunity in the female reproductive tract: the role of epithelial cells in balancing reproductive potential with protection against sexually transmitted pathogens. Am J Reprod Immunol. 2010;63:544–565. doi: 10.1111/j.1600-0897.2010.00842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue JC, Clayton MK. A similarity measure based on species proportions. Communications in Statistics - Theory and Methods. 2005;34:2123–2131. [Google Scholar]
- 46.Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]