Abstract
Aleutian mink disease virus (AMDV) is the only member in genus Amdovirus of the family Parvoviridae. During AMDV infection, six species of viral transcripts are generated from one precursor mRNA through alternative splicing and alternative polyadenylation. In addition to the large non-structural protein NS1, two small non-structural proteins, NS2 and NS3, are putatively encoded (Qiu J, et al: Journal of Virology, 80:654–62, 2006). However, these two proteins have not been experimentally demonstrated during virus infection, and nothing is known about their function. Here, we studied the nonstructural protein expression profile of AMDV, and for the first time, confirmed expression of NS2 and NS3 during infection, and identified their intracellular localization. More importantly, we provided evidence that both NS2 and NS3 are necessary for AMDV replication.
Introduction
Aleutian mink disease virus (AMDV), an autonomous parvovirus, belongs to genus Amdovirus, in the family Parvoviridae (Tijssen et al., 2012). AMDV infection is known only to produce clinical manifestation in mink and ferrets (Aasted, 1985; Porter et al., 1982). AMDV is the only classified member in the genus Amdovirus (Tijssen et al., 2012). However, identification of an Amdovirus sequence in grey fox (GFADV) (Li et al., 2011) suggests the likelihood of more members in this genus. Aleutian mink disease affects mink farming worldwide, and has become the most significant threat to mink production in some area (Newman and Reed, 2006). AMDV infection in neonatal mink kits causes acute, rapidly progressing interstitial pneumonia with a high mortality rate (Alexandersen, 1990); whereas, in adult mink, AMDV undergoes a persistent infection which manifests with symptoms of glomerulonephritis, plasmacytosis, hypergammaglobinemia, arteritis, decreased fertility, and spontaneous abortion (Alexandersen et al., 1994; Gorham et al., 1976). The hypergammaglobulinemia that is characteristic of Aleutian mink disease is partly caused by massively increased level of anti-AMDV antibodies in blood. The antibody and virus complex is deposited within tissues of multiple organs in the body causing inflammation, which also facilitates the entry of virus into certain target cell populations (Kanno et al., 1993a). In vitro infection confirmed the antibody-dependent enhancement of AMDV infection (Bloom et al., 2001; Dworak et al., 1997; Kanno et al., 1993b). Therefore, it is difficult to develop a vaccine to prevent AMDV infection (Aasted et al., 1998). Currently, there are also no prevention or treatment methods available for Aleutian mink disease.
The ferret is another host of AMDV (Porter et al., 1982). Ferrets have been widely bred as pets and laboratory animals, for example, as an animal model of influenza virus (Barnard, 2009) and cystic fibrosis (Sun et al., 2010), but the impact of AMDV infection on the pathogenesis of these other infections has not been studied. Clinical symptoms in ferrets are renal failure, weight loss, splenomegaly, neurological symptoms like seizures and clotting abnormalities, and disease progression will result in death of the ferret within a few months. Recently evidence of AMDV infection was noted in two mink farmers who displayed disease symptoms similar to those of Aleutian mink disease. Hence, AMDV may be a zoonotic disease rarely capable of infecting humans (Jepsen et al., 2009).
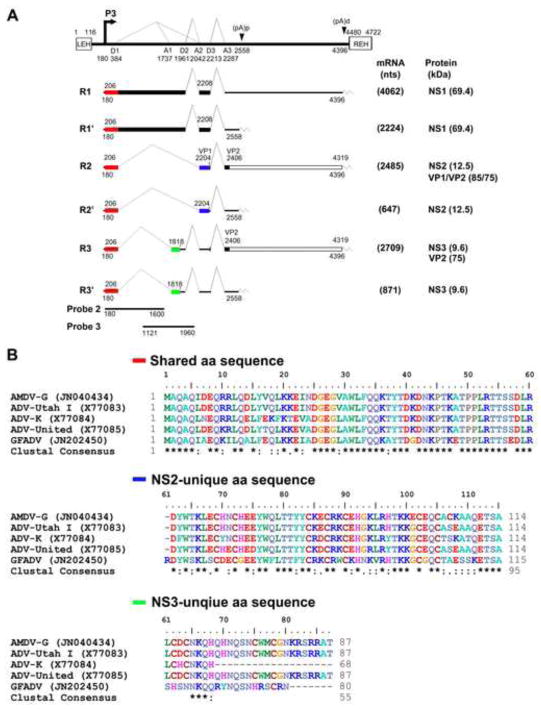
The transcription profile of AMDV during infection displays features similar to those of parvoviruses in genera Erythrovirus and Bocavirus (Chen et al., 2010a; Ozawa et al., 1987; Qiu et al., 2006a). The six species of AMDV mRNA transcripts are generated from a single promoter, and are processed through alternative splicing and alternative polyadenylation (Fig. 1A) (Qiu et al., 2006a). R1 and R2 mRNAs encode the large non-structural protein NS1. The NS1 protein is assumed to function similarly to that of other autonomous parvoviruses. Parvovirus NS1 contains motifs of DNA binding, ATPase and helicase activity that play key roles in viral DNA replication, viral promoter transactivation, and induction of cytopathic effects (Nüesch, 2006). Cleavage of the AMDV NS1 has been shown to be critical to AMDV replication (Best et al., 2003). The most abundant transcript is R2, which encodes structural proteins VP1 and VP2 as well as a non-structural protein NS2 (Qiu et al., 2007). NS2 is also potentially encoded by R2′ mRNA. However, NS2 was only shown to be expressed from transfection of R2 mRNA-transcribing plasmids in cells (Qiu et al., 2007). The dynamic expression of the NS2, its subcellular localization, and, more importantly, its function during virus infection have not been studied (Best and Bloom, 2005; Oleksiewicz et al., 1996). Notably, a third non-structural protein NS3 is putatively encoded by Rx and Rx′ mRNAs (Alexandersen et al., 1988). Expression of NS3 has not been experimentally confirmed either by transfection of Rx/Rx′-transcribing plasmid or during virus infection.
Fig. 1. Genetic map of AMDV and the amino acid sequence alignment of the AMDV NS2 and NS3 proteins among members in genus Amdovirus.
(A) Genetic map of AMDV. The genome of AMDV-G is depicted to scale with indicated transcription units, including the end hairpin termini (LEH and REH), P3 promoter, splice donor site (D1, D2, and D3), splice acceptor sites (A1, A2, and A3), internal proximal polyadenylation site [(pA)p], and internal distal polyadenylation site [(pA)d]. The major six mRNA transcripts are shown with R1-3 polyadenylated at the (pA)d site and R1′-3′ polyadenylated at the (pA)p site. Viral NS1, NS2, NS3 and VP1/VP2 proteins encoded from each mRNA are indicated. (B) Amino acid sequence alignment of AMDV NS2 and NS3 among members in genus Amdovirus. Amino acid sequence alignment of AMDV NS2 and NS3 among four AMDV isolates (AMDV-G, ADV-Utah, ADV-K, and ADV-United) and the newly identified GFADV were chosen to align using ClustalW2. The shared N-terminus, the unique C-termini of NS2 and NS3 are shown. Identical amino acids are shown as a “star” symbol, while homologous amino acids shown as two dots.
Due to the increasing importance of AMDV and its associated diseases in ferrets and potential for human infection, we studied the nonstructural protein expression profile of AMDV during infection. We demonstrated, for the first time, the expression of NS2 and NS3, defined their intracellular localization during infection, and characterized the role of NS2 and NS3 in AMDV DNA replication in the context of an AMDV infectious clone.
Results
AMDV NS2 protein is expressed at a level similar to that of NS1 during infection
To determine whether the putative NS2 and NS3 are expressed during AMDV infection, and their relative expression level compared to that of NS1, we examined NS proteins expression over a time course of 6 days during AMDV-G infection of CrFK cells at a multiplicity of infection (MOI) of 1 (Huang et al., 2012; Qiu et al., 2006a), employing a rabbit polyclonal antibody against the common N-terminal 60 aa shared by NS1, NS2 and NS3 (Fig. 1B). Both NS1 and NS2 were detected at 2 days post-infection (p.i.), reached the highest level at 3 day p.i., remained at a consistent level at 4–5 days p.i., and started to decrease at 6 days p.i. (Fig. 2A). The NS expression dynamics is consistent with the kinetics of the viral DNA replication and virus production during infection as we described previously (Qiu et al., 2006a). Notably, the level of NS2 expression was similar to that of the NS1 over the course of infection. However, the detected NS2 had a size of ~17 kDa (Fig. 2), which is larger than the predicted size at 12.5 kDa (Fig. 1A). We did not detect an obvious band at ~10 kDa (the putative size of NS3) in lysates of AMDV-infected cells resolved by electrophoresis in SDS through a 15% polyacrylamide gel electrophoresis (PAGE) gel, compared with the band from mock-infected cells (Fig. 2B). Thus, the result suggested that NS3 may be expressed at a level much lower than that of NS2.
Fig. 2. AMDV NS1, NS2 and NS3 expression during AMDV infection.
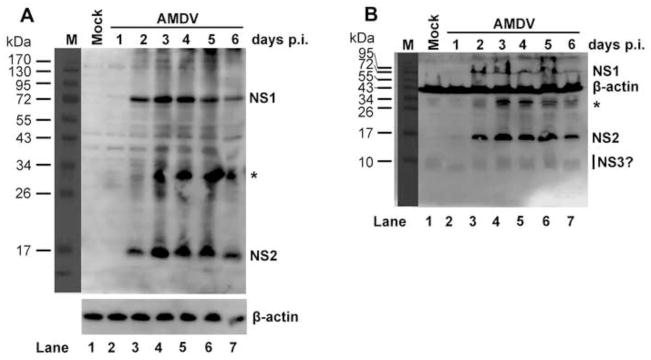
CrFK cells were infected with AMDV-G at an MOI of 1 or mock-infected. At the indicated days p.i., infected cells were harvested, lysed and resolved in SDS-8%-PAGE (panel A) or SDS-15%-PAGE (panel B) followed by Western blot analysis using a rabbit polyclonal anti-NS1/2/3 antibody. β-actin was blotted as a loading control. Asterisk, cleaved NS1 bands (Cheng et al., 2009).
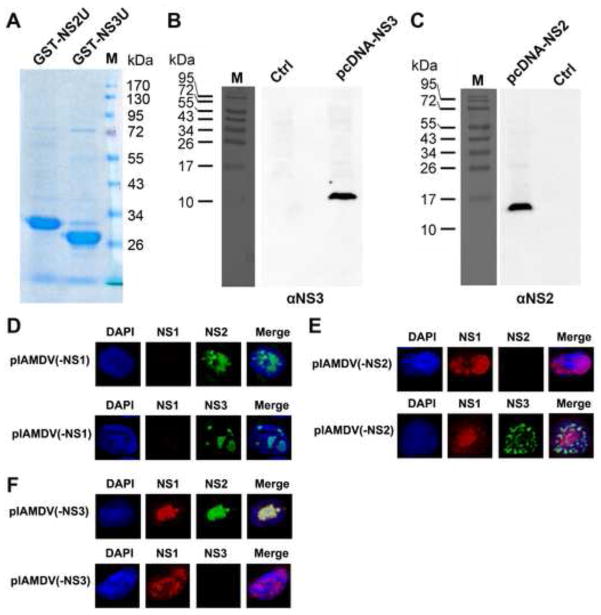
AMDV NS3 protein is expressed in the nucleus of AMDV-infected cells
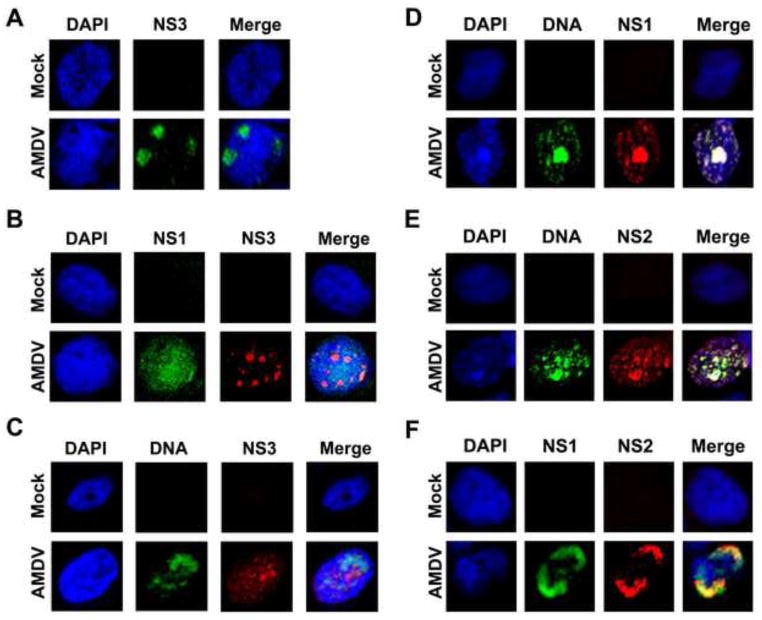
We have previously determined that NS3-encoding mRNAs (Rx/Rx′) are transcribed at a comparable level to that of the NS1-encoding mRNAs (R1/R1′) (Fig. 1A) (Qiu et al., 2006a). To prove that NS3 protein was expressed during AMDV infection, we generated an antibody against NS3-specific residues (NS3U, aa 61-87) by immunizing rats with purified GST-fused NS3U (Fig. 3A, GST-NS3U). The specificity of the anti-NS3 antibody was confirmed by Western blotting (Fig. 3B) and immunofluorescence (Fig. 3D–F). Next, we used this antibody to examine NS3 expression during AMDV infection of CrFK cells. At 4 days p.i., we detected NS3 in the nuclei of infected cells (Fig. 4A). Moreover, when AMDV-infected cells were co-stained with antibodies to both NS1 and NS3 (Fig. 4B), NS3 did not co-localize with the NS1. We further used fluorescent in situ hybridization (FISH)-immunofluorescence assay to examine an association of NS3 with viral DNA. The result showed that NS3 did not colocalize with viral DNA (Fig. 4C). Taken together, these results suggest that AMDV NS3 protein is expressed in the nucleus during AMDV infection, but does not localize within the viral DNA replication centers.
Fig. 3. Analyses of the specificity of the rat anti-NS2 and anti-NS3 antibodies.
(A) Coomassie blue staining of the purified GST-NS2U and GST-NS3U. ~2 μg of the purified GST-NS2U and GST-NS3U proteins were resolved in SDS-10%PAGE gel. The gel was stained using Coomassie blue. A molecule weight marker is shown. (B&C) Western blot analysis of NS2 and NS3 expression. CrFK cells were transfected with constructs pcDNA-NS2, pcDNA-NS3, and pcDNA3 (as a control, ctrl). At 2 days post-transfection, transfected cells were harvested, lysed and resolved in SDS-15%PAGE (panel B) or SDS-12%PAGE (panel C) followed by Western blot analysis using the rat anti-NS3 or anti-NS2 antibody. (D–F) Immunofluorescence analysis of NS expression. CrFK cells were transfected with pIAMDV(-NS1) (D), pIAMDV(-NS2) (E) and pIAMDV(-NS3) (F). At 4 days post-transfection, the transfected cells were analyzed by immunofluorescence using rabbit anti-NS1 and rat anti-NS2 antibodies or rabbit anti-NS1 and rat anti-NS3 antibodies, as indicated. Confocal images were taken at × 60 magnification (objective lens). Nuclei were stained with DAPI.
Fig. 4. Immunofluorescence analysis of AMDV NS2 and NS3 expression.
CrFK cells were infected with AMDV-G at an MOI of 1 or mock-infected. (A–C) Analysis of NS3 expression. At 4 days p.i., cells were stained with rat anti-NS3 (A), rabbit anti-NS1 and rat anti-NS3 (B), respectively. The cells were also analyzed by FISH-Immunofluorescence with a biotintylated AMDV-G DNA probe and a rabbit anti-NS3 antibody (C). (D–F) Analysis of NS2 expression. At 4 days p.i., cells were analyzed by FISH-Immunofluorescence with a biotintylated AMDV-G DNA probe and a rabbit anti-NS1 antibody (D) or a rat anti-NS2 antibody (E). The cells were also co-stained with rabbit anti-NS1 and rat anti-NS2 antibodies (F).
Confocal images were taken at × 60 magnification (objective lens). Nuclei were stained with DAPI.
AMDV NS1 and NS2 colocalize with each other within the viral DNA replication centers
To understand the function of NS2, we also examined the localization of the NS2 in relation to NS1 and the AMDV DNA replication centers using antibodies that recognize the unique regions of each protein. Anti-NS2 antibody was produced by immunizing rats with purified GST-fused NS2 unique aa 61-114 (Fig. 3A; GST-NS2U). The specificity of the anti-NS2 antibody was confirmed both by Western blotting (Fig. 3C) and immunofluorescence (Fig. 3D–F). Both NS1 (Fig. 4D) and NS2 (Fig. 4E) precisely localized within the viral DNA replication centers, as shown by the FISH-immunofluorescence assay. This result was also supported by the colocalization of the NS1 and NS2 in the nucleus (Fig. 4F). Thus, our results suggested a role for AMDV NS2 in viral DNA replication.
Both NS2 and NS3 are important to the production of infectious progeny virions
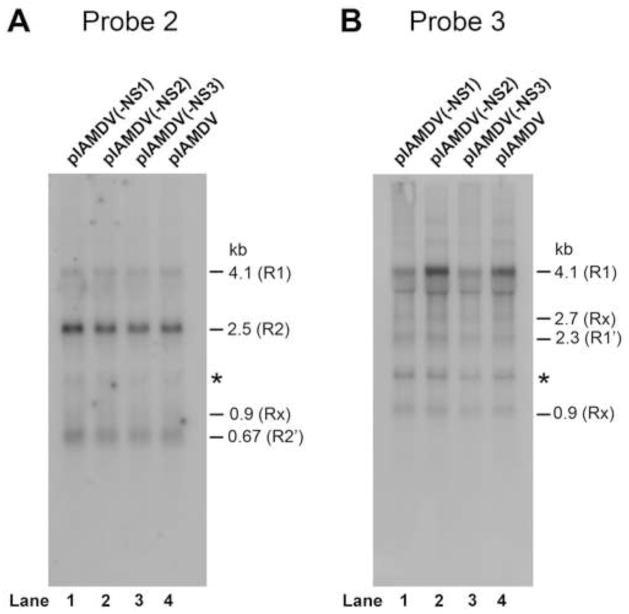
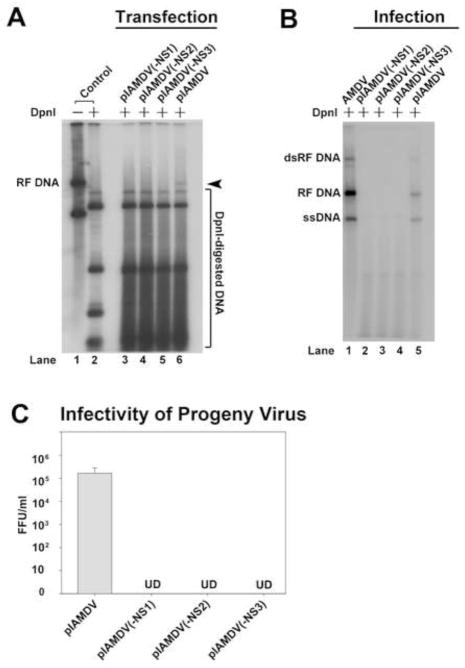
To explore a role of NS2 and NS3 in AMDV replication, we created NS protein knockout mutants in the context of an AMDV infectious clone (pIAMDV) and tested them for viral DNA replication as well as infectious virion production. The successful knockout of the NS1, NS2, and NS3 in their respective constructs was confirmed by immunofluorescence analysis (Fig. 3D–F). The mutations in the context of pIAMDV did not alter the levels of each viral transcript in the cytoplasm of the transfected cells as detected by Northern blotting using Probe 2 and Probe 3 (Qiu et al., 2006a), respectively (Fig. 5A&B). The results showed that NS1-knockout mutant [(pIADMV(-NS1)], as a control, did not produce replicative form viral DNA (RF DNA, DpnI digestion-resistant band) from transfection; neither did the NS2- and NS3-knockout mutants [(pIAMDV(-NS2) and pIAMDV(-NS3), respectively] (Fig. 6A). We further infected fresh CrFK cells with the lysate of pIAMDV(-NS1)-, pIAMDV(-NS2)-, pIAMDV(-NS3) and pIAMDV-transfected cells. The results showed that the RF DNA band was obviously detected in Hirt DNA samples prepared from cells infected with lysate of the pIAMDV-transfected cells, but not in samples from cells infected with lysates of the pIAMDV(-NS1)-, pIAMDV(-NS2)-, pIAMDV(-NS3)-transfected cells (Fig. 6B, compare lanes 2–4 with 5). Taken together, these findings suggested that all the NS1, NS2 and NS3 are indispensable for viral DNA replication.
Fig. 5. Northern blot analysis of AMDV mRNAs in the cytoplasm of the cells transfected with pIAMDV and the NS knockout mutants.
CrFK cells were transfected with pIAMDV, pIAMDV(-NS1), pIAMDV(-NS2) and pIAMDV(-NS3), respectively. At 4 days post-transfection, cytoplasmic RNA was extracted from transfected cells and analyzed by Northern blot using Probe 2 and Probe 3, which span a region of nt 180-1600 and nt 1121-1960, respectively, of the AMDV-G genome (diagramed in Fig. 1A). The identity and the size of detected viral mRNAs, which have been confirmed previously (Qiu et al., 2006a), are shown to the right of each panel.
Fig. 6. Analyses of the NS knockout mutant in the context of an AMDV infectious clone.
(A&B) Southern blot analysis of viral DNA replication. (A) CrFK cells were transfected with pIAMDV, pIAMDV(-NS1), pIAMDV(-NS2) and pIAMDV(-NS3), respectively. At 6 days post-transfection, transfected cells were collected and used to prepare Hirt DNA. (B) CrFK cells were infected with virus samples prepared from the above CrFK cells transfected with pIAMDV, or pIAMDV(-NS1), pIAMDV(-NS2), and pIAMDV(-NS3). At 6 days p.i., infected cells were collected and used to prepare Hirt DNA samples. The Hirt DNA samples were digested with DpnI for Southern blotting. The blots were probed with a full-length AMDV probe (Qiu et al., 2006a). The identity of major bands on the blots is indicated. RF DNA, replicative form viral genome; dRF DNA, double replicative form viral genome; ssDNA, single-stranded viral genome. Cells infected with AMDV-G were used a control (AMDV). (C) Titration of virus production in cells infected with progeny virus generated from transfection. CrFK cells were infected with virus samples prepared from the above CrFK cells transfected with pIAMDV, or pIAMDV(-NS1), pIAMDV(-NS2) and pIAMDV(-NS3), and were harvested at 6 days post-transfection. The cells were frozen and thawed three times, and briefly centrifuged. The supernatant was collected, serially diluted, and used to infect fresh CrFK cells in order to titrate FFU. Results shown represent the averages and standard deviation from at least three independent experiments. UD, undetectable.
We also looked for virus production from infection of progeny virus produced following transfection of each mutant. Transfection of the three mutants, pIAMDV(-NS1), pIAMDV(-NS2) and pIAMDV(-NS3), did not lead to production of any detectable infectious virions in CrFK cells; whereas reinfection of progeny virus produced from the wild-type control (pIAMDV) generated a titer of 2.0 × 105 fluorescence-focus forming unit (FFU)/ml (Fig. 6C). These results confirmed that all the NS1, NS2 and NS3 are essential to AMDV replication.
Discussion
In this study, we demonstrated the expression of the AMDV NS2 and NS3 during AMDV infection in CrFK cells, and provided evidence of the essential role of both NS2 and NS3 in progeny virus production.
The presence and the roles of small parvovirus non-structural proteins have received limited attention. Expression of small non-structural proteins by parvoviruses has only been demonstrated during B19V infection (Ozawa et al., 1987). B19V expresses two small nonstructural proteins, namely, 11-kDa and 7.5-kDa protein (Luo and Astell, 1993; St and Astell, 1993). The B19V 7.5-kDa is encoded from mRNAs spliced out of NS1-encoding mRNA in a similar manner to that of AMDV NS3. However, the 11-kDa is very different since it is encoded by mRNAs spliced out of VP1/2-encoding mRNA (Qiu et al., 2006b). The function of B19V 7.5-kDa during B19V infection is unknown yet.
Many parvoviruses express a second NS protein, which is smaller than NS1, e.g., MVM NS2 (MW=24 kDa) (Cotmore et al., 1983; Cotmore and Tattersall, 1990). AMDV NS2 is encoded by mRNAs that are spliced in a manner similar to that of the MVM NS2, in that NS2 shares an N-terminus with NS1 and a C-terminus overlapped with NS1 (Pintel et al., 1995). However, AMDV NS2 is much smaller with a predicted MW of only 12.5 kDa, and contains 54 unique amino acids. Notably, AMDV NS2 was detected at a size of ~17 kDa, indicating modifications of the protein, e.g., phosphorylation, as detected in the small nonstructural protein NP1 of bocavirus bovine parvovirus (Lederman et al., 1984). MVM NS2 can be phosphorylated in vitro (Nüesch, 2006). Amino acid alignment between AMDV NS2 and MVM NS2 does not reveal a significant similarity. MVM NS2 is essential to optimal virus replication (Naeger et al., 1990), and has been shown to play a role in capsid assembly (Cotmore et al., 1997) and in egress of progeny virus from the nucleus (Eichwald et al., 2002), but only in certain cell types. MVM NS1 and NS2 interact and colocalize in the APAR body (Young et al., 2002), and replication of MVM DNA is critically dependent on accumulated levels of NS2 at least in murine cells (Choi et al., 2005; Ruiz et al., 2006). The colocalization of AMDV NS2 with NS1 and viral DNA strongly supports a unique role of the AMDV NS2 during infection of CrFK cells.
AMDV NS3-encoding Rx/Rx′ mRNAs were generated at an abundance similar to that of the NS1-encoding R1/R1′ mRNAs (Qiu et al., 2006a). However, the expression level of NS3 protein during AMDV infection was extremely low, and it was not clearly detected by Western-blotting. Detection was possible only by immunofluorescence using a sensitive anti-NS3 antibody that we developed suggesting that either the NS3 protein is translated at low levels or is rapidly turned over or unstable. NS3 clearly did not colocalize with NS1 in the viral DNA replication centers, and was not required for viral DNA replication. It is intriguing that the NS3 is important for virus replication. The NS3 ORF is present in wild-type AMDV strains as well (Fig. 1B) (Gottschalck et al., 1994), suggesting an important role of the NS3 during AMDV infection in vivo. NS3 is a small protein with only 87 amino acids, in which 60 amino acids from the N-terminus are identical to those of the NS1 and NS2 (Fig. 1B). Hence, NS3 only has a unique sequence of 27 aa. Thus, we have demonstrated a critical role of an unusual small non-structural protein in autonomous parvovirus infection. Further mutagenesis study of the 27 aa of the NS3 is warranted to understand its functional motifs.
In conclusion, our study demonstrated multiple controls of AMDV replication by the three non-structural proteins NS1, NS2 and NS3 during infection. The function of the NS3 during virus replication is novel in that it is only contains unique 27aa. The mechanism underlying NS3-controlled AMDV replication warrants further investigation, which might provide insight in the function of the B19V 7.5-kDa protein.
Materials and Methods
Virus and infection
CrFK cells (Crandall feline kidney cell line, ATCC CCL-94) were purchased from the American Type Culture Collection (ATCC, Manassas, VA), and were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum at 37°C in 5% CO2. CrFK cells were infected with AMDV-G at an MO of 1 (FFU per cell) and kept at 31.8°C as described previously (Bloom et al., 1980; Huang et al., 2012; Porter et al., 1977).
Transfection
Two μg of DNA was transfected CrFK cells on 60-mm plate with Lipofectamine and Plus reagent (Invitrogen, Carlsbad, CA) as described previously (Huang et al., 2012).
Plasmid construction
pIAMDV and its mutants: The parent AMDV infectious clone, pIAMDV, has been previously described (Cheng et al., 2009; Huang et al., 2012). pIAMDV-based NS protein knockout mutants, pIAMDV(-NS1), pIAMDV(-NS2) and pIAMDV(-NS3), were constructed by mutating nucleotide G(404)T and G(407)T, T(2049)A and A(2056)T, and A(1753)T and C(1756)T, respectively, in the AMDV-G genome of pIAMDV, to early terminate translation of NS1, NS2 and NS3. pcDNA-NS2 and pcDNA-NS2 were constructed by inserting the NS2 ORF and NS3 ORF in pcDNA3 (Invitrogen, Grand Island, NY).
Constructs for glutathione S-transferase (GST)-fusion protein expression: The DNA sequences to express the unique C-terminus of the AMDV NS2 (aa 61-114) and NS3 (aa 61-87) were cloned into BamHI/XhoI-digested pGEX4T3 (GE Healthcare, Piscataway, NJ) as pGEX-NS2U and pGEX-NS3U, respectively. All the nucleotide or amino acid numbers shown were referred to the AMDV-G genome sequence (accession no.: JN040434). The sequences of all the constructs were confirmed at MCLAB (South San Francisco, CA).
Southern blot and Northern blot analyses
Low-molecular-weight DNA (Hirt DNA) was isolated by a modified Hirt extraction method from either transfected or infected cells as previously described (Qiu et al., 2006a). Samples were run on a 1% agarose gel, followed by Southern blotting as described previously (Huang et al., 2012; Naeger et al., 1990; Tullis et al., 1993).
Cytoplasmic RNA was isolated from transfected cells using RNeasy Mini Kit (Qiagen, Valencia, CA) following the supplementary protocol for purification of cytoplasmic RNA, and was analyzed following Northern blotting as described previously (Pintel et al., 1983; Qiu et al., 2006a).
Blots were exposed to a GE Health phosphor imaging screen, and analyzed using a Typhoon FLA 9000 Phosphor Imager and ImageQuant™ TL (version 4.2.2) imaging software (GE Heath).
Antibody production
Rat anti-NS2 and NS3 antibodies: GST-fused AMDV NS2- and NS3-unique amino acid sequences (Fig. 1B) were expressed in Escherichia coli BL21 cells transformed with pGEX-NS2 and pGEX-NS3, respectively, and were purified using a GSTrap FF 1-ml column (GE Healthcare) on the Biologic LP system (Bio-Rad, Hercules, CA). Purified proteins were examined for a purity of >90% on SDS-PAGE. Purified proteins (500 μg/ml) were emulsified with TiterMax® Gold adjuvant (TiterMax® USA, Norcross, GA) and used to immunize rats following an animal protocol that has been approved by the KUMC Institutional Animal Care and Use Committee, as previously described (Sun et al., 2009).
Rabbit ant-NS1 and anti-shared NS domain antibodies: NS1-specific antibody (anti-NS1; R2799) and NS1/2/3-reacted antibody (anti-NS1/2/3; R3810) were raised in rabbit by immunizing NS1-unique residues aa 173 to 464 and NS1 peptides aa 2 to 61 (Fig. 1B), respectively, and have been reported previously (Best et al., 2003).
Western-blot analysis
Infected cells were harvested, lysed and resolved by SDS-PAGE. Western blotting was performed as previously described (Huang et al., 2012).
Fluorescence in situ hybridization (FISH) and immunofluorescence assay
An AMDV DNA fragment (nt 1761-2033) was biotinylated using the Label IT FISH (Biotin) Labeling Kit (Mirus, Madison, WI). FISH-immunofluorescence assay was performed as described previously (Luo et al., 2011). Briefly, CrFK cells in wells on 4-well chamber slide (Nunc. Lab-Tek, Thermo Scientific) were infected with AMDV-G. Cells were fixed in 3.7% paraformaldehyde for 16 min, and permeablized with 0.2% Triton X-100 for 10 min. Slides were then treated with 5 μg/ml RNase A at 37°C for 1 hr and washed with 2 × SSC buffer. Next, the slides were dehydrated with 70%, 85% and 100% ethanol in order for 2 min at room temperature, and denatured with 70% formamide for 2 min. Sequentially, the slides were dehydrated with 70% pre-chilled ethanol, 85% and 100% ethanol at room temperature in order for 2 min each. Finally, the slides were hybridized with the biotintylated AMDV probe at 37°C overnight. After hybridization, the slides were incubated with mouse anti-biotin (Abcam, Cambridge, MA) and rat anti-NS1, anti-NS2 or anti-NS3 antibodies followed by incubation with FITC-conjugated anti-mouse and TexRed-conjugated anti-rat secondary antibodies for immunofluorescence analysis as previously described (Chen et al., 2010b). Confocal images were taken at a magnification of 60 × (objective lens), with an Eclipse C1 Plus confocal microscope (Nikon) controlled by Nikon EZ-C1 software.
Research Highlights.
Two small nonstructural proteins, NS2 and NS3, are expressed during AMDV infection.
AMDV NS2 colocalizes with NS1 and viral DNA in the viral replication centers; whereas NS3 does not.
Both AMDV NS2 and NS3 play important roles in the production of infectious progeny virions.
Acknowledgments
This work was supported by PHS grant R01 AI070723 from NIAID. We thank all the members of the Qiu lab for valuable discussions. The work is supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasted B. Aleutian disease of mink. Virology and immunology. Acta Pathol Microbiol Immunol Scand Suppl. 1985;287:1–47. 1–47. [PubMed] [Google Scholar]
- Aasted B, Alexandersen S, Christensen J. Vaccination with Aleutian mink disease parvovirus (AMDV) capsid proteins enhances disease, while vaccination with the major non-structural AMDV protein causes partial protection from disease. Vaccine. 1998;16:1158–1165. doi: 10.1016/s0264-410x(98)80114-x. [DOI] [PubMed] [Google Scholar]
- Alexandersen S. Pathogenesis of disease caused by Aleutian mink disease parvovirus. APMIS Suppl. 1990;14:1–32. 1–32. [PubMed] [Google Scholar]
- Alexandersen S, Bloom ME, Perryman S. Detailed transcription map of Aleutian mink disease parvovirus. J Virol. 1988;62:3684–3694. doi: 10.1128/jvi.62.10.3684-3694.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersen S, Larsen S, Aasted B, Uttenthal A, Bloom ME, Hansen M. Acute interstitial pneumonia in mink kits inoculated with defined isolates of Aleutian mink disease parvovirus. Vet Pathol. 1994;31:216–228. doi: 10.1177/030098589403100209. [DOI] [PubMed] [Google Scholar]
- Barnard DL. Animal models for the study of influenza pathogenesis and therapy. Antiviral Res. 2009;82:A110–A122. doi: 10.1016/j.antiviral.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best SM, Bloom ME. Aleutian mink disease parvovirus. In: Kerr JR, Cotmore SF, Bloom ME, Linden ME, Parrish CR, editors. The parvoviruses. Hodder Arnold; London, United Kingdom: 2005. pp. 457–471. [Google Scholar]
- Best SM, Shelton JF, Pompey JM, Wolfinbarger JB, Bloom ME. Caspase cleavage of the nonstructural protein NS1 mediates replication of Aleutian mink disease parvovirus. J Virol. 2003;77:5305–5312. doi: 10.1128/JVI.77.9.5305-5312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ME, Best SM, Hayes SF, Wells RD, Wolfinbarger JB, McKenna R, Agbandje-McKenna M. Identification of aleutian mink disease parvovirus capsid sequences mediating antibody-dependent enhancement of infection, virus neutralization, and immune complex formation. J Virol. 2001;75:11116–11127. doi: 10.1128/JVI.75.22.11116-11127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom ME, Race RE, Wolfinbarger JB. Characterization of Aleutian disease virus as a parvovirus. J Virol. 1980;35:836–843. doi: 10.1128/jvi.35.3.836-843.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. Characterization of the gene expression profile of human bocavirus. Virology. 2010a;403:145–154. doi: 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Luo Y, Cheng F, Sun Y, Qiu J. Bocavirus infection induces a mitochondrion-mediated apoptosis and cell cycle arrest at G2/M-phase. J Virol. 2010b;84:5615–5626. doi: 10.1128/JVI.02094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Chen AY, Best SM, Bloom ME, Pintel D, Qiu J. The capsid proteins of Aleutian mink disease virus (AMDV) activate caspases and are specifically cleaved during infection. J Virol. 2009;84:2687–2696. doi: 10.1128/JVI.01917-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EY, Newman AE, Burger L, Pintel D. Replication of minute virus of mice DNA is critically dependent on accumulated levels of NS2. J Virol. 2005;79:12375–12381. doi: 10.1128/JVI.79.19.12375-12381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, D’Abramo AM, Jr, Carbonell LF, Bratton J, Tattersall P. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology. 1997;231:267–280. doi: 10.1006/viro.1997.8545. [DOI] [PubMed] [Google Scholar]
- Cotmore SF, Sturzenbecker LJ, Tattersall P. The autonomous parvovirus MVM encodes two nonstructural proteins in addition to its capsid polypeptides. Virology. 1983;129:333–343. doi: 10.1016/0042-6822(83)90172-1. [DOI] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology. 1990;177:477–487. doi: 10.1016/0042-6822(90)90512-p. [DOI] [PubMed] [Google Scholar]
- Dworak LJ, Wolfinbarger JB, Bloom ME. Aleutian mink disease parvovirus infection of K562 cells is antibody-dependent and is mediated via an Fc(gamma)RII receptor. Arch Virol. 1997;142:363–373. doi: 10.1007/s007050050082. [DOI] [PubMed] [Google Scholar]
- Eichwald V, Daeffler L, Klein M, Rommelaere J, Salome N. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J Virol. 2002;76:10307–10319. doi: 10.1128/JVI.76.20.10307-10319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham JR, Henson JB, Crawford TB, Padgett GA. The epizootiology of aleutian disease. Front Biol. 1976;44:135–58. 135–158. [PubMed] [Google Scholar]
- Gottschalck E, Alexandersen S, Storgaard T, Bloom ME, Aasted B. Sequence comparison of the non-structural genes of four different types of Aleutian mink disease parvovirus indicates an unusual degree of variability. Arch Virol. 1994;138:213–231. doi: 10.1007/BF01379127. [DOI] [PubMed] [Google Scholar]
- Huang Q, Deng X, Best SM, Bloom ME, Li Y, Qiu J. Internal polyadenylation of parvoviral precursor mRNA limits progeny virus production. Virology. 2012;426:167–177. doi: 10.1016/j.virol.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen JR, d’Amore F, Baandrup U, Clausen MR, Gottschalck E, Aasted B. Aleutian mink disease virus and humans. Emerg Infect Dis. 2009;15:2040–2042. doi: 10.3201/eid1512.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno H, Wolfinbarger JB, Bloom ME. Aleutian mink disease parvovirus infection of mink macrophages and human macrophage cell line U937: demonstration of antibody-dependent enhancement of infection. J Virol. 1993a;67:7017–7024. doi: 10.1128/jvi.67.12.7017-7024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno H, Wolfinbarger JB, Bloom ME. Aleutian mink disease parvovirus infection of mink macrophages and human macrophage cell line U937: demonstration of antibody-dependent enhancement of infection. J Virol. 1993b;67:7017–7024. doi: 10.1128/jvi.67.12.7017-7024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman M, Patton JT, Stout ER, Bates RC. Virally coded noncapsid protein associated with bovine parvovirus infection. J Virol. 1984;49:315–318. doi: 10.1128/jvi.49.2.315-318.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Pesavento PA, Woods L, Clifford DL, Luff J, Wang C, Delwart E. Novel amdovirus in gray foxes. Emerg Infect Dis. 2011;17:1876–1878. doi: 10.3201/eid1710.110233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Astell CR. A novel protein encoded by small RNAs of parvovirus B19. Virology. 1993;195:448–455. doi: 10.1006/viro.1993.1395. [DOI] [PubMed] [Google Scholar]
- Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol. 2011;85:8046–8055. doi: 10.1128/JVI.00831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Cater J, Pintel DJ. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J Virol. 1990;64:6166–6175. doi: 10.1128/jvi.64.12.6166-6175.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SJ, Reed A. A national survey for Aleutian disease prevalence in ranch mink herds in Canada. Scientiffur. 2006;30:33–40. [Google Scholar]
- Nüesch J. Regulation of non-structural protein functions by differential synthesis, modification and trafficking. In: Kerr JR, Cotmore SF, Bloom ME, Linden ME, Parrish CR, editors. Parvoviruses. Hodder Arnold; London, United Kingdom: 2006. pp. 275–286. [Google Scholar]
- Oleksiewicz MB, Costello F, Huhtanen M, Wolfinbarger JB, Alexandersen S, Bloom ME. Subcellular localization of Aleutian mink disease parvovirus proteins and DNA during permissive infection of Crandell feline kidney cells. J Virol. 1996;70:3242–3247. doi: 10.1128/jvi.70.5.3242-3247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol. 1987;61:2395–2406. doi: 10.1128/jvi.61.8.2395-2406.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintel D, Dadachanji D, Astell CR, Ward DC. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 1983;11:1019–1038. doi: 10.1093/nar/11.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintel DJ, Gersappe A, Haut D, Pearson J. Determinants that govern alternative splicing of parvovirus pre-mRNAs. Seminars in Virology. 1995;6:283–290. [Google Scholar]
- Porter DD, Larsen AE, Cox NA, Porter HG, Suffin SC. Isolation of Aleutian disease virus of mink in cell culture. Intervirology. 1977;8:129–144. doi: 10.1159/000148888. [DOI] [PubMed] [Google Scholar]
- Porter HG, Porter DD, Larsen AE. Aleutian disease in ferrets. Infect Immun. 1982;36:379–386. doi: 10.1128/iai.36.1.379-386.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Burger LR, Pintel D. The transcription profile of Aleutian Mink Disease Virus (AMDV) in CRFK cells is generated by alternative processing of pre-mRNAs produced from a single promoter. J Virol. 2006a;80:654–662. doi: 10.1128/JVI.80.2.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cheng F, Pintel D. The abundant R2 mRNA generated by aleutian mink disease parvovirus is tricistronic, encoding NS2, VP1, and VP2. J Virol. 2007;81:6993–7000. doi: 10.1128/JVI.00244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Yoto Y, Tullis GE, Pintel D. Parvovirus RNA processing strategies. In: Kerr JR, Cotmore SF, Bloom ME, Linden ME, Parish CR, editors. Parvoviruses. Hodder Arnold; London, UK: 2006b. pp. 253–274. [Google Scholar]
- Ruiz Z, D’Abramo A, Jr, Tattersall P. Differential roles for the C-terminal hexapeptide domains of NS2 splice variants during MVM infection of murine cells. Virology. 2006;349:382–395. doi: 10.1016/j.virol.2006.01.039. [DOI] [PubMed] [Google Scholar]
- St AJ, Astell CR. Identification and characterization of a family of 11-kDa proteins encoded by the human parvovirus B19. Virology. 1993;192:121–131. doi: 10.1006/viro.1993.1014. [DOI] [PubMed] [Google Scholar]
- Sun X, Sui H, Fisher JT, Yan Z, Liu X, Cho HJ, Joo NS, Zhang Y, Zhou W, Yi Y, Kinyon JM, Lei-Butters DC, Griffin MA, Naumann P, Luo M, Ascher J, Wang K, Frana T, Wine JJ, Meyerholz DK, Engelhardt JF. Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis. J Clin Invest. 2010;120:3149–3160. doi: 10.1172/JCI43052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol. 2009;83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijssen P, Agbandje-McKenna M, Almendral JM, Bergoin M, Flegel TW, Hedman K, Kleinschmidt J, Li Y, Pintel DJ, Tattersall P. Family Parvoviridae. In: King AM, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; 2012. pp. 405–425. [Google Scholar]
- Tullis GE, Burger LR, Pintel DJ. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J Virol. 1993;67:131–141. doi: 10.1128/jvi.67.1.131-141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PJ, Jensen KT, Burger LR, Pintel DJ, Lorson CL. Minute virus of mice small nonstructural protein NS2 interacts and colocalizes with the Smn protein. J Virol. 2002;76:6364–6369. doi: 10.1128/JVI.76.12.6364-6369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]