Abstract
Introduction
Functional genomic screens apply knowledge gained from the sequencing of the human genome toward rapid methods of identifying genes involved in cellular function based on a specific phenotype. This approach has been made possible through the use of advances in both molecular biology and automation. The utility of this approach has been further enhanced through the application of image-based high content screening, an automated microscopy and quantitative image analysis platform. These approaches can significantly enhance acquisition of novel targets for drug discovery.
Areas covered
Both the utility and potential issues associated with functional genomic screening approaches are discussed along with examples that illustrate both. The considerations for high content screening applied to functional genomics are also presented.
Expert opinion
Functional genomic and high content screening are extremely useful in the identification of new drug targets. However, the technical, experimental, and computational parameters have an enormous influence on the results. Thus, although new targets are identified, caution should be applied toward interpretation of screening data in isolation. Genomic screens should be viewed as an integral component of a target identification campaign that requires both the acquisition of orthogonal data, as well as a rigorous validation strategy.
Keywords: Genome-wide siRNA screening, High content screening, Target identification, Target deconvolution
1. Introduction
In order to identify the cause and possible treatment of human disease, it is critical to understand the underlying genes, proteins and processes involved in its etiology. Until modern molecular technologies became generally available, genes and their function were generally defined using classical genetics, an approach that requires alternate phenotypes that segregate in genetic crosses. Frequently, model organisms were necessary in order to perform the mutagenesis required to alter or abrogate gene function. Using these laborious systems, many genes of interest have been identified and exploited. However, with the sequencing of the human genome and the advent of a wide variety of new molecular tools, the entire paradigm of genetic analysis has changed enabling the application of ‘reverse genetics’. In classical ‘forward genetics’ the analysis is initiated with a phenotype from which the mutated gene causing that phenotype is identified. In contrast, reverse genetics requires a priori knowledge of the gene that will be perturbed to alter its structure or expression and cause the biologically relevant phenotype.
High throughput transfection technologies combined with the ability to produce cDNA and short double-stranded RNA libraries at large scale have enabled current high-throughput loss- or gain-of-function studies using these short RNA or cDNA overexpression libraries in mammalian cells. Furthermore, the application of high content screening for functional genomic analysis has been facilitated by use of automated microscopy and quantitative image analysis. However, as with all cell-based screens, artifacts can be observed and care must be taken in analysis and interpretation of the screening data. In addition, all confirmed screening hits must be validated using alternative assays to enhance confidence in any new biological information obtained.
2. Functional genomic screens
Numerous approaches are available to rapidly interrogate gene function at the level of the genome. All are dependent on the availability of genomic sequencing that allows identification and prediction of expressed genes. Currently, it is estimated that the human genome contains ~21,000 genes that express proteins, although this counts alternative spliced transcripts as a single gene. In addition, this estimate only includes genes that are translated into proteins, and it is now well established that much of the genome is transcribed into non-coding RNA, which also have important regulatory roles1. It is well recognized that an understanding of the function of the expressed genome is a requirement for a better understanding of both normal and pathological conditions.
The combination of improved understanding of biological processes, and new or improved technologies, has facilitated a systematic examination of gene function at the genome level. Of particular relevance to this discussion are gains made in the manipulation of mammalian cells and development of high throughput transfection technologies2,3. These advances have enabled the large-scale introduction of arrayed libraries into mammalian cells. Relevant libraries include cDNA collections and those composed of small regulatory RNAs. These libraries can be used to interrogate any cellular process with a defined molecular or cellular phenotype under the in vitro cell culture procedures being used.
Many cellular processes have benefited from genome-wide functional genomic screens including studies of genes involved in proliferation, apoptosis, differentiation and oncogenesis, as well as other therapeutically relevant areas such as inflammation4,5. We have chosen to focus on examples in the area of virology to illuminate the utility of these approaches as well as potential associated issues.
Gain-of-function screens are most often performed with cDNA libraries to define which ectopically expressed proteins overcome or cause the phenotype being studied2,3,6. These cDNA libraries are derived from genome sequencing and are designed to encode proteins expressed by most of the known open reading frames (ORFs), and can include 5′ and 3′ UTRs, or just coding sequencing (termed the ORFeome)7,8. These cDNAs are cloned into the desired vectors downstream of strong mammalian promoters to enhance expression9. Initially most scientists used plasmid vector systems, but these studies were restricted to cell types easily transfected with plasmids as well as by the transient nature of the expression system. Using retroviral or lentiviral cDNA libraries overcomes this limitation since these can be engineered to infect a very wide variety of cells and integration of the virus into the cellular genome produces extended expression of the cloned gene10. Moreover, lentiviral vectors integrate in both dividing and non-dividing cells, further expanding their utility11.
An example of a cDNA-based gain-of-function screen can be found in a study performed by Stremlau et al.12. In this report, the authors elucidated host cell barriers to human immunodeficiency virus type 1 (HIV-1) replication that were present in the cells of Old World Monkeys but absent in human cells. It was known that a dominant repressive factor that acted on the incoming capsid caused the block in viral replication. The study was performed by cloning a cDNA library from primary rhesus monkey lung fibroblasts (3.2 × 106 independent clones) into a murine leukemia virus vector (MLV) that was subsequently used to transduce human HeLa cells. The cells transduced with the MLV cDNA library were grown and infected with HIV-1 at a multiplicity of infection (MOI) sufficient to infect > 99% of the cells. The virus used for infection was a recombinant HIV-1 pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G) to efficiently infect the HeLa cells. The virus expressed green fluorescent protein (GFP) as a reporter that allowed tracking of the HIV infection. FACS sorting for non-infected, GFP-negative cells resulted in the isolation of 0.5% of the cells. These cells were grown and subjected to a second round of HIV-1 infection. Colonies were selected for an absence of fluorescence by microscopy, cloned and expanded. Two independent HeLa clones were found that were resistant to infection by HIV-1, but could be infected by simian immunodeficiency virus (SIV), which is not restricted in either human or monkey cells. The only insert that was common to both HIV-1-resistant clones was a cDNA sequence predicted to encode a protein called TRIM5α, a molecule that had no known function at the time. The authors went on to demonstrate that it was indeed monkey TRIM5α expression that was specifically responsible for blocking HIV-1 infection while having only a slightly inhibitory effect on SIV infection. Through the use of chimeric SIV/HIV viruses it was shown that viral capsid sequences influenced restriction by TRIM5α. The study further demonstrated that sequence differences between human and monkey TRIM5α were responsible for the restriction, and that reducing TRIM5α expression in monkey cells abrogated the restriction on HIV-1 infection. Given the appropriate biological system with a clear phenotype, this study demonstrates the power of forced cDNA expression to identify factors of significant biological interest. However, it must be recognized that cDNA gain-of-function screening can lead to identification of artifactual protein activities that are not physiologically relevant, and are caused solely by the overexpression of a protein.
To demonstrate the utility, as well as the caveats, associated with modern genome-wide loss-of-function screens, four screens for host factors required for HIV-1 replication will be considered13–15. (Though not further discussed in this review, the use of siRNA-based loss-of function screening to identify cellular factors restricting HIV replication has recently been described as well16). Due to their limited genome size, viruses utilize cellular processes to accomplish most of what is required for their replication. For example, the HIV genome only encodes 15 proteins17 and thus requires the host to provide most of the factors required for its replication14. Therefore elucidation of all of the host factors involved is essential for a full understanding of the HIV life cycle. This understanding could also lead to the identification of many new host-directed antiviral targets that would significantly reduce the likelihood of resistance, since these host-directed antiviral therapies would be less susceptible to the highly error-prone replication of the virus.
All of the studies to be considered use RNA interference (RNAi) for gene silencing. RNAi relies on sequence-specific double stranded RNAs complementary to a target gene to achieve silencing18. The use of small RNAs as genetic tools stems from observations originally made in plants and extended to model organisms such as Caenorhabditis elegans,19 Drosophila melanogaster 20. Although long double-stranded RNAs (dsRNAs) can be used to silence genes in C. elegans or D. melanogaster, they can not be used in mammalian cells due to the induction of an interferon response leading to inhibition of translation and cell death21. The direct introduction of double-stranded RNAs 21–23 nucleotides in length (small interfering RNAs or siRNAs), however, avoids this limitation and successfully leads to specific gene silencing. To achieve RNAi silencing, double-stranded RNA is processed by the RNase III-like enzyme Dicer to form small interfering RNAs (siRNAs), 21–23 basepairs in length22. These siRNAs are then incorporated into the RNA-induced silencing complex (RISC) that degrades target mRNAs with sequences corresponding to that of the siRNA. The degradation of the target mRNA subsequently leads to a reduction in protein synthesis. siRNAs can be introduced directly into cells by transfection23 and are then unwound and associated with the RISC, leading to selective mRNA targeting and silencing. As with cDNAs, transient silencing can be achieved with direct transfection of siRNAs while sustained gene silencing can be observed with the use of appropriate viral vectors. In addition, viral vectors are generally used for expression of small hairpin RNAs (shRNAs). shRNAs are synthesized from the vector’s promoter to form RNAs of about 70 nucleotides in length that are subsequently processed by Dicer to form active siRNAs24.
Three RNAi library formats can be utilized to conduct loss of function screens. The first type of format is an array of individual siRNAs in each assay well. The standard practice in the field is that the resulting phenotype needs to be confirmed with at least two distinct, non-overlapping siRNAs targeting the same gene25. Therefore, multiple active siRNAs are readily identifiable from this type of array without having to de-convolute the dataset. Data can also be analyzed using the Redundant siRNA Activity (RSA) analysis methodology which decreases the impact of off-target effects associated with RNAi screens26. However, as a disadvantage, the cost for utilizing this type of array can be compounded when running replicate experiments. In the second type of RNAi library format multiple siRNAs targeting one gene are pooled in one assay well. A genome-wide siRNA library contains typically 4–6 sequence independent siRNAs targeting one gene. These siRNAs can be combined in single or multiple pools. If multiple pools per gene are desired, typically 2 siRNAs will be pooled in one well, which results in 2–3 assay wells representing each gene27. The use of pooled libraries is less cost-prohibitive and if multiple pools per gene are available, data collected from this type of assay format is also suitable for RSA analysis. The described assay formats utilizing individual and pooled siRNA reagents are relevant for shRNA libraries as well, however libraries of individually arrayed shRNAs are less common due to the considerable cost and effort involved in the propagation and normalization of the individual lentiviral constructs. Finally, the third type of library format combines an entire shRNA library into one single pool of vector-encoded shRNAs. This format is only applicable if the loss-of-function phenotype confers a selectable property such as an impact on cell growth/survival or an effect on a fluorescent reporter. The shRNA-encoding vector requires special design, such as a selection marker, as well as a short unique sequence that allows the identification of the active shRNA through sequencing28. Following transduction and stable expression of the pooled shRNA library the cells undergo multiple rounds of selection, e.g. by survival due to a growth advantage of the desired phenotype or by fluorescence-activated cell sorting. This ultimately leads to an enrichment of cells expressing the phenotype-inducing shRNAs (positive selection). The enriched shRNAs, and thus the targeted genes, can then be identified by next-generation sequencing or microarray technology. A negative selection strategy that determines the loss of specific shRNAs by comparison to a control population can be utilized to identify genes conveying a phenotype that negatively affects cell viability4.
The screens for host factors required for HIV replication reviewed here all utilized genome-wide RNAi libraries but the libraries differ in format and design. Three libraries are comprised of chemically synthesized siRNAs (Brass et al.29; König et al.27; Zhou et al.30) that are transfected into the host cells, resulting in a transient knockdown of the targeted genes. While generally most siRNAs in a library achieve a knockdown efficiency of 70% or more, there is a distribution resulting in distinct knockdown levels for each targeted gene. Due to the different design algorithms used to create the siRNA libraries, the knockdown efficiencies for specific genes are likely to differ between screens. While all three siRNA libraries consist of siRNA pools, Brass et al. and Zhou et al. use one single pool per gene with 4 and 3 siRNAs per pool, respectively, König et al. utilizes an arrayed library that consists of 2–3 siRNA pools per gene with 2 siRNAs per pool. The fourth RNAi library is based on a lentiviral vector expressing shRNAs (Yeung et al.31). This lentiviral library targeted over 50,000 transcripts with 3–5 shRNAs per gene. Since a selectable phenotype (cell survival) was investigated, it was possible to combine all shRNA constructs in one single pool.
Although the endpoints of these four screens were similar, the experimental and analytical approaches used by each group were widely divergent. For example, the studies used different cell backgrounds. Approaches using synthetic oligonucleotides require cell lines that are transfected with high efficiency such as HeLa or HEK293. Unfortunately, none of those cell lines can be efficiently infected with HIV. To overcome this obstacle, Brass et al. and Zhou et al. used HeLa-based cell lines that stably express receptors and co-receptors required for viral entry. König et al., by contrast, relied on an envelope-deleted virus pseudotyped with VSV-G that efficiently infects HEK293 cells. The use of a lentiviral vector for delivery of the shRNA constructs in the screen by Yeung et al. allowed the transduction of a more physiologically relevant T cell line.
The timing of each step in these studies also differed. The time to achieve gene silencing depends on protein half-life and the amount of protein required to yield a phenotype. With the time between siRNA transfection and HIV infection ranging from 24 to 72 hours, a significant bias toward genes with shorter or longer half-lives will appear between these studies. In addition, time between HIV infection and assay readout also varied from 24 to 96 hours. Lentiviral shRNAs stably integrate into the genome, thus enabling a long-lived screen. Thus, Yeung et al. allowed 3 weeks between transducing the lentivirus library and HIV infection and another four weeks until readout.
Furthermore, the readout used in each screen varied and included detection of the HIV p24 protein by high-content imaging, reporter assays that directly measure viral gene expression through incorporation of a reporter gene into the virus, or indirectly through expression of a reporter gene that is integrated in the host cell and is dependent on the presence of the HIV tat protein. It should be noted that due to the use of an envelope-deleted, VSV-G pseudotyped virus by König et al., this study was limited to the early steps of HIV infection following viral entry. The other screens were designed to interrogate all steps of the infection cycle from viral entry to egress. The readout of the lentiviral shRNA screen identified shRNAs in cells that survived 4 weeks post-infection and would also be expected to identify genes acting at any stage of the viral reproductive process. In summary, when selecting the model system, screening readout, and timeframes it is critical that the assay design specifically recapitulates the clinical phenotype of interest.
Finally, the algorithms used by each group to analyze the screening data and identify hits also varied. Two groups normalized results using a plate median, which assumes that most samples have no effect, while a third group used dedicated negative controls composed of non-targeting siRNAs. The use of a plate median to normalize screening results is appropriate if, as in the studies discussed here, large (genome-wide) libraries are used that are arrayed without a particular order. In these cases, a random distribution of positive and negative effects, and thus comparable plate medians between assay plates, can be assumed. If, however, targeted libraries or siRNAs selected for their likelihood to cause a phenotype are screened, dedicated negative controls are preferred.34 The same applies for RNAi libraries that are arrayed in a particular order (e.g. a grouping of siRNAs that target specific functional classes, such as kinases, in the same plates).
How a positive hit is defined by each screen also varied. Brass et al. selected siRNA pools for further study where the duplicate pools both had a value of at least two standard deviations below the plate mean. Zhou et al. used the ‘strictly standardized mean difference’ approach to calculate a threshold for host factor identification. König et al. applied a different approach called ‘redundant siRNA activity’ in which the probability of a gene being involved in the process studied is calculated using a statistical methodology that considers the activities of all siRNAs targeting the same gene (4–6 siRNAs/gene). The results of this analysis were then combined with additional lines of evidence including single siRNA activity rankings and results from database searches to define genes for follow-up characterization. Yeung et al. selected genes with a two-fold enrichment over background. Generally, assay robustness and repeatability is typically evaluated for statistical separation of positive (chemical compound or RNAi or cDNA with known activity) and negative (vehicle treated, scrambled si/shRNA) control conditions using the Z-Prime value for single parameter32 or, if applicable, for multi-parameter33 read-outs. Additionally, testing multiple scrambled siRNAs provides insight into the number of off-target effects or false positives expected in the screen.
In conclusion, no standard operating procedure for hit selection exists and the analytical strategy needs to be chosen in accordance with the overall screening strategy34 (Table 1). A key factor impacting the analytical strategy is the capacity of the follow-up assays. If the throughput of the planned follow-up assays is high and the cost of processing a larger set of candidates is justifiable, a higher error rate among the selected hits of the primary screen can be accepted. Follow-up assays with a lower throughput, by contrast, will require a stricter candidate selection at the price of an increased number of false-negatives.
Table 1.
Overview of options available for RNAi screening.
| Options | Advantages | Disadvantages | |
|---|---|---|---|
| RNAi reagent | siRNA |
|
|
| shRNA |
|
|
|
| Cell lines | Primary cells |
|
|
| Immortalized cell lines |
|
|
|
| Library format | Multiple arrayed siRNAs/shRNAs per gene |
|
|
| Single siRNAs/ shRNA pool per gene |
|
|
|
| Pooled shRNA library |
|
|
|
| Readout | Whole-well readout |
|
|
| Single-cell readout (HCS, FACS) |
|
|
|
| Normalization | By plate median |
|
|
| Dedicated negative control |
|
|
|
| Analysis method | Standard deviation from mean (Z score) |
|
|
| Median absolute deviation from median (robust Z score) |
|
||
| SSMD |
|
|
|
| RSA |
|
|
|
| Additional details and analysis strategies are reviewed in Birmingham et al34. | |||
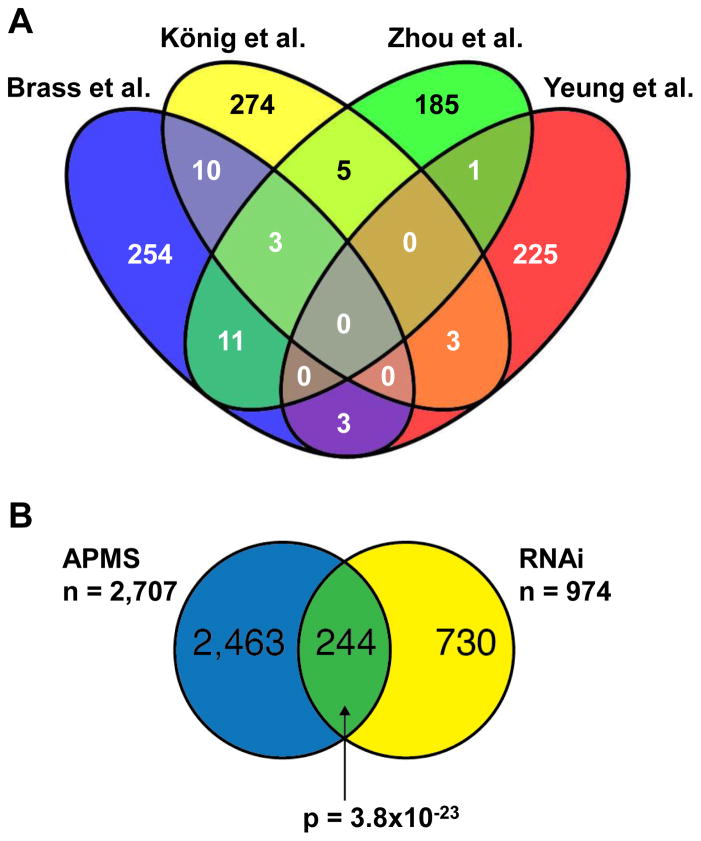
Although all four of these RNAi screens were designed to identify host factors required for HIV replication, the overlap between these studies was surprisingly low. The three siRNA screens each identified between 200 and 300 genes that play a role in HIV infection. However, there were only three genes common to all three screens. When looked at in pair-wise fashion, there was also a limited though significant overlap: 13 common genes between König et al. and Brass et al. (p < 0.001); 8 genes in common between König et al. and Zhou et al. (p < 0.05) and 14 common genes between Brass et al. and Zhou et al. (p < 0.001) (Figure 1A). Perhaps the most surprising result is the limited overlap in genes identified between Brass et al. and Zhou et al. since the two screens were, in broad strokes, quite similar. The lentiviral shRNA screen performed by Yeung et al. identified 252 gene candidates. None of these genes were identical to the three genes overlapping between the three siRNA screens. There were three common genes found with König et al. and with Zhou et al., respectively; none were common with Brass et al. However, despite the modest overlap between screens, many genes mapped to the same cellular pathways identified in the other screens. Five pathways, represented by 42 genes identified by Yeung et al., overlapped with the König screen. The overlap between Yeung et al. and Brass et al. was even greater with 7 common pathways (identified by 41 genes in Yeung et al.). For example, Brass et al. identified the gene AKT while Yeung et al. found the factors PI3K, PTEN and RAS that act immediately upstream of AKT. Thus, while the specific results of each screen might be different, the identified pathways are in good agreement, demonstrating the power of this approach.
Figure 1. Venn diagrams representing the overlaps between HIV-1 screening datasets.
(A) The overlaps between four published RNAi screens identifying HIV host factors are small. No genes were found in all four screens, though the three siRNA-based screens had three genes in common. Venn diagram was generated from supplementary data of using Venny49. (B) The overlap between cellular proteins interacting with HIV proteins (APMS) and HIV host factors identified by the four genome-wide RNAi screens (RNAi) is highly significant.
Interestingly, a very similar situation was observed when multiple genome-wide screens to identify host factors required for influenza replication were compared35–37. Again, very limited overlap for specific genes was observed38. Taken together, the limited concordance between individual screens may indicate that the set of cellular factors impacting viral replication differs with experimental settings39. However, in both the screens for host factors involved in HIV and influenza virus replication, despite the limited overlap of specific genes in these assays, numerous common pathways were identified13,40. Moreover, the individual genes, once validated with wild-type virus in relevant cell backgrounds offered more than five times as many potential targets for drug discovery as were known at the time. Thus, despite the variance seen that is likely due to the differences in cell backgrounds, RNAi, viruses and procedures, the application of functional genomic screens to the identification of genes can have great utility in the identification of new targets for drug discovery.
The RNAi approach has certain limitations. To induce phenotypic effects, siRNAs or shRNAs used should meet the following requirements: (i) sufficient reduction in cognate mRNA to observe biological effect; (ii) the half-life of the target protein must be short enough to observe the effect during the experimental time course; (iii) RNAs with toxic or off-target activities in the chosen cell background should be avoided; (iv) RNAi-induced knockdown cannot be observed if the function of interest is supplied by redundant genes. Of these requirements, the two that can potentially be controlled are (i) and (iii), though verification of knockdown levels is difficult at a large scale. Advances in technology and approaches have enhanced the ability to reduce gene expression. These changes include using pools of multiple siRNAs targeting the same gene41 as well as the use of improved bioinformatics to better predict the most potent siRNA sequences. More troubling have been off-target effects associated with siRNA treatment in which genes other than the target gene are modulated42. In general, off-target effects can be attributed to three broad phenomena: sequence-specific silencing of mRNA that is imperfectly matched with the siRNA; activation of the innate immune system by either the siRNA itself or the delivery vehicles used to allow entry of the siRNA into the cell; or saturation of the RNAi machinery. Examples of off-target effects with siRNAs screens are common. In one study, siRNA targeting green fluorescent protein (GFP) was tested in mammalian cells where GFP is not naturally expressed43. In multiple cell lines frequently used for siRNA studies, including HeLa, U-2 OS and HEK cells, the authors found that siRNA targeting GFP consistently reduced expression of multiple genes. Of greater importance to those running phenotypic genome-wide siRNA screens, it was found that using a randomly selected collection of siRNAs, a fraction of them altered cell viability in a sequence-independent fashion44. Multiple strategies have been developed to mitigate the effects of off-target silencing, including siRNA redundancy and pooling of siRNAs at reduced concentrations to avoid sequence-specific effects, elimination of certain sequences, and chemical modification to avoid immune-mediated effects. Moreover, new generations of siRNA libraries feature chemical modifications that promise to considerably reduce off-target effects45. Recently bioinfomatic strategies have been proposed to identify common off-target transcripts that occur in RNAi screens due to seed sequence mismatches46,47. Nevertheless, off-target silencing remains a potential issue that needs to be addressed by a suitable hit selection approach that takes siRNA redundancy into account, such as RSA 26,34, or by an appropriate validation strategy as part of the follow-up analysis.
One approach to avoid these issues and increase confidence in genes selected using functional genomic screens is through performing additional validation assays. To minimize the false-positive rate due to the common occurrence of off-target effects among siRNAs/shRNAs it is advisable to require two or more distinct, non-overlapping siRNAs/shRNAs targeting the gene in question to cause the same phenotype25. A library comprised of multiple RNAi reagents for each gene provides this redundancy already at the primary screening level while libraries limited to a single siRNA pool per gene require additional follow-up experiments to deconvolute the pools. To further validate an observed phenotype, the specific knockdown of the target mRNA should be verified by quantitative PCR, although this is generally done for only a selected number of genes, as shown by Brass et al. and Zhou et al. The specificity of the observed siRNA/shRNA-induced phenotype can further be validated by co-expression of RNAi-resistant cDNAs. The cDNA should rescue the loss-of-function phenotype as observed by RNAi knockdown. However, such rescue experiments require considerable effort and cannot be carried out at a large scale. For example, Zhou et al. demonstrated a rescue of viral replication for the four host factors CD97, SERPINB6, BMP2K, and NEIL3. Typically, genome-wide siRNA screens are conducted in highly transfectable cell lines and attenuated pathogens or small molecules are employed as ligands to modulate signaling responses. While these parameters are amenable for high throughput screen, they do not necessarily reflect the true biological system. Phenotypes identified from a primary screen require validation under a physiological relevant context. For example, to study immune responses, the phenotypes should be confirmed in immune cell such as monocyte derived dendritic cells (MDDC) or macrophages. Furthermore, to study the host-pathogen interactions, cells can be challenged with wild type or virulent strains of the pathogen37.
Additionally, it is becoming apparent that the integration of orthogonal large-scale data sets, preferably those that are experimentally derived within an RNAi assay system, can provide a powerful tool to further validate and characterize putative targets identified through genetic screens. For example, a comprehensive proteomic analysis to identify all host proteins that interact with any HIV protein was recently published48. Using affinity tagging and purification mass spectrometry (APMS), the authors identified a comprehensive set of HIV-human protein-protein interactions involving more than 2700 individual human proteins. Importantly, of the more than 900 proteins that were identified in the four HIV RNAi screens described above, 244 are identical to proteins identified in this proteomic screen (p = 3.8×10−23) (Figure 1B). These results indicate that approximately 10% of the collectively identified HIV host factors both directly interact with the virus, and are required for replication. It is likely that the remaining factors may act as either genetic (indirect) regulators of the viral life cycle, function redundantly in virally associated biochemical complexes, or represent false positive activities attributed to the experimental platforms. Through juxtaposition of these two systems-level platform technologies, as well as the application of sophisticated bioinformatics approaches, genetic networks can be constructed that incorporate data from these, and additional, data sets. Because application of these orthogonal data sets identifies key pathways and biochemical complexes involved in disease pathology, this approach can provide greatly improved confidence in the selection of new targets and pathways for drug discovery.
The utility of functional genomic screening approaches for drug discovery is not limited to the identification of new targets. A second very useful application is the use of functional genomic screens to determine the mechanism of action of a compound identified in a phenotypic screen. For example, Luesch et al.50 used a functional genomic screen to determine the mechanism of action of the cyanobacterial metabolite apratoxin A, a potent cytotoxin for tumor cell lines that acts through G1 phase cell cycle arrest and induction of apoptosis. After identifying a tumor cell line that was both very sensitive to apratoxin A as well as highly transfectable, the authors performed a genome-wide cDNA overexpression screen for genes whose expression inhibited the apratoxin A-induced apoptosis. After eliminating cDNAs with non-specific effects, they identified 46 genes that conferred resistance to apratoxin A. They then separated those that acted directly on apratoxin-A induced cell cycle arrest from those that inhibited apoptosis without affecting cell cycle arrest or those genes that caused cell cycle arrest independent of apratoxin A. Of the 22 cDNAs remaining, five encoded fibroblast growth factor receptors (FGFRs). After showing that apratoxin A-resistant tumor cell lines overexpressed FGFR they hypothesized that FGFR was responsible for the activity of apratoxin A and in fact demonstrated that apratoxin A inhibited FGFR-mediated tyrosine phosphorylation of STAT3, a gene critically involved in tumor cell growth. Using a very different approach, Eggert et al.51 combined a genome-wide RNAi screen with chemical genetics to identify target genes for compounds that directly affect cytokinesis. In Drosophila cells, the authors first screened over 50,000 compounds for those that inhibited cytokinesis and followed up on 25 of the most potent and readily available compounds. Many compounds acted through pathways known to inhibit cytokinesis through binding actin and inhibiting its polymerization. Using a genome-wide RNAi screen for genes that caused a binucleate phenotype in Drosophila cells, they identified 214 genes related to cytokinesis, 20% of which were previously identified. The authors then subcategorized the the phenotypes induced with the compounds and the RNAis, compared them in detail and identified new pathways involved in cytokinesis, including a new protein in the Aurora B kinase pathway. Given the importance of phenotypic small molecule screens in drug discovery today, these examples show the power of functional genomic screens to determine the mechanism of action of compounds identified in cell-based phenotypic assays.
3. High Content Screening
There are significant limitations to functional genomic screens that depend on endpoints such as reporter activity quantified at the well level. Some of these limitations are obviated through the application of phenotypic image-based high content screening (HCS). In addition to interrogating a specific endpoint based on intensity of fluorescently labeled endogenous proteins or reporters, HCS offers the opportunity to simultaneously examine multiple biological or phenotypic parameters at the single cell level. HCS also allows quantitative evaluation of spatially-derived cellular changes and cell population heterogeneity. This enables rapid elimination of a variety of non-specific and/or toxic effects and enhances confidence in the genes selected in the screen.
Using an image-based multi-parametric approach to identify genes that influence a cellular phenotype allows detection of not only changes in endogenous protein level, but also in sub-cellular (co-)localization or spatial distribution of proteins or cellular constituents. Therefore, subtle differences in cellular phenotypes can be measured as well as temporal changes, e.g. changes in cell cycle and mitosis extracted from time-lapse images to identify genes involved in cell division52,53. As a result of this phenotypic analysis, image-based high content assays can provide more detailed information than simple end-point single parameter assays. Some phenotypic changes can only be quantified by image-based assays, including phenotypes based on morphological changes, e.g. changes of cell shape or cytoskeletal organization, such as encountered during differentiation of stem cells into different lineages54, invadopodia formation55, ciliogenesis56. Due to the ability to multiplex fluorescent labels and extract multiple parameters per label, toxicity or other pleotropic effects can be extracted in parallel with the specific assay read-out. Image-based high content assays also allow investigation of heterogenous cell populations, e.g. cells containing reporter genes with less than optimal transfection efficiency or cells requiring co-culture with another cell type (e.g. Kaltenbach et al.57). Because the details of the phenotypic signature achieved through loss-of-function or gain-of-function genomic methodologies can vary due to on-target or off-target effects, image-based high content assays that account for single-cell heterogeneity are often the assay type of choice for functional genomic screens58.
Care needs to be taken to design the assay and the detection method and readouts to achieve reproducible and robust results of the functional genomics imaging assay. Details on image-based high content assay design considerations can be found elsewhere (e.g. Conrad and Gerlich59, Haney60, Shariff61). For functional genomic screens, the assay time point has to be carefully selected by balancing the time needed to achieve gene knock-down with optimal cell density and systemic effects caused by long incubation times in the small media volumes of multi-well plates. Multiple time point live cell assays are possible for screening in lower throughput, if the typical image acquisition, if the long imaging times of 15 minutes to 2 hours per 384-well plate do not cause significant changes within the plate or plate set. Most frequently, an HCS system with an environmental control module is employed for such live cell assays to minimize phenotypic cellular changes due to the change in environmental conditions. For larger scale screens, for example genome-wide screens often resulting in more than a hundred 384-well plates, fixed endpoint assays are often desirable to decouple the plate preparation from the image acquisition and achieve the high throughput needed for rapid project execution. With high content screening systems now being widely used in drug discovery, a large number of image analysis algorithms are readily available in high content analysis software packages, which only have to be adapted and combined into an assay-specific protocol (e.g. Haney60, Niederlein et al.62).
Although a large-scale image-data storage infrastructure is required, typically containing several terabytes of storage space, it is good practice to save the images acquired during the screen for later review. The ability to visually evaluate some of the images is paramount for quality control of all aspects of the assay, including removal of false positives due to fluorescent artifacts, and for confirmation of the computer-generated phenotypic signature by manual image review of the hits selected for follow-up. Review of some selected images can also lead to observation of more subtle differences in phenotype and subsequent adjustment of the image analysis protocol to detect them. For example, in a druggable genome-wide ciliogenesis functional genomics screen aimed at quantifying the number of ciliated cells, a phenotype with extended cilia was observed and the average length of detected cilia was added to the measured parameters for the follow-up experiments56 to distinguish the cilia stabilizing phenotype. This resulted in identification of the inhibitory role of actin-related protein ACTR3 and branched actin network formation in ciliogenisis.
To use the full potential of the multi-parametric data generated by image-based high content functional genomics studies, several analysis approaches have been applied to best mine the data for relevant information. The most frequent use of the multi-parametric data focuses on features in the analysis that allow elimination of possible toxic or non-specific effects of the RNAis or cDNAs. This approach was chosen by Brass et al.29 to identify toxic siRNAs at the same time as determining expression levels of the HIV p24 protein, thereby avoiding the need for a separate cell viability counter-screen as used by König et al.37 and Zhou et al.30. The simplest parameter to determine toxicity is the cell count of the imaged region, but more complex analysis is easily obtained from a multiplexed nuclei stain, such as Hoechst 33342 or DAPI, which allows quantification of the chromatin condensation state, DNA content, nuclei size and shape, etc. (e.g. Dull et al.63, Kim et al.56, Young et al.64). For example, in a screen aimed at identifying ligands of estrogen receptor alpha using a nuclear translocation assay63, cytotoxic perturbagens were identified using average nuclear size of the cell population and nuclei count of the imaged region. Perturbagens resulting in a small average nuclear area or low cell counts were flagged as cytotoxic and subsequently removed from the hit list. With recent advances in ease-of-use of computational methods, phenotypes are more commonly defined using more than one feature and combining them using multi-parametric data analysis techniques, such as distance-based parameter combinations, principal components of feature sets, or other advanced classification techniques (e.g. Duda et al.65, Horvath et al.66, Misselwitz et al.67, Zhang and Pham68). To better describe the heterogeneity of the phenotypes observed within the cell populations, often the percentage of cells displaying a particular phenotype is used as the primary assay read-out, rather than using the cell populations mean values of a parameter or a combination of parameters. A variety of classification techniques, such as Bayesian approaches, discriminant functions, support vector machines, neural networks etc. (e.g. Duda et al.65, Hudson & Cohen69, Kotsiantis70, Theodoridis & Koutroumbas71), can be employed to aid in distinction of subtle or complex phenotypes. For example, a population composition approach was utilized to evaluate phenotypic outcomes of cell-cycle modulators72. Briefly, HCT-116 cells were treated with 41 cell cycle modulators (10-point concentration curves), fixed and fluorescently labeled with Hoechst to stain the nuclei and antibodies against Cyclin B1, phospho-histone H3, and alpha-tubulin. Multiple features were extracted from the images and a decision tree-based classifier was designed using eight reference compounds to distinguish nine cellular phenotypes (eight defined phenotypes plus “other”). The percentage of cells corresponding to each phenotype was then calculated for each cell cycle modulator at each concentration to produce a phenotypic signature. If the cellular phenotypes are not known a priori, non-supervised classifiers, such as self-organizing networks or clustering algorithms, can be used to distinguish the varying cellular responses (e.g. Duda et al65, Theodoridis & Koutroumbas71). However, care needs to be taken to choose a classification technique applicable to the biological problem and to validate the biological relevance of the resulting classes. The generated phenotypic signatures can then be investigated further using clustering techniques to correlate similar signatures with gene-focused groups of siRNAs, shRNAs, or cDNAs, to enable investigation of the underlying mechanism of action64.
In summary, the high content data derived from image-based assays can lead to improved validation and interpretation of the results of RNAi or cDNA library screens as compared to single-parameter whole well endpoint assays. However, these image-based high content assays require more complex assay development due to the multi-label and multi-parameter optimization, generally have a higher cost associated with them due to multiplexed fluorescent labels combined with multi-step wash and stain protocols, are slower due to long imaging times, and require specialized expertise for the image and data analysis. This additional cost and time is often well justified, since the multiplexed multi-parametric nature of the image-based high content assays combined with modern data mining tools allows execution of cellular genomics analyses in a more complex biological setting, which better mimics biological systems in living beings.
4. Conclusions
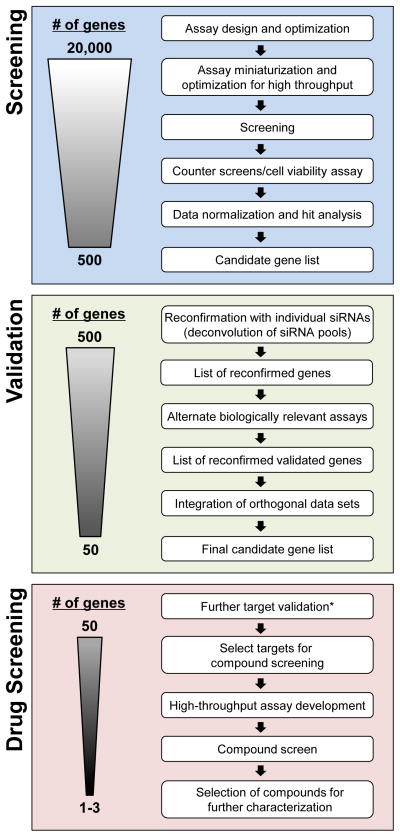
With the loss of patent protection for numerous blockbuster medicines the necessity for new significant drug discovery targets is becoming ever more critical. Numerous approaches have been used in the identification of these new targets over time. Recently, advances in molecular biology, automation and data acquisition and analysis have enabled the rapid and comprehensive analyses of novel targets using clinically relevant cellular assays. This is likely to create an explosion in potential new targets in virtually every therapeutic area. Given the large number of genes identified in these screens, the success of a target identification campaign will not only rely on the establishment of appropriate experimental and computational systems-based strategies, but will also require extensive validation pipelines to triage putative targets (Figure 2). In addition, it is important to recognize that, although a particular functional genomics screen may identify potentially important new genes, based on the results of the screens for host cellular factors required for HIV replication described above, it is highly likely that any screen will only identify a subset of critical factors, and the implementation of a combination of appropriate experimental, technical, and computational approaches will ensure a higher likelihood of success. In addition, the use of high content screening may improve the reproducibility of results in functional genomic screens as these imaging approaches enable detailed quantification of the heterogeneous responses of cells within a population. For each cell, a multiparametric readout is provided that includes cytological changes enabling both observation of the desired endpoint and potential correlation of that endpoint to a variety of factors, including toxicity. Additionally, parallel systems-level efforts, including (but not limited to) proteomics, metabolomics, and transcriptomics, will provide additional value to function genomics screening datasets. Despite the specific caveats pointed out in this and other reviews, cellular genomics and high content screening provides a relatively rapid route toward new target identification, and has the potential for driving novel drug discovery projects to address unmet medical needs.
Figure 2. Workflow for a genomic screening approach to identify targets for drug development.
*Further target validation can be achieved using numerous approaches including transgenic or knock-out/null mice, disease-related genetics (mutations or SNPs), gene expression pattern changes, proteomics and model organisms (e.g. C. elegans, Zebrafish, Drosophilia)
5. Expert opinion
Functional genomic and high content screening offer a route to greater understanding of fundamental biological processes and the role of changes in these processes in human disease. For those interested in the development of new medicines to treat these diseases, cellular genomics and high content screening provide a valuable addition to the drug discovery process. These technologies provide relatively rapid methodologies toward the identification of novel drug discovery targets. However, unlike traditional approaches toward new target identification, functional genomic and high content screening can simultaneously yield hundreds of new targets for a particular endpoint. Thus, efficient approaches toward validation of these candidate targets are necessary in order to sort through these candidate genes and select those most likely to progress through the drug discovery pipeline.
The four different functional genomic screens described above designed to identify host cellular factors required for HIV replication identified numerous potential novel targets. However, based on the limited overlap observed in the four screens, caution in the interpretation of the results is required. First, given that all of the screens identified 200–300 individual genes, any particular functional genomic screen is likely to miss many potential candidate genes. This is despite the fact that the RNAi systems used are designed to interrogate virtually all expressed genes and in part due to the different design algorithms of the RNAi libraries that lead to differences in knockdown efficiencies of specific target genes. In addition, given the way the assays are designed, with a single gene being depleted in each well, a screen is unlikely to find genes whose function is redundant with other genes. Second, but perhaps less important to most current approaches to drug discovery, is the fact that functional genomic gain- or loss-of-function screens cannot identify some non-coding RNAs involved in a phenotype. Thus, in order to enhance confidence in the results of a functional genomic screen multiple parameters must be considered that include: (a) the type of RNAi system used (siRNA or shRNA), (b) the vector and system used to introduce the RNAi into the cell, (c) choice of endpoint to define a phenotype, (d) timing for each step, (e) data analysis incorporating normalization and hit definition and (f) validation. Since validation can be both time-consuming and costly it is critical to reduce the number of genes for which validation is required. The acquisition of orthogonal data sets through approaches such as proteomic screening, as described above, will be an extremely useful complement to the data obtained in a genomic screen and will both add confidence to the data obtained in each screen and make the validation process considerably less onerous.
The combination of functional genomic screens with high content imaging enhances the reliability of screening data based on a number of advantages that are unique to the platform. First, HCS enables the interrogation of endogenous cellular and protein markers as assay endpoints. In addition, the ability to examine individual cells allows normalization of the desired endpoint to a secondary cellular readout for each cell and enables the extraction of activities that may be masked when analyzing a population average. Finally, the multiparametric nature of high content screening allows examination and normalization of several endpoints for a single phenotype, thus further increasing confidence in the candidate genes selected. Taken together, while this technology requires considerable investment in capital and expertise, it represents an invaluable component of a systems-based target identification platform.
Important new targets are critical for the continued growth of the pharmaceutical industry. The genomic screening approaches addressed in this review will play a critical role in these efforts. With proper attention to the details of running functional genomic high content screens, numerous new targets for drug discovery in a wide variety of therapeutic areas can be identified, providing the opportunity for the discovery of important new medicines.
Article Highlights.
Advances in molecular biology, automation and data analysis have allowed the development of new screening systems for functional genomics.
Through the use of cDNA overexpression or RNA interference (RNAi) both gain-of-function and loss-of-function studies can be performed in mammalian cells at the level of the genome.
In an example of an RNAi study to identify host genes required for human immunodeffiency virus (HIV) replication, approximately five times as many genes were identified as were previously known.
Similar genomic screens for HIV host factors were performed in multiple laboratories, resulting in very limited overlap in specific genes, although similar pathways were often identified. Therefore even relatively minor differences in a variety of assay parameters and computation strategies appear to have a significant influence on results.
Functional genomic screens utilizing high content screening systems enable the interrogation of multiple phenotypic endpoints at the level of a single cell, and may circumvent pleotropic effects of RNAi reagents, thus improving the consistency of the results.
Functional genomic screens can also be applied to the identification of the mechanism of action of a small molecule modulator found in a phenotypic screen.
Footnotes
Declaration of Interest: The authors declare that they have no conflict of interest and have received no payment for the preparation of this manuscript.
References
- 1.Gerstein MB, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 2*.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. This study demonstrates the applicability of high-throughput transfection on a glass slide format. [DOI] [PubMed] [Google Scholar]
- 3*.Chanda SK, et al. Genome-scale functional profiling of the mammalian AP-1 signaling pathway. Proc Natl Acad Sci U S A. 2003;100:12153–12158. doi: 10.1073/pnas.1934839100. This manuscript provides a first description of 384-well ‘reverse-transfection’, which is currently the standard for high-throughput genetic screens in mammalian cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 5.Gewurz BE, et al. Genome-wide siRNA screen for mediators of NF-kappaB activation. Proc Natl Acad Sci U S A. 2012;109:2467–2472. doi: 10.1073/pnas.1120542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Harada JN, et al. Identification of novel mammalian growth regulatory factors by genome-scale quantitative image analysis. Genome Res. 2005;15:1136–1144. doi: 10.1101/gr.3889305. The first example of combining genome-scale genetic analysis with high content screening in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rual JF, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strausberg RL, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skalamera D, et al. A high-throughput platform for lentiviral overexpression screening of the human ORFeome. PLoS One. 2011;6:e20057. doi: 10.1371/journal.pone.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blesch A. Lentiviral and MLV based retroviral vectors for ex vivo and in vivo gene transfer. Methods. 2004;33:164–172. doi: 10.1016/j.ymeth.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 13**.Bushman FD, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. Explores the issues of non-overlap between published RNAi screens, and provides possible explanations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 15.Pache L, Konig R, Chanda SK. Identifying HIV-1 host cell factors by genome-scale RNAi screening. Methods. 2011;53:3–12. doi: 10.1016/j.ymeth.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, et al. A whole genome screen for HIV restriction factors. Retrovirology. 2011;8:94. doi: 10.1186/1742-4690-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel AD, Young JA. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Montgomery MK. The use of double-stranded RNA to knock down specific gene activity. Methods Mol Biol. 2004;260:129–144. doi: 10.1385/1-59259-755-6:129. [DOI] [PubMed] [Google Scholar]
- 19.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 20.Clemens JC, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 22.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 23**.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. Proof of concept for RNA interference in mammalian cell culture systems. [DOI] [PubMed] [Google Scholar]
- 24.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. Provides important caveats for RNAi screens. [DOI] [PubMed] [Google Scholar]
- 26.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 27.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 29.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 33*.Kummel A, et al. Integration of multiple readouts into the z’ factor for assay quality assessment. J Biomol Screen. 2010;15:95–101. doi: 10.1177/1087057109351311. Evaluation of assay performance when using multi-parametric assay read-outs often employed in high content screening. [DOI] [PubMed] [Google Scholar]
- 34**.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. An important reference for approaches towards analysis of large-scale RNAi datasets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 37.Konig R, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stertz S, Shaw ML. Uncovering the global host cell requirements for influenza virus replication via RNAi screening. Microbes Infect. 2011;13:516–525. doi: 10.1016/j.micinf.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houzet L, Jeang KT. Genome-wide screening using RNA interference to study host factors in viral replication and pathogenesis. Exp Biol Med (Maywood) 2011;236:962–967. doi: 10.1258/ebm.2010.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittler R, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- 42*.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. An important discussion on the impact of off-target activities in siRNA screens. [DOI] [PubMed] [Google Scholar]
- 43.Tschuch C, et al. Off-target effects of siRNA specific for GFP. BMC Mol Biol. 2008;9:60. doi: 10.1186/1471-2199-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fedorov Y, et al. Off-target effects by siRNA can induce toxic phenotype. Rna. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bramsen JB, Kjems J. Chemical modification of small interfering RNA. Methods Mol Biol. 2011;721:77–103. doi: 10.1007/978-1-61779-037-9_5. [DOI] [PubMed] [Google Scholar]
- 46.Sigoillot FD, et al. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat Methods. 2012 doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marine S, Bahl A, Ferrer M, Buehler E. Common seed analysis to identify off-target effects in siRNA screens. J Biomol Screen. 2012;17:370–378. doi: 10.1177/1087057111427348. [DOI] [PubMed] [Google Scholar]
- 48**.Jager S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. This report provides important support that the integration of orthogonal systems-level datasets can be beneficial in target identification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 < http://bioinfogp.cnb.csic.es/tools/venny/index.html>.
- 50*.Luesch H, et al. A functional genomics approach to the mode of action of apratoxin A. Nat Chem Biol. 2006;2:158–167. doi: 10.1038/nchembio769. An example for utilizing mammalian genetics to identify the mechanism of action for a small molecule. [DOI] [PubMed] [Google Scholar]
- 51.Eggert US, et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harder N, et al. Automatic analysis of dividing cells in live cell movies to detect mitotic delays and correlate phenotypes in time. Genome Res. 2009;19:2113–2124. doi: 10.1101/gr.092494.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. This is an excellent example of live cell high content imaging and phenotypic data mining applied to functional analysis of cell division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Treiser MD, et al. Cytoskeleton-based forecasting of stem cell lineage fates. Proc Natl Acad Sci U S A. 2010;107:610–615. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55**.Quintavalle M, Elia L, Price JH, Heynen-Genel S, Courtneidge SA. A cell-based high-content screening assay reveals activators and inhibitors of cancer cell invasion. Sci Signal. 2011;4:ra49. doi: 10.1126/scisignal.2002032. An excellent recent example of the application of HCS to cancer drug discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Kaltenbach LS, et al. Composite primary neuronal high-content screening assay for Huntington’s disease incorporating non-cell-autonomous interactions. J Biomol Screen. 2010;15:806–819. doi: 10.1177/1087057110373392. Good example for use of primary co-cultures (multiple cell types) in image-based screening. [DOI] [PubMed] [Google Scholar]
- 58.Snijder B, et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- 59.Conrad C, Gerlich DW. Automated microscopy for high-content RNAi screening. J Cell Biol. 2010;188:453–461. doi: 10.1083/jcb.200910105. See 61. * Good overview of image-based screening strategies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haney S. High Content Screening: Science, Techniques and Applications. Wiley; 2008. See 61. [Google Scholar]
- 61*.Shariff A, Kangas J, Coelho LP, Quinn S, Murphy RF. Automated image analysis for high-content screening and analysis. J Biomol Screen. 2010;15:726–734. doi: 10.1177/1087057110370894. These three referneces provide excellent reviews of the considerations required for of image-based HCS. [DOI] [PubMed] [Google Scholar]
- 62.Niederlein A, Meyenhofer F, White D, Bickle M. Image analysis in high-content screening. Comb Chem High Throughput Screen. 2009;12:899–907. doi: 10.2174/138620709789383213. [DOI] [PubMed] [Google Scholar]
- 63.Dull A, Goncharova E, Hager G, McMahon JB. Development of an image analysis screen for estrogen receptor alpha (ERalpha) ligands through measurement of nuclear translocation dynamics. J Steroid Biochem Mol Biol. 2010;122:341–351. doi: 10.1016/j.jsbmb.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Young DW, et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat Chem Biol. 2008;4:59–68. doi: 10.1038/nchembio.2007.53. An example of the application of clustering techniques to determine mechanism of action. [DOI] [PubMed] [Google Scholar]
- 65.Duda RO, HP, Stork DG. Pattern Classification. Wiley; 2000. [Google Scholar]
- 66.Horvath P, Wild T, Kutay U, Csucs G. Machine learning improves the precision and robustness of high-content screens: using nonlinear multiparametric methods to analyze screening results. J Biomol Screen. 2011;16:1059–1067. doi: 10.1177/1087057111414878. [DOI] [PubMed] [Google Scholar]
- 67.Misselwitz B, et al. Enhanced CellClassifier: a multi-class classification tool for microscopy images. BMC Bioinformatics. 2010;11:30. doi: 10.1186/1471-2105-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang B, Pham TD. Phenotype recognition with combined features and random subspace classifier ensemble. BMC Bioinformatics. 2011;12:128. doi: 10.1186/1471-2105-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.ME, HDaC . Neural networks and artificial intelligence for biomedical engineering. IEEE Press; 2000. [Google Scholar]
- 70.Kotsiantis S. Supervised machine learning: A review of classification techniques. Informatica. 2007;31:249–268. [Google Scholar]
- 71.KTSaK . Pattern Recognition. 4. Academic Press; 2009. [Google Scholar]
- 72.Sutherland JJ, et al. A robust high-content imaging approach for probing the mechanism of action and phenotypic outcomes of cell-cycle modulators. Mol Cancer Ther. 2011;10:242–254. doi: 10.1158/1535-7163.MCT-10-0720. [DOI] [PubMed] [Google Scholar]