Abstract
Rationale:
MicroRNAs (miRs) are small, non-coding RNAs that function to post-transcriptionally regulate gene expression. First transcribed as long primary miR transcripts (pri-miRs), they are enzymatically processed in the nucleus by Drosha into hairpin intermediate miRs (pre-miRs) and further processed in the cytoplasm by Dicer into mature miRs where they regulate cellular processes following activation by a variety of signals such as those stimulated by β-adrenergic receptors (βARs). Initially discovered to desensitize βAR signaling, β-arrestins are now appreciated to transduce multiple effector pathways independent of G protein-mediated second messenger accumulation, a concept known as biased signaling. We previously showed that the β-arrestin-biased βAR agonist carvedilol activates cellular pathways in the heart.
Objective:
Here, we tested whether carvedilol could activate β-arrestin-mediated miR maturation, thereby providing a novel potential mechanism for its cardioprotective effects.
Methods and Results:
In human cells and mouse hearts, carvedilol upregulates a subset of mature and pre-miRs but not their pri-miRs in β1AR-, G protein-coupled receptor kinase 5/6- and β-arrestin1-dependent manner. Mechanistically, β-arrestin1 regulates miR processing by forming a nuclear complex with hnRNPA1 and Drosha on pri-miRs.
Conclusions:
Our findings indicate a novel function for β1AR-mediated β-arrestin1 signaling activated by carvedilol in miR biogenesis, which may be linked, in part, to its mechanism for cell survival.
Keywords: β-arrestin-biased β-adrenergic receptor signaling, carvedilol, heart disease, microRNA biogenesis
Introduction
MicroRNAs (miRNAs or miRs), a class of ~22 nucleotide non-coding RNAs, govern post-transcriptional repression of target mRNAs. Various roles of miRs in normal cardiac physiology have been reported including the control of myocyte growth, contractility, and the maintenance of cardiac rhythm 1. Furthermore, gain- and loss-of-function studies of a selective group of miRs suggested that aberrant expression of the miRs could be necessary and sometimes even sufficient for the pathogenesis of various heart diseases 2, 3, pointing towards miRs as new regulatory mechanisms and potential therapeutic targets for heart disease to complement pharmacological approaches 1.
MiR biogenesis is regulated in a complex manner, involving numerous protein-protein and protein-RNA interactions 4. Both miR-regulator and miR-target availability often differ among cell types, tissues and especially during disease initiation and progression, responding to different upstream signaling pathways to activate distinct downstream targets. It is understood that miRs are influenced at the transcriptional level but are also regulated during further downstream steps in which two RNase III enzymes, Drosha and Dicer, play dominant roles in the control of miR maturation. Several post-transcriptional regulatory mechanisms of miR maturation have been identified. For example, several proteins including Smads and E2-ERα modulate miR processing in a RNA helicase-dependent or -independent manner 5-7. Interestingly, a proteomic analysis assessing the global cellular interactions of the G protein-coupled receptor (GPCR) signaling mediators, β-arrestin1 and β-arrestin2, identified that β-arrestins may play regulatory roles in miR processing 8.
β-arrestin1 and β-arrestin2 were initially discovered to desensitize GPCR signaling in response to agonist stimulation. However, it is now appreciated that β-arrestins can also transduce multiple effector pathways independent of G protein signaling when receptors are stimulated by certain ligands, a concept known as biased signaling 9-13. The proposed mechanism for this signaling bias is based on the bar-code hypothesis where unbiased and β-arrestin-biased ligands impart distinct patterns of receptor phosphorylation by specific GPCR kinases (GRKs), thus converting ligand-induced conformation of the receptor into selective β-arrestin functions 14-16. For example, ligands that promote GRK2/3-mediated receptor phosphorylation lead to desensitization and internalization whereas ligands, such as the β-adrenergic receptor (βAR) antagonist (i.e. β-blocker) carvedilol (Carv), that promote GRK5/6-mediated receptor phosphorylation stimulate β-arrestin signaling 14-16. Indeed, Carv is one of three β-blockers approved for heart failure and has many documented actions including antagonism of β1AR, β2AR and α1AR as well as antioxidant effects 17, 18. We previously showed that Carv stimulates β-arrestin-mediated β1AR cardioprotective signaling without activating G proteins, providing an additional mechanism for its clinical efficacy 9. However, our understanding of whether β-arrestin-biased signaling regulates nuclear processes remains limited.
We postulated that miR could in part explain how GPCR-mediated β-arrestin signaling pathways confer physiological outcomes such as anti-apoptosis. Although β-arrestins are known to be involved in multiple cytoplasmic signaling networks 19, 20, it is increasingly appreciated that β-arrestins also play important roles in the nucleus 21, 22. Of the two non-visual and ubiquitous arrestins, β-arrestin1 is thought to be the major isoform involved in nuclear signaling since, unlike β-arrestin2, it lacks a nuclear export signal 23. Here, we investigate whether stimulation of βARs by the β-arrestin-biased agonist Carv, can regulate miR expression in both cultured cells and the heart. Out of 9 human and 1,040 mouse miRs examined, we found that human miR-190 and five human/mouse miRs (125a-5p, 125b-5p, 150, 199a-3p and 214) were upregulated by Carv stimulation and that this effect was absent in cells or mice lacking either β1AR, GRK5/6 or β-arrestin1. While Carv did not increase the expression of pri-miRs, it enhanced expression of pre-miRs by promoting the interaction of β-arrestin1 with components of the nuclear Drosha microprocessor complex. Our data provide evidence that the biased β-blocker Carv stimulates β-arrestin1-mediated miR processing which may be an important mechanism for its cardioprotective effects.
Methods
Details of cell culture, siRNA experiments, immunoprecipitation, immunoblotting, immunofluorescence staining, quantitative real-time RT-PCR, Northern blot, RNA-CHIP, treatment protocol for mice, βAR radioligand binding, microRNA microarray analysis, luciferase-based microRNA processing assay, and statistical analysis are provided in online supplement.
Results
A β-arrestin-biased βAR ligand, carvedilol induces the expression of human miR-190 in HEK293 cells
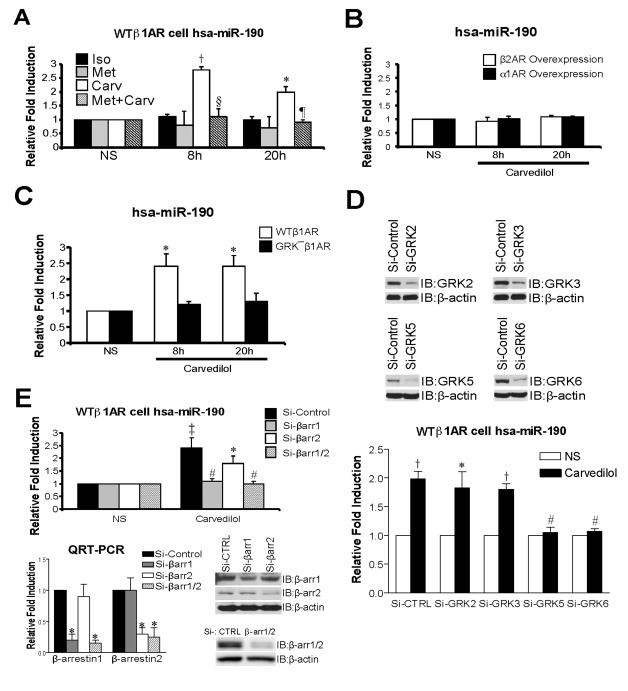
To test whether the β-arrestin-biased β-blocker carvedilol (Carv) can regulate miR expression, we used HEK293 cells stably expressing the wild-type β1AR (WTβ1AR cells). WTβ1AR cells were treated with 1μM of the βAR agonist isoproterenol (Iso, unbiased agonist), the β1AR antagonist metoprolol (Met, neutral unbiased β-blocker), or Carv (β-arrestin-biased β-blocker). We assessed the expression of 4 miRs (miR-1, -21, -190 and -221) based on their known association with GPCR signaling pathways 24-26. Among the 4 human miRs examined, only hsa (homo sapiens, human)-miR-190 was activated at 8h and 20h after Carv stimulation (Figure 1A and Online Figure I). In our Carv time-course experiments, 8h and 20h were the time points showing significant activation of miR-190 expression (Online Figure II). The increase in miR-190 was not seen with either Iso or Met stimulation, but Met pretreatment was able to block the increase with Carv (Figure 1A). In addition, Carv did not upregulate miR-190 expression in HEK293 cells overexpressing either β2AR or α1AR (Figure 1B). Lastly, treatment with 10μM of antioxidants (α-tocopherol and ebselen) failed to affect miR-190 expression (Online Figure III). Collectively, these results indicate a β1AR-mediated mechanism of Carv action.
Figure 1. Carvedilol stimulation induces upregulation of human miR-190, which is dependent on β1AR, GRK5/6 and β-arrestin1.
A, HEK293 cells stably expressing WTβ1ARs (WTβ1AR cells) were treated with 1μM of isoproterenol (Iso), metoprolol (Met) or carvedilol (Carv) for 8h or 20h. Expression of mature human (hsa: homo sapiens)-miR-190 was detected using the TaqMan miR assay. Among the 3 βAR ligands tested, only the β-arrestin-biased ligand Carv activated expression of hsa-miR-190. The pretreatment of 10μM Met for 4h blocked Carv-mediated hsa-miR-190 activation. B, HEK293 cells overexpressing β2ARs or α1ARs were treated with 1μM Carv and the expression of hsa-miR-190 was measured as described above. Carv did not induce miR-190 expression in β2AR- or α1AR-overexpressing cells. C, HEK293 cells stably expressing WTβ1AR or GRK−β1AR were treated with 1μM Carv. WTβ1AR induced an increase in hsa-miR-190 expression following Carv treatment, while GRK−β1AR lacked this effect. D, WTβ1AR cells were transfected with either scrambled siRNA (Si-Control or Si-CTRL) or siRNAs targeting GRKs. Hsa-miR-190 activation was abolished in cells transfected with siRNAs targeting GRK5 or GRK6. E, WTβ1AR cells were transfected with either Si-Control or siRNAs targeting β-arrestin1/2 (Si-βarr1/2), β-arrestin1 (Si-βarr1) or β-arrestin2 (Si-βarr2). Carv-mediated hsa-miR-190 activation was diminished in cells transfected with Si-β-arrestin1 or Si-β-arrestin1/2. Knockdown of β-arrestins was confirmed by both QRT-PCR and IB. NS: no stimulation with Carv (vehicle, 0.1% [v/v] DMSO). IB: immunoblotting. Data are shown as mean ± SEM for n=4 independently obtained biological samples (A, C and D) and n=5 independently obtained biological samples (B and E). *, P < 0.05 vs. NS, Iso, Met, GRK− β1AR or Si-Control; †, P < 0.01 vs. NS, Iso or Met; ‡, P < 0.001 vs. NS; #, P < 0.05 vs. Si-Control; §, P < 0.01 vs. Carv; ¶, P < 0.001 vs. Carv.
Since Iso did not stimulate miR-190 expression (Figure 1A), we tested the effect of treatment with the activator of adenylyl cyclase, forskolin (Forsk). Interestingly, the upregulation of miR-190 by Carv was blocked by pretreatment with 10μM forskolin (Online Figure IV), suggesting inhibition of miR-190 expression by Gαs protein-mediated signaling. Since Carv is dissolved in DMSO, we treated WTβ1AR cells with DMSO (0.1% [v/v]) alone for 8h or 20h and found no activation of miR-190 expression (Online Figure V).
Increase in miR-190 levels elicited by β-arrestin-biased β1AR stimulation requires GRK5/6 phosphorylation and β-arrestin1
We next tested whether β-arrestin signaling is required for Carv-induced miR-190 expression. Since GRK-mediated phosphorylation of the receptor promotes the recruitment of β-arrestins to the ligand-activated receptor 9, 14, 27, we examined the involvement of GRK phosphorylation in Carv-mediated miR-190 activation. Cells stably expressing WTβ1ARs or mutant GRK− β1ARs (that lack GRK phosphorylation sites on the β1AR c-terminal tail) were treated with 1μM Carv. WTβ1AR cells showed an increase in miR-190 expression upon Carv stimulation but GRK− β1AR cells lacked this effect (Figure 1C). To test which of the GRKs are involved in miR-190 upregulation, we performed knockdown experiments using siRNAs targeting the individual GRKs. WTβ1AR cells transfected with either scrambled siRNA (Si-Control) or siRNAs individually targeting GRK2, GRK3, GRK5 or GRK6 were stimulated with 1μM Carv. Carv-mediated miR-190 upregulation was abrogated in cells transfected with siRNAs targeting GRK5 or GRK6, but not GRK2 or GRK3 (Figure 1D), consistent with the hypothesis that phosphorylation-specific GRK sites on the c-terminal tail of the β1AR are required to promote Carv-mediated signaling 9, 15, 16. To test the role of β-arrestins in this process, we treated WTβ1AR cells with Carv in the presence of siRNAs targeting β-arrestin1 (Si-βarr1), β-arrestin2 (Si-βarr2) or β-arrestin1/2 (Si-βarr1/2). Knockdown of β-arrestin1 abrogated the increase in miR-190 expression (Figure 1E), indicating its central role in this β1AR-mediated process.
Induction of miR-190 by β-arrestin1-biased β1AR agonism occurs at a post-transcriptional step
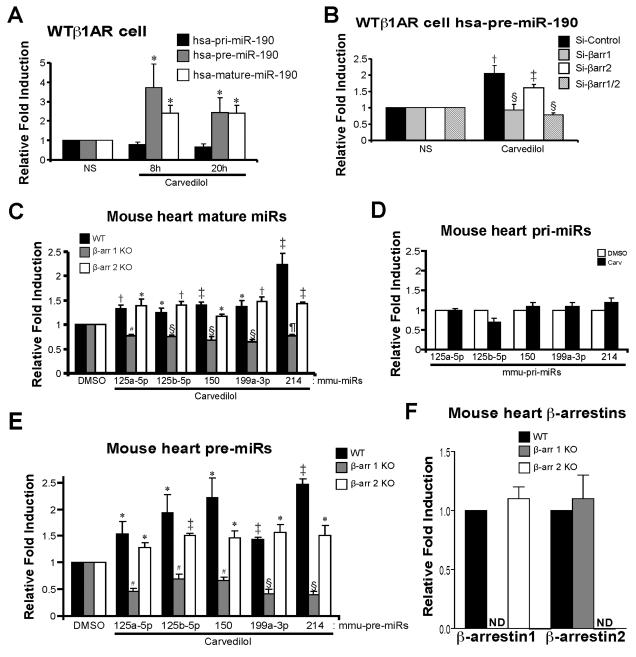
To examine which step of miR biogenesis is regulated by β-arrestin1, we measured the expression of primary transcript (pri)-, premature (pre)- and mature-miR-190. WTβ1AR cells were treated with 1μM Carv for 8h or 20h and the expression of pri-, pre- or mature-miR-190 was detected using QRT-PCR and Northern blot analysis. While Carv increased the expression of pre- or mature-miR-190, it did not increase the expression of pri-miR-190 at any of the time-points examined (Figure 2A and Online Figure VI), suggesting that β-arrestin1 is involved in miR-190 processing. Supporting this idea, we observed that Carv-mediated upregulation of pre-miR-190 was prevented by treatment with siRNAs directed against β-arrestin1 or β-arrestin1/2 but not with siRNAs directed against β-arrestin2 (Figure 2B). Altogether, we demonstrate that Carv stimulation requires β-arrestin1 to activate human miR-190 processing.
Figure 2. β-arrestin1 is required for Carv-mediated miR activation, which occurs at a post-transcriptional step.
A, WTβ1AR cells were treated with 1μM Carv for 8h or 20h. Expression of primary (pri), premature (pre), or mature hsa-miR-190 was detected using TaqMan miR assay for mature and pri-miRs and using Power SYBR Green PCR assay with pre-miR primers. Carv stimulation did not activate hsa-pri-miR-190 expression but resulted in mature or pre-hsa-miR-190 activation. B, WTβ1AR cells were transfected with siRNAs as described in Figure 1E. Carv-mediated hsa-pre-miR-190 activation was diminished in cells transfected with siRNAs targeting β-arrestin1/2 or β-arrestin1. C and E, WT, β-arrestin1 knockout (KO) or β-arrestin2 KO mice were infused with DMSO (vehicle control) or Carv (19mg/Kg/day) for 7 days by using micro-osmotic pumps. QRT-PCR experiments were performed on RNAs from mouse hearts. Five mature (C) or pre- (E) miRs were elevated upon Carv stimulation in both WT and β-arrestin2 KO mice. However, this induction was completely abolished in β-arrestin1 KO mice, indicating an essential role of β-arrestin1 in the synthesis of pre-miRs. D, WT mice were infused with DMSO or Carv as above. QRT-PCR experiments were performed in mouse hearts using Taqman pri-miR assays. Expression of pri-miRs was not changed significantly upon Carv stimulation. F, QRT-PCR analysis was performed in mouse hearts using Taqman gene expression assays for β-arrestins. NS: no stimulation. ND: not detected. Data are shown as mean ± SEM for n=7 independently obtained biological samples (A), n=5 independently obtained biological samples (B) and n=8 independent mice per group (C, D, E and F). *, P < 0.05 vs. NS, hsa-pri-miR-190 or DMSO; †, P < 0.01 vs. NS or DMSO; ‡, P < 0.001 vs. NS or DMSO; #, P < 0.05 vs. WT or β-arrestin2 KO; §, P < 0.01 vs. Si-Control, WT or β-arrestin2 KO; ¶, P < 0.01 vs. WT or β-arrestin2 KO. Notably, the levels of mature miR-214 (C) and pre-miR-214 (E) are reduced in β-arrestin2 KO compared to WT (P < 0.001) and the level of pre-miR-150 (E) is reduced β-arrestin2 KO compared to WT (P < 0.05).
Carvedilol-mediated β-arrestin-biased agonism of βAR induces unique miR signatures in mouse hearts
Based on our cell data, we hypothesized that in the mammalian heart β-arrestin1 may promote the processing of a specific subset of pri-miRs into pre-miRs by the nuclear Drosha microprocessor complex. We tested this hypothesis by performing miR microarray profiling in mouse hearts to identify miR signatures regulated by stimulation with Carv. We used 8 to 12-week-old WT mice and infused them with DMSO (vehicle control) or Carv (19mg/kg/day) for 7 days based on our time-course experiments with 2 cardiac-enriched miRs (data not shown). Among 1,040 mmu (mus musculus, mouse)-miRs that we profiled, 21 miRs were upregulated and 13 miRs were downregulated upon stimulation with the β-arrestin-biased ligand Carv (Online Figure VII and Online Table I). Interestingly, we found that the expression level of miR-190, which was regulated by Carv in HEK293 cells, was not detectable in the mouse heart and only approximately 10% of profiled miRs were detectable in the hearts, indicating tissue- or cell type-specific miR expression patterns.
We next sought to validate the 11 miRs with a minimum intensity of 500 (Online Table I, top panel). Using Taqman miR QRT-PCR analysis, we found that only 5 miRs were verified to be upregulated by Carv (Online Table II, shown in red color). Time-course experiments from 1 to 7 days of Carv treatment showed that the relative expression levels of the 5 regulated miRs were highest at 7 days (Online Figure VIII). Importantly, Iso or Met did not significantly activate the expression of these 5 miRs (Online Figure IX) in agreement with our HEK293 cell data (Figure 1A). In summary, we found that the expression of 5 mouse miRs (125a-5p, 125b-5p, 150, 199a-3p, and 214) and human miR-190 is upregulated upon stimulation with the β-arrestin-biased βAR ligand Carv.
β-arrestin1 and GRK5/6 phosphorylation of β1AR post-transcriptionally induce in vivo miR expression by promoting miR processing
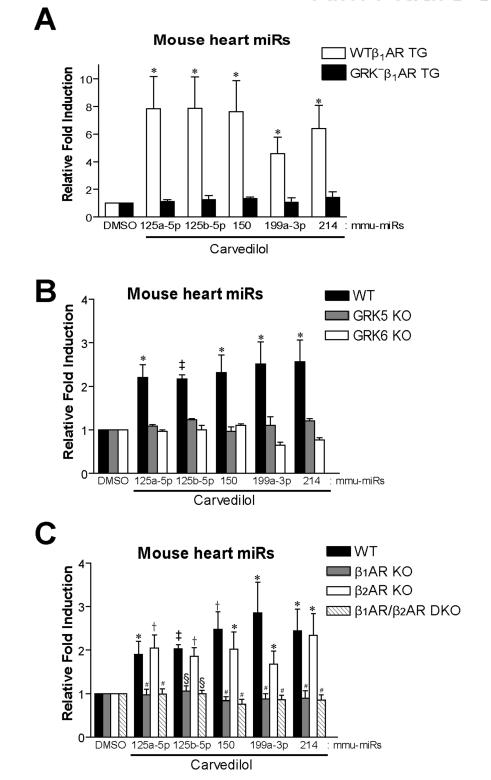
We next tested whether Carv-mediated induction of the five mouse miRs occurs post-transcriptionally and whether it requires β-arrestins, GRKs and two βAR subtypes. We measured the expression level of the 5 verified miRs using QRT-PCR and Northern blot analysis in hearts from WT, β-arrestin1 knockout (KO) and β-arrestin2 KO mice infused with DMSO or Carv. The Carv-mediated activation of five miRs occurred in both WT (Figure 2C and Online Figure X) and β-arrestin2 KO mice (Figure 2C and F), but was not observed in hearts from β-arrestin1 KO mice (Figure 2C and F). Carv did not increase the expression of pri-miRs (Figure 2D and Online Figure X) although levels of pre-miRs were increased upon Carv stimulation in WT (Figure 2E and Online Figure X) and β-arrestin2 KO mice (Figure 2E-F), and these increases were blunted in β-arrestin1 KO mice (Figure 2E-F). While Carv stimulation of transgenic (TG) mice overexpressing WTβ1ARs induced an increase in expression of pre- and mature miRs, hearts overexpressing a receptor that lacks GRK phosphorylation sites (GRK−β1AR TG) or hearts lacking either GRK5, GRK6 or β1AR, showed no induction of these 5 miRs (Figure 3A-C and Online Figure XI). These in vivo data are consistent with the cellular data and support the concept that Carv stimulates β1AR-mediated miR biogenesis in β-arrestin1- and GRK5/6-dependent manner.
Figure 3. Carv-mediated in vivo miR activation requires GRK5/6 phosphorylation of β1AR.
A, Cardiac specific transgenic (TG) mice expressing WTβ1AR or GRK− β1AR were treated with Carv as shown in Figure 2. WTβ1AR induced an increase in mature miR expression following Carv treatment, while GRK− β1AR lacked this effect. B-C, WT and various KO mice were treated with Carv as aforementioned. Carv-mediated miR activation, which is seen in WT and β2AR KO mice, was abolished in GRK5 KO, GRK6 KO, β1AR KO and β1AR/β2AR double KO (DKO) mice. Data are shown as mean ± SEM for n=6 independent mice per group. *, P < 0.05 vs. DMSO or GRK− β1AR TG; †, P < 0.01 vs. DMSO ; ‡, P < 0.001 vs. DMSO; #, P < 0.05 vs. WT or β2AR KO; §, P < 0.01 vs. WT or β2AR KO.
To test whether the upregulation of the 5 miRs found in the in vivo experiments also occurs in Carv-stimulated WTβ1AR cells, we performed QRT-PCR and Northern blot analysis after 20hr treatment and showed the induction of 5 miRs (Online Figure XII), suggesting that the newly identified miR regulatory mechanism exists in both HEK293 cells and mouse hearts.
We next investigated whether the β1AR-mediated mechanism of miR regulation is confined to Carv. We measured the expression level of six identified pri-, pre- and mature miRs in the hearts from WT mice and WTβ1AR cells treated with the βAR antagonist Alprenolol (Alp), which has also been shown to be a weak β-arrestin-biased ligand of β1AR 28. Similar to Carv, Alp increased the levels of pre- and mature miRs without affecting the expression of pri-miRs in both Alp-treated mouse hearts and WTβ1AR cells (Online Figure XIII). Taken together, these data indicate that β-arrestin1-biased signaling of β1AR stimulates the processing of a subset of miRs.
β-arrestin1 interacts with the nuclear Drosha microprocessor complex in a Carv-dependent manner
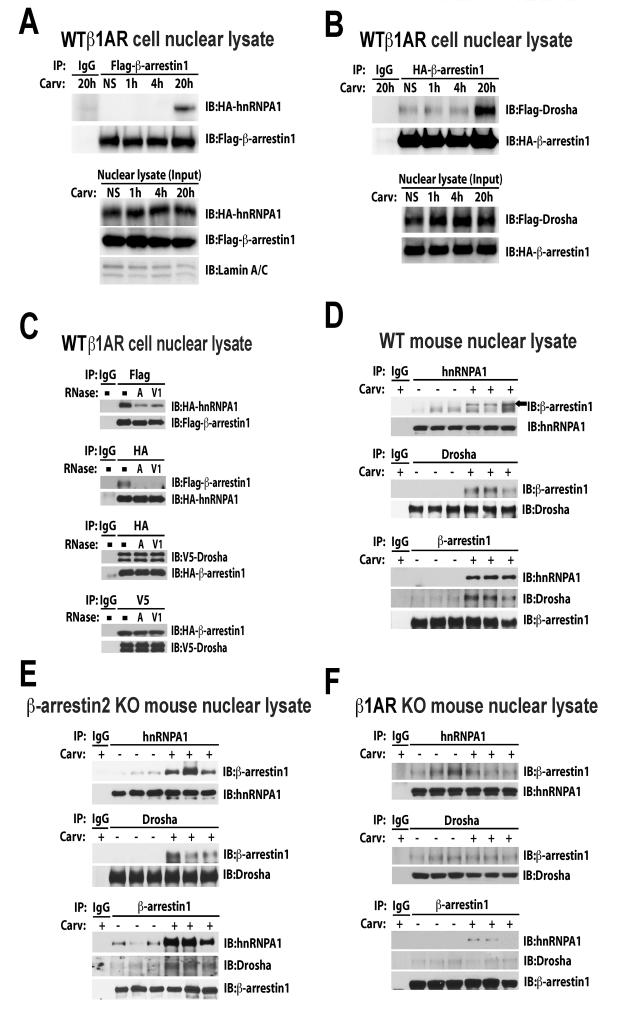
Based on the nuclear localization of β-arrestin1 23 and its potential interaction with two components of the nuclear Drosha microprocessor complex (DDX5 or hnRNPA1) 8, we tested whether β-arrestin1 may regulate miR processing in the nucleus by interacting with the Drosha microprocessor complex. We performed co-immunoprecipitation experiments in the nuclear lysates of both WTβ1AR cells transiently overexpressing tagged-plasmids and mouse hearts without and with treatment of Carv. We observed that Carv induced a time-dependent association of β-arrestin1 with both hnRNPA1 (a RNA binding protein involved in RNA helicase-independent miR processing 29) and Drosha in the nuclear lysates of WTβ1AR cells overexpressing β-arrestin1 but not β-arrestin2 (Figure 4 A-B and Online Figure XIVA-D). We also demonstrate that β-arrestin1 co-localizes with endogenous Drosha and hnRNPA1 in the nucleus, by performing immunofluorescence staining on WTβ1AR cells that contain overexpressed GFP-β-arrestin1 following stimulation with Carv (Online Figure XV).
Figure 4. Carv induces the RNA-dependent nuclear interaction of β-arrestin1 with hnRNPA1, an important regulator of RNA helicase-independent miR processing by Drosha.
A-B, WTβ1AR cells were transfected with Flag-β-arrestin1 and HA-hnRNPA1 (A) or HA-β-arrestin1 and Flag-Drosha constructs (B). After Carv treatment, nuclear extracts (NEs) were prepared and subjected to immunoprecipitation (IP) with anti-Flag, anti-HA, or non-specific IgG (control). NEs were immunobloted with lamin A/C antibody for nuclear marker. Interaction of hnRNPA1 or Drosha with β-arrestin1 was examined by immunoblotting (IB) with anti-HA and anti-Flag. C, RNA dependence of interaction of β-arrestin1 with hnRNPA1 and Drosha. WTβ1AR cells transfected with tagged plasmids were serum-starved for 4h and stimulated with Carv for 20h. NEs were treated with RNase A (single-stranded RNA nuclease) or RNase V1 (double-stranded RNA nuclease) prior to IP. Immunoprecipitates were subjected to IB. D-F, WT (D), β-arrestin2 KO (E) and β1AR KO (F) mice were infused with Carv or vehicle control and then NEs were prepared from three independent mice per group. Endogenous interaction was confirmed using indicated antibodies. NS: no stimulation.
The Carv-mediated association of β-arrestin1 with hnRNPA1 was markedly decreased by treatment with RNase A [single-stranded RNA nuclease] and RNase V1 [double-stranded RNA nuclease] whereas the interaction of β-arrestin1 with Drosha was not affected by these RNases, indicating that β-arrestin1 interacts with hnRNPA1 via RNA molecules (Figure 4C). Importantly, we demonstrate that Carv stimulated the nuclear interaction of β-arrestin1 with hnRNPA1 and Drosha in endogenous systems using WT, β-arrestin2 KO and β1AR TG mouse hearts (Figure 4 D-E and Online Figure XIVE), but was lost in Carv-treated β1AR KO hearts (Figure 4F). No interaction with β-arrestin1 was found with the two RNA helicases: DDX5 and DDX17. Taken together, our data suggest that Carv stimulation of the β1AR promotes β-arrestin1 translocation to the nucleus where it interacts with hnRNPA1 and Drosha of the microprocessor complex to process a subset of pri-miRs.
Carvedilol induces the association of β-arrestin1 with primary transcripts of β-arrestin1-regulated miRs (β1-miRs)
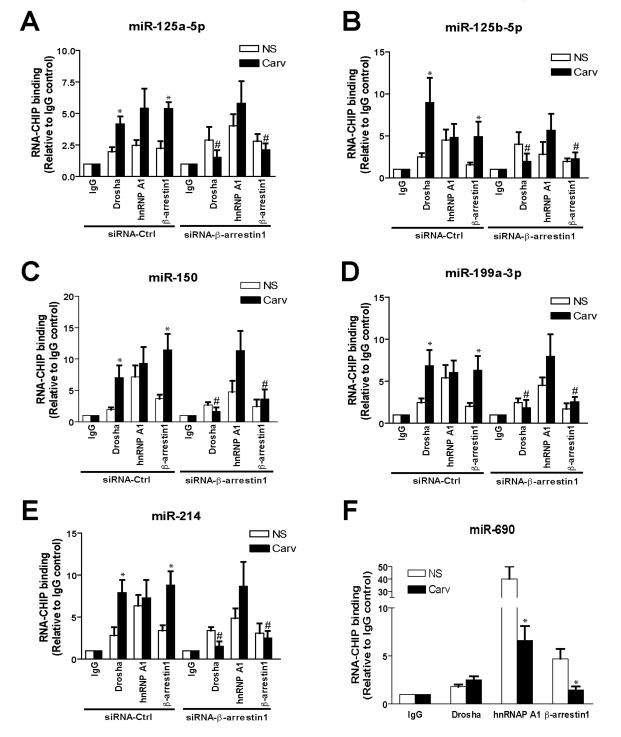
To test whether the β-arrestin1-hnRNPA1-Drosha complex assembles specifically on pri-β1-miRs after Carv stimulation, we performed RNA-chromatin immunoprecipitation (ChIP) analysis on WTβ1AR cells co-transfected with pCMV-β1-miRs and tagged β-arrestin1, hnRNPA1, or Drosha along with siRNAs targeting β-arrestin1. The association of β-arrestin1 or Drosha with pri-β1-miRs was induced on Carv stimulation for 20h, whereas a RNA binding protein, hnRNPA1 constitutively associated with pri-β1-miRs. Knockdown of β-arrestin1 abrogated the Carv-mediated increase in association of the β-arrestin1-Drosha complex with pri-β1-miRs (Figure 1E and 5A-E). We detected a constitutive association of pri-miR-690 with β-arrestin1-hnRNPA1-Drosha complex while the nuclear interaction was not induced by Carv (Figure 5F), confirming that miR-690 is not regulated by Carv stimulation (Online Supplementary Table 2). Thus, formation of β-arrestin1-hnRNPA1-Drosha complex is pri-miR-specific.
Figure 5. β-arrestin1 associates with pri-β1-miRs in a Carv-dependent manner.
WTβ1AR cells were transfected with pCMV-β1-miRs (A-E) or pCMV-miR-690 [control miR] (F), tagged plasmids (HA-β-arrestin1, Flag-hnRNPA1 or V5-Drosha) and control siRNA (siRNA-Ctrl) or siRNA-β-arrestin1 (A-E, as described in Figure 1E), followed by Carv treatment (20h). RNA-ChIP was performed with HA, V5, hnRNPA1 antibody or non-specific IgG (control), followed by PCR amplification with β1-miR primers (A-E) or miR-690 primers (F). Data are shown as mean ± SEM for n=4 independently obtained biological samples. *, P < 0.05 vs. NS; #, P < 0.05 vs. Carv of siRNA-Ctrl.
Carvedilol induces Drosha-mediated microRNA processing by β-arrestin1
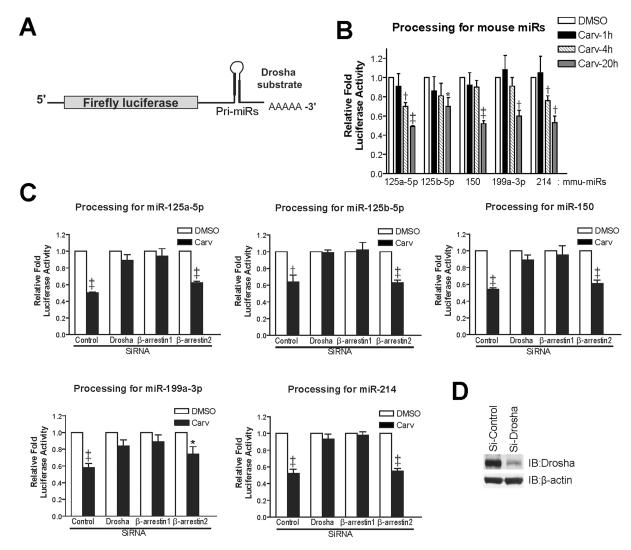
To directly demonstrate a role of β-arrestin1 in pri-miR processing by the Drosha microprocessor complex, we performed pri-miR processing assays in WTβ1AR cells as described previously 30. We fused luciferase (LUC) reporters to pri-miRs that are regulated by β-arrestin1 [β-arrestin1-regulated miRs or β1-miRs] (Figure 6A) and monitored the loss of LUC activity as a measure of Drosha-dependent processing of pri-miRs into pre-miRs 30. Carv treatment resulted in a time-dependent 30-50% fall in LUC activity due to cleavage of the pri-miRs (Figure 6B), which was prevented in the presence of siRNA targeting either β-arrestin1 or Drosha, but not β-arrestin2 (Figure 1E and 6C-D). Similar results were obtained for Carv- induced processing of the human pri-miR-190 by β-arrestin1 (Online Figure XVI). Taken together, these data indicate that β-arrestin1 can regulate the post-transcriptional processing of miRs through its nuclear interaction with pri-miRs and the Drosha microprocessor complex by stimulating the β1AR with the biased-ligand Carv.
Figure 6. Carv facilitates Drosha-mediated miR processing by β-arrestin1.
A, Pri-miRs of five mouse β-arrestin1-regulated miRs were cloned into 3’UTR of luciferase construct. B-D, In vivo pri-miR processing assay measures pri-miR cleavages by Drosha. WTβ1AR cells were transfected with mock (B), control siRNA or siRNAs directed against Drosha, β-arrestin1 or β-arrestin2 (C-D). At the same time, cells were transfected with CMV-LUC empty or CMV-LUC-pri-miR constructs together with pRL-CMV for transfection efficiency control. After 48h, cells were serum-starved for 4h and stimulated with Carv for either 1-20h (B) or 20h (C). Firefly LUC activity was normalized to Renilla LUC activity using dual LUC assays. The relative fold induction of LUC activity was calculated by normalizing to the CMV-LUC empty plasmid control. Efficiency of Drosha, β-arrestin1 or β-arrestin2 interference was confirmed by IB (D and Figure 1E). Data are shown as mean ± SEM for four independent experiments. *, P < 0.05 vs. DMSO; †, P < 0.01 vs. DMSO; ‡, P < 0.001 vs. DMSO.
Discussion
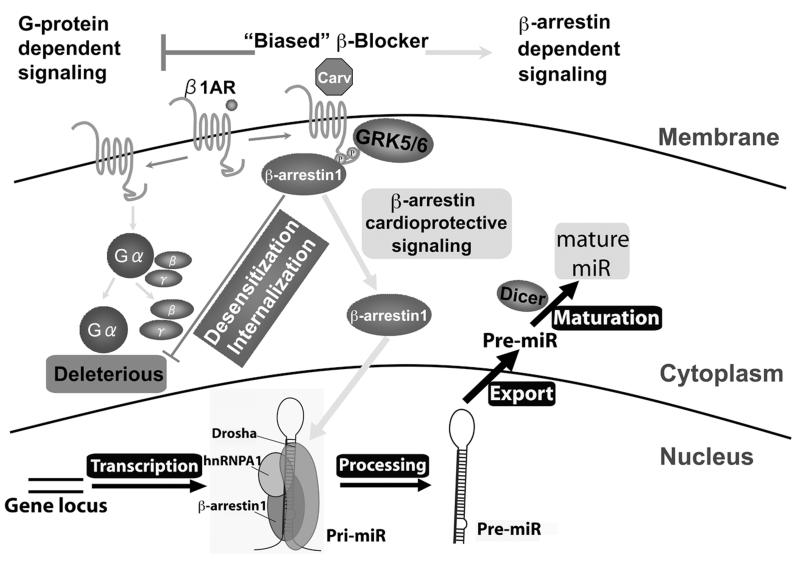
In this study, we show an essential role of β-arrestin1 in miR processing following stimulation by the β-arrestin-biased βAR agonist carvedilol. We demonstrate that this process results from stimulation of the β1AR and requires β-arrestin1 to promote the processing of a subset of miRs in murine hearts and human cells. The molecular mechanism for this β-arrestin1-mediated miR processing function involves the formation of a nuclear complex of hnRNPA1 and Drosha with β-arrestin1 to activate RNA helicase-independent miR processing (Figure 7). Our working hypothesis for the mechanism by which β-arrestin1 enhances miR processing is that GRK5/6 phosphorylation of the β1AR mediates the recruitment of β-arrestin1 to the ligand-occupied receptor, resulting in translocation of β-arrestin1 to the nucleus where it interacts with a subset of pri-miRs and components of the Drosha microprocessor complex. Supporting this hypothesis is the miR processing data showing that knockdown of β-arrestin1 or Drosha prevented pri-miR processing of the 6 identified β-arrestin1-regulated miRs (β1-miRs) (Figure 6C and Online Figure XVI) and co-immunoprecipitation data showing that the interaction of β-arrestin1 with hnRNPA1 is sensitive to RNase treatment (Figure 4C).
Figure 7. β-arrestin1 stimulates the processing of a subset of miRs in the mouse heart and human cells.
The β-arrestin-biased β-blocker Carv, which was shown to stimulate β-arrestin-mediated cardioprotective signaling in the absence of G protein activation 9, induces the expression of a selective group of miRs in a β1AR-, GRK5/6- or β-arrestin1-dependent manner (Figures 1-3). Our data produced using both HEK293 cells stably expressing WTβ1AR and mouse hearts suggest that β-arrestin1 promotes RNA helicase-independent processing of primary miR transcript (pri-miR) into precursor miR (pre-miR) by forming a complex with components (eg. hnRNPA1 or Drosha) of the nuclear miR microprocessor complex (Figures 4-6).
Sequence motif detection analysis using 6 identified miRs suggests that β1-miRs may have potentially conserved sequence motifs in their stem regions although additional profiling analyses in different tissues or cells will be required to definitely identify a β-arrestin1 sequence motif. This sequence analysis together with the RNA-CHIP data (Figure 5) suggest that β-arrestin1 promotes miR processing by translocating to the nucleus and directly associating with pri-miRs. While we believe this to be the most plausible mechanism, it is possible that activation of β1AR signaling pathways downstream of β-arrestin1 (eg. EGFR, ERK or AKT) could regulate miR processing and thus indirectly exert regulatory effects on the activation of miR processing 31. It is also possible that the regulation of β-arrestin1 in the Drosha step is indirect through interaction with hnRNPA1 or other RNA-binding proteins although our sequence analysis showed that β1-miRs have no consensus sequences for direct hnRNPA1 binding. Additional studies will be needed to further clarify the mechanism of β-arrestin1 in miR processing.
β-arrestins not only desensitize G protein signaling but also activate a number of signaling networks by scaffolding a diverse group of signaling proteins at the GPCRs 32, 33. The important roles of β-arrestins in regulating cytoplasmic signaling networks are now well recognized 19, 20. However, the role of β-arrestins in modulating nuclear function is less well studied 8, 21, 22. Our data identifies a new nuclear function of β-arrestin1, the isoform of β-arrestins known to translocate to the nucleus 23, as a regulator of miR processing in β1AR. It is interesting that only β-arrestin1 regulates miR processing while both β-arrestins are involved in β1AR-mediated cardioprotective signaling 9, 27. This likely reflects the fact that β-arrestin1 lacks a nuclear export sequence allowing for its retention in the nucleus after activation and translocation 23. However, we can not rule out the possibility that β-arrestin2 regulates other miR biogenesis steps (eg. miR degradation, nuclear export and dicing) rather than Drosha processing because we observed that the levels of pre-miR-150, pre-miR-214 and mature miR-214 are reduced in β-arrestin2 KO mouse hearts compared to WT (Figure 2C and E).
β-arrestin1 functioning in the nucleus has recently been reported. β-arrestin1 is a nuclear transcriptional regulator of endothelin type A receptor-mediated β-catenin signaling 34, and shown to be important for tumor progression and stem cell regulation. β-arrestin1 also functions as an important regulator of polycomb group proteins (transcriptional repressors), suggesting its involvement in epigenetic regulation 22 as well as in the regulation of histone acetylation in human neuroblastoma cells following δ-opioid receptor stimulation 21.
We previously showed that β1AR uses GRK5/6 and β-arrestins to promote cardiomyocyte survival pathways against chronic catecholamine stimulation in the absence of G protein activation 27. Our subsequent study suggested that Carv functions as a β-arrestin-biased ligand to promote cardioprotective signaling 9, providing a possible mechanism for its clinical efficacy. Interestingly, a recent meta-analysis showed that Carv did not reduce patient readmissions compared with other β-blockers although it led to less sudden cardiac death and all-cause mortality in patients with acute myocardial infarction and those with heart failure 35. Therefore, identifying additional beneficial downstream signaling pathways activated by Carv should lead to a better understanding of how biased ligands exert their cardioprotective effects.
Consistent with our previous findings, we show that the unbiased agonist Iso and unbiased antagonist Met did not activate the expression of β1-miRs (Figure 1A and Online Figure IX) and that Forsk, which activates Gαs protein signaling, blocked Carv-mediated miR-190 activation (Online Figure IV). These data, in addition to data in Figure 1D and 3B, are consistent with the receptor phosphorylation bar-code hypothesis where distinct phosphorylation patterns of the c-terminal tail of the receptor encodes for different function of β-arrestin 14. In particular, it was shown for the β2AR that Carv induced a phosphorylation pattern distinct from that of Iso. Notably, Carv only induced an increase in phosphorylation on two GRK5/6 sites while Iso triggered a change in the phosphorylation status of 13 sites including PKA, GRK2 and GRK5/6 sites 14. This selective phosphorylation profile of Carv is consistent with the β-arrestin-biased signaling induced by this ligand. Surprisingly, recent studies demonstrated that GRK2, which requires G proteins for its activation, exerts a strong negative effect on β-arrestin-dependent signaling through its competition with GRK5 and GRK6 for receptor phosphorylation 14, 16, which in turn mediates the balance between G protein and β-arrestin signaling. These previous studies are again in agreement with our data showing that Iso did not activate the expression of β1-miRs and that Met or Forsk blocked Carv-mediated activation of β1-miRs.
Carvedilol is a nonselective βAR antagonist with high affinity for both β1ARs and β2ARs. However, we show that only β1AR stimulation by Carv mediates miR processing. A possible mechanism for the βAR subtype specificity is the requirement of a unique β-arrestin conformation for activating miR processing. This idea is supported by our recent study showing that recruitment of the β-arrestin to β1AR, but not β2AR induces a β-arrestin conformation that promotes a stable complex between the β1AR, β-arrestin and CaMKII to activate signaling 36. Moreover, recent work has shown that catecholamine stimulation of β2ARs promotes DNA damage in a β-arrestin1-dependent manner 37. Taken together, our findings suggest that carvedilol-stimulated β1ARs require GRK5/6 to promote β-arrestin1-mediated miR processing in the nucleus and ultimately cardioprotective signaling.
Additional discussion of the therapeutic potential and the in vivo relevance of targeting the β-arrestin1-mediated miR regulatory mechanism in cardiac dysfunction is provided in online supplement.
In conclusion, our data identify β-arrestin1 as an important mediator of Drosha function to regulate miR biogenesis in the heart and provide new insights into our understanding of how selective ligands for the β1AR may modulate the metabolism of specific miRs. We postulate that the development of high affinity β1AR-biased ligands, that display better efficacy for this newly discovered β-arrestin1-mediated miR regulatory network, may provide a new class of drugs for the treatment of cardiovascular diseases.
Supplementary Material
Novelty and Significance.
What Is Known?
β-arrestin-mediated β1-adrenergic receptor (β1AR) signaling confers cardiac protection.
The β-blocker carvedilol is a weak activator (biased ligand) of the β1AR/β-arrestin pathway.
A role for β-arrestin in activating cellular signaling networks is increasingly being recognized as an important modulator of normal physiology and disease.
What New Information Does This Article Contribute?
β-arrestin1-biased β1AR signaling induced by the β-blocker, carvedilol regulates the biogenesis of a subset of microRNAs in the mouse heart and human cells.
Carvedilol-mediated microRNA processing requires G protein-coupled receptor (GPCR) kinase (GRK) 5/6, β-arrestin1 and the β1AR.
This effect of carvedilol is mediated by the formation of a nuclear complex of β-arrestin1 with the Drosha and hnRNPA1, critical components for RNA helicase-independent microRNA processing.
β-arrestin-biased agonism is an emerging concept in the GPCR signaling field in which unique ligand-activated conformational states can selectively stimulate GPCRs to signal through β-arrestin in the absence of G protein activation. Since GPCRs are the target of approximately 40% of all modern medicinal drugs, β-arrestin-biased ligands have been considered to have important therapeutic utility. Here, , we report an essential role for β-arrestin1 in microRNA processing following stimulation by the β-arrestin-biased βAR agonist carvedilol. While carvedilol did not increase the expression of primary microRNA transcripts, it enhanced expression of premature microRNAs by promoting the interaction of β-arrestin1 with components of the nuclear Drosha microprocessor complex. We believe that modulation of the β1AR/β-arrestin1/microRNA pathway by carvedilol could lead to the development of pharmacological strategies (i.e. β-arrestin1-biased ligands), allowing effective modulation of microRNAs that may be important in regulating the progression of cardiac disease and cardiac remodeling.
Acknowledgements
We thank Drs. David Fulton, David Stepp, David Pollock, Sangmi Kim, Ganesan Ramesh, and Ruth Caldwell for critically reviewing the manuscript and sharing their equipment, and Weili Zou for excellent technical assistance.
Sources of Funding
This work was supported by Georgia Regents University (GRU, previously known as Georgia Health Sciences University) Departmental Start-Up Fund and American Heart Association (AHA) Greater Southeast Affiliate (GSA) Grant-in-Aid 12GRNT12100048 to IK, GRU Diabetes and Obesity Discovery Institute Scholar Program to JV, AHA National Scientist Development Grant 11SDG6960011 to HS, AHA GSA Grant-in-Aid 13GRNT17080109 to JAJ, and National Institutes of Health grant P01HL075443 to WJK and HAR.
Non-Standard Abbreviations and Acronyms
- Alp
Alprenolol
- βARs
β-adrenergic receptors
- β-blockers
β-adrenergic receptor antagonists
- β1-miRs
β-arrestin1-regulated miRs
- Carv
carvedilol
- Forsk
forskolin
- GPCR
G protein-coupled receptor
- GRK
G protein-coupled receptor kinase
- Iso
isoproterenol
- KO
knockout
- LUC
luciferase
- Met
metoprolol
- MiRNAs or MiRs
microRNAs
- Pre-miRs
hairpin intermediate microRNAs or premature microRNAs
- Pri-miRs
long primary microRNA transcripts
- TG
transgenic
- WT
wild-type
- WTβ1AR cells
HEK 293 cells stably overexpressing wild-type β1ARs
Footnotes
I.K. and Y.W. contributed equally to this study.
Disclosures
HAR is a scientific cofounder of Trevena Inc., a company that is developing G protein-coupled receptor targeted drugs.
References
- 1.Quiat D, Olson EN. Micrornas in cardiovascular disease: From pathogenesis to prevention and treatment. The Journal of clinical investigation. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu H, Fan GC. Role of micrornas in the reperfused myocardium towards post-infarct remodelling. Cardiovasc Res. 2011 doi: 10.1093/cvr/cvr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krol J, Loedige I, Filipowicz W. The widespread regulation of microrna biogenesis, function and decay. Nature reviews. Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC. Posttranscriptional regulation of microrna biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Davis BN, Hilyard AC, Lagna G, Hata A. Smad proteins control drosha-mediated microrna maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O'Malley BW, Kato S. Maturation of microrna is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The rna-binding protein ksrp promotes the biogenesis of a subset of micrornas. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, 3rd, Lefkowitz RJ. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci U S A. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim IM, Tilley DG, Chen J, Salazar NC, Whalen EJ, Violin JD, Rockman HA. Beta-blockers alprenolol and carvedilol stimulate beta-arrestin-mediated egfr transactivation. Proc Natl Acad Sci U S A. 2008;105:14555–14560. doi: 10.1073/pnas.0804745105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisler JW, DeWire SM, Whalen EJ, Violin JD, Drake MT, Ahn S, Shenoy SK, Lefkowitz RJ. A unique mechanism of beta-blocker action: Carvedilol stimulates beta-arrestin signaling. Proc Natl Acad Sci U S A. 2007;104:16657–16662. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei H, Ahn S, Shenoy SK, Karnik SS, Hunyady L, Luttrell LM, Lefkowitz RJ. Independent beta-arrestin 2 and g protein-mediated pathways for angiotensin ii activation of extracellular signal-regulated kinases 1 and 2. Proc Natl Acad Sci U S A. 2003;100:10782–10787. doi: 10.1073/pnas.1834556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Violin JD, DeWire SM, Yamashita D, Rominger DH, Nguyen L, Schiller K, Whalen EJ, Gowen M, Lark MW. Selectively engaging beta-arrestins at the angiotensin ii type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther. 2010;335:572–579. doi: 10.1124/jpet.110.173005. [DOI] [PubMed] [Google Scholar]
- 13.Patel CB, Noor N, Rockman HA. Functional selectivity in adrenergic and angiotensin signaling systems. Mol Pharmacol. 2010;78:983–992. doi: 10.1124/mol.110.067066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang TY, Bressler EA, Hara MR, Shenoy SK, Gygi SP, Lefkowitz RJ. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular mechanism of beta-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitzler D, Durand G, Gallay N, Rizk A, Ahn S, Kim J, Violin JD, Dupuy L, Gauthier C, Piketty V, Crepieux P, Poupon A, Clement F, Fages F, Lefkowitz RJ, Reiter E. Competing g protein-coupled receptor kinases balance g protein and beta-arrestin signaling. Mol Syst Biol. 2012;8:590. doi: 10.1038/msb.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruffolo RR, Jr., Feuerstein GZ. Pharmacology of carvedilol: Rationale for use in hypertension, coronary artery disease, and congestive heart failure. Cardiovasc Drugs Ther. 1997;11(Suppl 1):247–256. doi: 10.1023/a:1007735729121. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki M, Yano M, Oda T, Tateishi H, Kobayashi S, Yamamoto T, Ikeda Y, Ohkusa T, Ikemoto N, Matsuzaki M. Scavenging free radicals by low-dose carvedilol prevents redox-dependent ca2+ leak via stabilization of ryanodine receptor in heart failure. J Am Coll Cardiol. 2007;49:1722–1732. doi: 10.1016/j.jacc.2007.01.064. [DOI] [PubMed] [Google Scholar]
- 19.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 20.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 21.Kang J, Shi Y, Xiang B, Qu B, Su W, Zhu M, Zhang M, Bao G, Wang F, Zhang X, Yang R, Fan F, Chen X, Pei G, Ma L. A nuclear function of beta-arrestin1 in gpcr signaling: Regulation of histone acetylation and gene transcription. Cell. 2005;123:833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Yue R, Kang J, Zhao C, Hu W, Tang Y, Liu X, Pei G. Beta-arrestin1 regulates zebrafish hematopoiesis through binding to yy1 and relieving polycomb group repression. Cell. 2009;139:535–546. doi: 10.1016/j.cell.2009.08.038. [DOI] [PubMed] [Google Scholar]
- 23.Ma L, Pei G. Beta-arrestin signaling and regulation of transcription. J Cell Sci. 2007;120:213–218. doi: 10.1242/jcs.03338. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Zhang Y, Shan H, Pan Z, Li X, Li B, Xu C, Zhang B, Zhang F, Dong D, Song W, Qiao G, Yang B. Microrna-1 downregulation by propranolol in a rat model of myocardial infarction: A new mechanism for ischaemic cardioprotection. Cardiovasc Res. 2009;84:434–441. doi: 10.1093/cvr/cvp232. [DOI] [PubMed] [Google Scholar]
- 25.Carrillo ED, Escobar Y, Gonzalez G, Hernandez A, Galindo JM, Garcia MC, Sanchez JA. Posttranscriptional regulation of the beta2-subunit of cardiac l-type ca2+ channels by micrornas during long-term exposure to isoproterenol in rats. J Cardiovasc Pharmacol. 2011;58:470–478. doi: 10.1097/FJC.0b013e31822a789b. [DOI] [PubMed] [Google Scholar]
- 26.Zheng H, Zeng Y, Zhang X, Chu J, Loh HH, Law PY. Mu-opioid receptor agonists differentially regulate the expression of mir-190 and neurod. Mol Pharmacol. 2010;77:102–109. doi: 10.1124/mol.109.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noma T, Lemaire A, Naga Prasad SV, Barki-Harrington L, Tilley DG, Chen J, Le Corvoisier P, Violin JD, Wei H, Lefkowitz RJ, Rockman HA. Beta-arrestin-mediated beta1-adrenergic receptor transactivation of the egfr confers cardioprotection. J Clin Invest. 2007;117:2445–2458. doi: 10.1172/JCI31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim IM, Tilley DG, Chen J, Salazar NC, Whalen EJ, Violin JD, Rockman HA. Beta-blockers alprenolol and carvedilol stimulate beta-arrestin-mediated egfr transactivation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14555–14560. doi: 10.1073/pnas.0804745105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guil S, Caceres JF. The multifunctional rna-binding protein hnrnp a1 is required for processing of mir-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 30.Allegra D, Mertens D. In-vivo quantification of primary microrna processing by drosha with a luciferase based system. Biochem Biophys Res Commun. 2011;406:501–505. doi: 10.1016/j.bbrc.2011.02.055. [DOI] [PubMed] [Google Scholar]
- 31.Monk CE, Hutvagner G, Arthur JS. Regulation of mirna transcription in macrophages in response to candida albicans. PLoS ONE. 2010;5:e13669. doi: 10.1371/journal.pone.0013669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luttrell LM, Gesty-Palmer D. Beyond desensitization: Physiological relevance of arrestin-dependent signaling. Pharmacol Rev. 2010;62:305–330. doi: 10.1124/pr.109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: Biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosano L, Cianfrocca R, Tocci P, Spinella F, Di Castro V, Spadaro F, Salvati E, Biroccio AM, Natali PG, Bagnato A. Beta-arrestin-1 is a nuclear transcriptional regulator of endothelin-1-induced beta-catenin signaling. Oncogene. 2012 doi: 10.1038/onc.2012.527. [DOI] [PubMed] [Google Scholar]
- 35.Dinicolantonio JJ, Lavie CJ, Fares H, Menezes AR, O'Keefe JH. Meta-analysis of carvedilol versus beta 1 selective beta-blockers (atenolol, bisoprolol, metoprolol, and nebivolol) Am J Cardiol. 2013 doi: 10.1016/j.amjcard.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 36.Mangmool S, Shukla AK, Rockman HA. Beta-arrestin-dependent activation of ca(2+)/calmodulin kinase ii after beta(1)-adrenergic receptor stimulation. J Cell Biol. 2010;189:573–587. doi: 10.1083/jcb.200911047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hara MR, Kovacs JJ, Whalen EJ, Rajagopal S, Strachan RT, Grant W, Towers AJ, Williams B, Lam CM, Xiao K, Shenoy SK, Gregory SG, Ahn S, Duckett DR, Lefkowitz RJ. A stress response pathway regulates DNA damage through beta2-adrenoreceptors and beta-arrestin-1. Nature. 2011;477:349–353. doi: 10.1038/nature10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.