Abstract
BRI2, a protein mutated in Familial British and Familial Danish Dementias, interacts with Amyloid Precursor Protein (APP) and reduces the levels of secreted APPβ (sAPPβ), which derives from APP cleavage by β-secretase (BACE1). Exploring the mechanisms of this effect, we obtained data that BRI2 decreases the cellular levels of BACE1 thus reducing the β-cleavage of APP. Deletion of N-terminal cytoplasmic or C-terminal extracellular sequences of BRI2 neither affected its interaction with BACE1 or APP (Fotinopoulou et al., 2005) nor the reduction in the levels of BACE1 and sAPPβ. These results suggest that BRI2 may prevent access of BACE1 to APP and the BRI2/BACE1 interaction may mediate the reduction in BACE1 levels. In support, BRI2 expression induced lysosomal but not proteasomal degradation of BACE1. In parallel, BRI2 expression was also found to reduce BACE1 mRNA levels by 50%. This study adds novel information regarding the mechanism by which BRI2 affects APP processing and BACE1 levels.
Keywords: Familial British dementia, Familia Danish dementia, Alzheimer’s disease, neurodegeneration, BRI2, APP, BACE1
INTRODUCTION
Familial British Dementia (FBD) and Familial Danish Dementia (FDD) are two neurodegenerative diseases genetically linked with two different mutations in the gene that encodes for BRI2 protein: a stop codon mutation [1] and a duplication of ten nucleotides [2], respectively. These mutations disrupt the open reading frame of the BRI2 gene and result in the production of the mutated proteins ABriPP or ADanPP that contain 11 more aminoacids than the wild type protein. BRI2 is cleaved in its extracellular sequence by furin and releases a secreted peptide of 23 amino acids [3–5]. In patients suffering from FBD/FDD the mutated BRI2 proteins are cleaved at the same site as wild type BRI2 and release the ABri or ADan peptides. These peptides are common in part with the wild type peptide, but due to the BRI2 mutations they also contain an aminoacid extension in the C-terminus, which makes them prone to form cerebral and systemic amyloid deposits.
Recent interest in FBD/FDD has aroused because they present striking similarities with the most common form of dementia, Alzheimer’s disease (AD). Among these similarities is the presence of neurofibrillary tangles (NFTs), composed of paired helical filaments of hyperphosphorylated tau protein [6, 7] and inflammation, manifested by the activation of astrocytes and microglial cells, as well as the presence of complement components. Another similarity between FBD/FDD and AD is the deposition of amyloid peptides in the brain. In the case of AD the peptide deposited is Aβ and derives from the proteolytic processing of APP while in FBD and FDD the peptides deposited derive from the proteolytic processing of BRI2. Most importantly, in all three diseases, there is extensive loss of neurons in the brain regions exhibiting the above pathologies.
The neuropathological similarities in FBD/FDD and AD prompted us, previously, to hypothesize that common cellular mechanisms leading to neuronal loss might exist in these conditions. Therefore, we examined the possibility that the two central proteins in the pathogenesis of those diseases, BRI2 and APP, physically or functionally interact. We and others [8, 9] showed that BRI2 and APP physically interact. Also, BRI2 was found to affect APP processing by reducing both secretion of sAPPα (secreted form of APP that derives from its processing by α-secretase in the non-amyloidogenic pathway) and production of Aβ. In the amyloidogenic pathway that produces Aβ, APP is firstly cleaved by β-secretase (Beta-Site Amyloid Beta A4 Precursor Protein-cleaving Enzyme1, BACE1) to release a secreted fragment (sAPPβ) and to produce a membrane-bound C-terminal fragment. This fragment is subsequently subjected to regulated intramembrane proteolysis by γ-secretase to release extracellularly Aβ and intracellularly AICD (APP Intracellular Domain) [10–12]. BRI2 expression in mice reduces Aβ deposition [13–15] and knockdown of BRI2 in cells enhances the levels of Aβ production and sAPPβ [13]. Furthermore, transgenic mice that express one allele of BRI2 carrying the Danish mutation and one wild type allele show increased levels of Aβ and sAPPβ suggesting a loss of function effect of this mutation [16–18]. In addition to its effect on APP processing, BRI2 has been found to increase secretion of Insulin Degrading Enzyme (IDE) and to promote Aβ degradation [15]. Taken together, these results bring up BRI2 as an important physiological regulator of APP processing, Aβ production, and Aβ degradation.
In this study, we obtained data indicating that BRI2 reduces APP cleavage by BACE1. Furthermore, we found that BRI2 physically interacts with BACE1 and reduces its cellular levels by promoting its lysosomal degradation and by reducing its mRNA levels. Our data provides important insight into common neurodegenerative mechanisms of three demented syndromes.
MATERIALS AND METHODS
Chemicals
All chemicals were purchased from Sigma–Aldrich, Greece, unless otherwise stated.
DNAs and Plasmids
Human BRI2 cDNA was subcloned in myc-pRK5 vector to express the myc-tagged BRI2 protein (mycBRI2). The deletion constructs of mycBRI2 (designated mycBRI2Δ1-45, mycBRI2Δ107-266, mycBRI2Δ147-266, mycBRI2Δ187-266 and mycBRI2Δ227-266) were described previously [8]. The BRI2 construct with an internal deletion between aminoacids 107 and 146 (mycBRI2Δ107-146) was generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene, CA, USA). The mutagenic primers are available upon request. The cDNAs of BACE1 and APP695 were subcloned in the pcDNA3.1 vector (Invitrogen, CA, USA). Human Homer3 cDNA subcloned in myc-pRK5 vector has been described previously [19].
Antibodies
The anti-myc monoclonal antibodies 9E10 (Iowa University Hybridoma Bank, Iowa, IA, USA) and 9B11 (Cell Signaling Technology, MA, USA) were used for western blot and immunoprecipitation of myc-tagged BRI2, respectively. The anti-BACE1 antibodies 7523 (N-terminal) and 7520 (C-terminal) were a kind gift of Professor Christian Haass (Ludwig Maximilians University, Munich, Germany) and were used for western blot and immunoprecipitation of BACE1, respectively. R1(57) that recognizes the last 20 amino acids of APP cytoplasmic tail was used for the detection of full-length APP and was a kind gift of Dr. P. Mehta (Institute for Basic Research in Developmental Disabilities, Staten Island, NY). The antibody T-1515, raised in chicken, against the N-terminus of BRI2 was purchased by BMA Biomedicals (Augst, Switzerland). The anti-sAPPβ specific antibody 192wt was a generous gift of Dr. Dale Schenk (ELAN pharmaceuticals, San Francisco, CA, USA).
Cell Culture and Transfections
Human Embryonic Kidney cells (HEK293) were purchased from ATCC (Middlesex, UK) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with heat inactivated 10% Fetal Bovine Serum (both from PAA laboratories GmbH, Linz, Austria), and Penicillin/Streptomycin (Sigma–Aldrich, Athens, Greece). Transfections of the cDNAs were performed with Escort IV reagent (Sigma–Aldrich, Athens, Greece), used according to manufacturer’s instructions. The human neuroblastoma cell line SH-SY5Y was obtained from ATCC and cultured in RPMI medium (PAA laboratories GmbH, Linz, Austria). Validated siRNA oligos (Stealth RNAiTM siRNA) to downregulate BRI2(Itm2b) gene (NM_021999.4) expression were ordered from Invitrogen (Carlsbad, CA, USA). The siRNA oligos were transfected into SH-SY5Y cells using the HiPer-Fect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Protein Extraction and Western Blotting
Forty-eight hours after transfection, conditioned medium was removed. Cells were lysed at 4 °C in lysis buffer [50mM Tris–HCl, 150mM NaCl, 2mM EDTA, pH 7.6, 1% Triton X-100 (v/v)] supplemented with complete protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany). Samples were incubated for 30 min on ice and protein extracts were clarified by centrifugation at 16,000×g for 30 min at 4 °C. The supernatants were quantified for protein content using the BCA assay kit (Pierce, Rockford, IL, USA). Samples from cell extracts were prepared in SDS Laemmli sample buffer (3% SDS, 0.4M Tris pH 6.7, 40mM EDTA, 50% glycerol, bromophenol blue) containing 5% β-mercaptoethanol. Extracts were separated by SDS-PAGE, transferred to polyvinyldiene difluoride membranes (Roth GmbH, Karlsruhe, Germany) and analyzed by western blot as previously described [8].
Co-Immunoprecipitations (co-IPs)
For the immunoprecipitation experiments, cell extracts (1000–1500 µg of total protein) were incubated with appropriate dilutions of the IP antibody overnight at 4°C. Antibody-bound protein complexes were collected with protein G agarose beads for monoclonal antibodies, or protein A agarose beads for polyclonal antibodies (Millipore, Billerica, USA) following incubation for 1 h at 4°C. Pellets were washed three times with washing buffer containing 50mM Tris, 150mM NaCl, 2mM EDTA, pH 7.6, 0.5% Triton X-100. The beads were resuspended in SDS Laemmli sample buffer and the recovered proteins analyzed by immunoblot.
Immunofluorescence Confocal Microscopy
24 hours post-transfection the supernatant medium was removed, cells were rinsed twice with PBS and fixed for 15 minutes with cold methanol (−20°C). After extensive washing the cells were incubated with blocking solution for 30 minutes and overnight at 4°C with the primary antibodies. Following washing, cells were incubated with the secondary antibodies goat anti-mouse Alexa Fluor 488 or goat anti-rabbit Alexa Fluor 546 (Invitrogen) for 2 hours at RT and mounted with Vectashield mounting medium containing DAPI (Vector Laboratories, Inc. Burlingame, CA, USA) for nuclear staining. The fluorescent signal was visualized with the use of a Leica TCS SP5 confocal microscope.
Preparation of mRNA, Synthesis of cDNA and Quantitative Real Time PCR
Total mRNA was extracted using Magnapure Compact RNA Isolation kit on a Magnapure DNA/RNA extractor (F. Hoffmann-La Roche AG, Basel, Switzerland) according to the manufacturer’s instructions. cDNA synthesis was performed using the Superscript II Reverse Transcriptase and random primers both from Invitrogen (Carlsbad, CA, USA) according to the manufacturer’s instructions. Initially, using the same primers to be used in real time PCR but not the labeled probes, we performed PCR reactions, purified the corresponding BACE1 and GAPDH fragments and generated DNA solutions covering a range of concentrations from 25ng/ul to 25×10−9 ng/ul. These solutions were used to generate by real time PCR a standard curve for BACE1 and GAPDH. The real time PCR reactions performed in a total volume of 20 ul using TaqMan Master Mix (F. Hoffmann-La Roche AG, Basel, Switzerland) and 0,5 pmoles of primers for BACE1 or GAPDH and 0,2 pmoles of dual labelled probes for BACE1 or GAPDH [20]. The primers and the conditions of the real time PCR were as previously described [20]. Specifically for BACE1, the primers are designed based on BACE1 mRNA with reference number: NM_138972.3. The sense primer spans the sequence between nucleotides 1582 and 1603 of exon 9 (5’-TGGAGGGCTTCTACGTTCTCTT-3’) while the antisense primer spans the sequence between nucleotides 1647 and 1667 that covers the junction of exon 9 and exon 10 (5’-CCTGAACTCATCGTGCACATG-3’). Following completion of the real time PCR reactions, the absolute concentration (ngr/ul) of the mRNA of BACE1 and GAPDH and their ratios were calculated. Then we calculated the average percentage of the ratio of the mRNA of BACE1 and GAPDH in cells transfected with pRK5 vector and the cDNA for BACE1 relative to the ratio of the mRNA of BACE1 and GAPDH in cells transfected with the cDNAs for BRI2 and BACE1. The data was analyzed and plotted on Microsoft Excel. The statistical significance between samples was calculated by a student’s t-test in R language 2.10.2, setting the significance level at p<0.01. The data was also analyzed using Relative Expression Software Tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. This analysis confirmed the results obtained using Student’s t-test in R language 2.10.2.
RESULTS
BRI2 Inhibits Processing of APP by BACE1
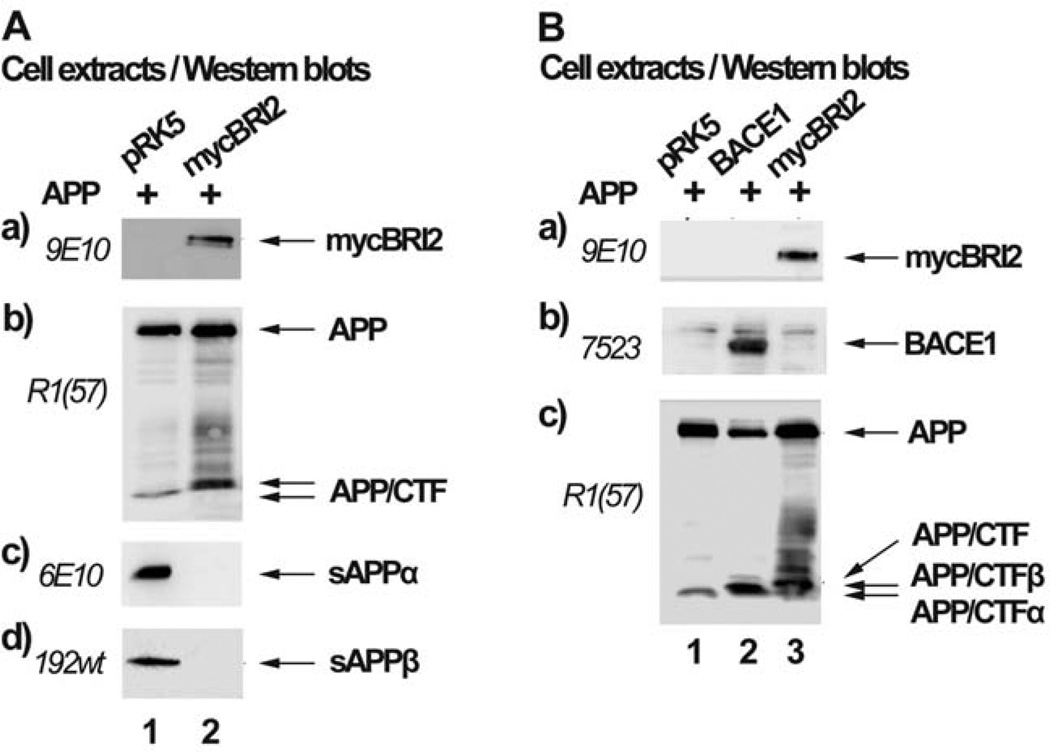
BRI2 inhibits processing of APP by α-secretase and results in reduced levels of sAPPα and increased levels of APP C-terminal fragments (APP/CTFs) [8, 9], (Fig. 1A, panel b and c). To examine the effect of BRI2 on the processing of APP by BACE1, we overexpressed APP or APP and BRI2 in HEK293 cells and assayed for sAPPβ by an antibody that specifically recognizes the last methionine exposed after β-secretase cleavage of APP. Our data indicates that overexpression of BRI2 (Fig. 1A, panel a) results in reduced secretion of sAPPα (panel c) and sAPPβ (panel d). In support of the latter, it has been shown that BRI3, a member of the BRI family of proteins, inhibits processing of APP by BACE1 [21]. In parallel, we observed that BRI2 caused accumulation of APP/CTFs (panel b), which are unlikely to be those derived from the activity of α-secretase (APP/CTFα) or β-secretase (APP/CTFβ), since reduction of sAPPα and sAPPβ should be accompanied by a concomitant reduction in APP/CTFα and APP/CTFβ. To clarify whether the APP/CTFs produced in cells transfected with APP and BRI2, derive by the activity of α-secretase, β-secretase or another enzyme, we compared their molecular mass (Fig. 1B, panel c, lane 3) with that of APP/CTFβ produced in cells transfected with APP and BACE1 (panel c, lane 2), as well as with that of APP/CTFα produced from cells transfected with APP only (panel c, lane 1) (APP is mainly processed by α-secretase in the absence of overexpressed BACE1). Fig. 1B shows that the APP/CTFs that accumulate in the presence of BRI2 have molecular mass higher than that of APP/CTFα or APP/CTFβ. This data suggests that BRI2 inhibits cleavage of APP by α- and β-secretase and allows its cleavage by another protease upstream of the β-site.
Fig. (1). BRI2 inhibits processing of APP by BACE1.
(A) Extracts of HEK293 cells transfected with the cDNA encoding for APP and the myc-tag bearing vector, pRK5 (lane 1) or the cDNA for mycBRI2 (lane 2) were assayed by western blot for the expression of mycBRI2 (panel a), APP (panel b), sAPPα (panel c) and sAPPβ (panel d) using the antibodies 9E10, R1(57), 6E10 and 192wt respectively. (B) Extracts of HEK293 cells transfected with the cDNA encoding for APP and pRK5 vector (lane 1), the cDNA encoding for BACE1 (lane 2) or the cDNA encoding for mycBRI2 (lane 3) were analyzed by western blot for the expression of mycBRI2 (panel a), BACE1 (panel b) and APP (panel c), using the antibodies 9E10, 7523 and R1(57) respectively.
BRI2 Interacts with BACE1
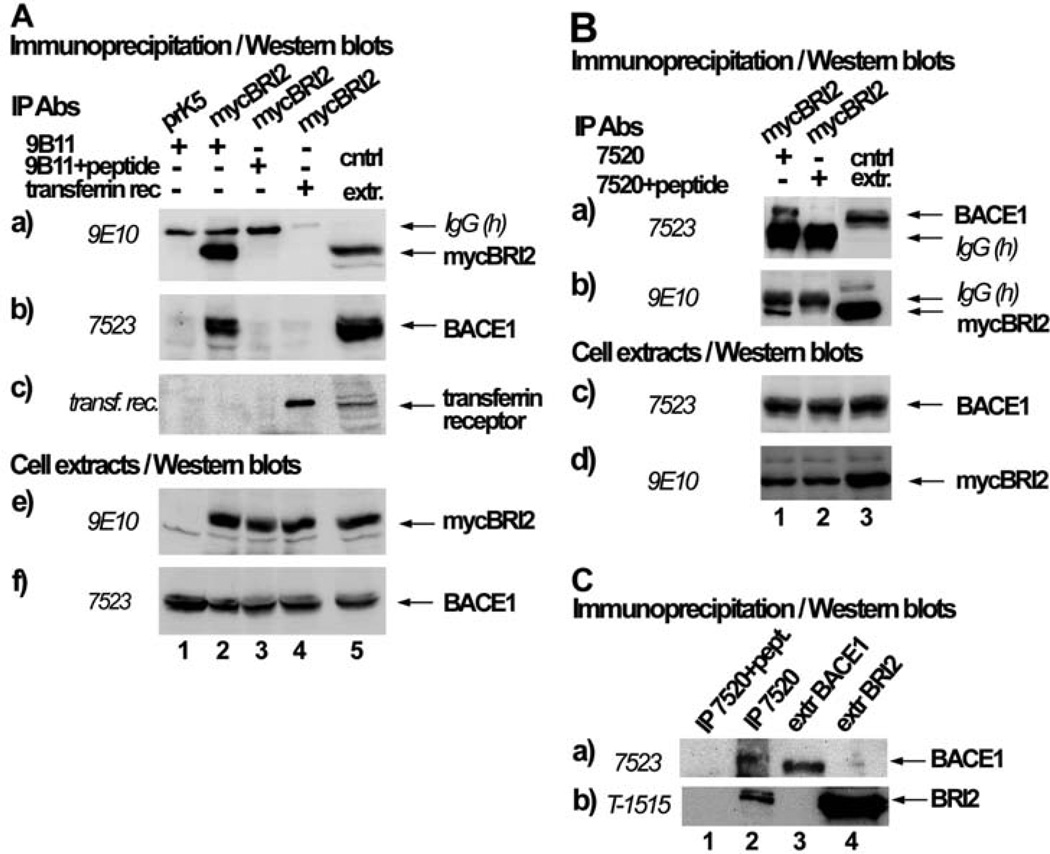
The inhibitory effect of BRI2 on APP processing by BACE1 could be mediated by the physical interaction of BRI2 with BACE1. To explore this possibility, we expressed in HEK293 cells mycBRI2 and BACE1 and performed immunoprecipitation of mycBRI2 from the cell extracts, using the antibody 9B11 against the myc epitope of BRI2 (Fig. 2A, panel a, lane 2). Western blot analysis of the precipitates showed that immunoprecipitation of mycBRI2 resulted in co-precipitation of BACE1 (Fig. 2A, panel b, lane 2). Immunoprecipitation with the 9B11 antibody pre-absorbed with its antigenic peptide (Fig. 2A, lane 3) or immunoprecipitation with an antibody recognizing the transferrin receptor (Fig. 2A, lane 4) did not result in non-specific precipitation of mycBRI2 (panel a) or co-precipitation of BACE1 (panel b). In the reverse independent experiment (Fig. 2B) immunoprecipitation of BACE1 (panel a, lane 1) with the antibody 7520 resulted in co-precipitation of mycBRI2 (panel b, lane 1). When the 7520 antibody was pre-incubated with its antigenic peptide, BACE1 (panel a, lane 2) was not precipitated neither did BRI2 (panel b, lane 2). This indicates that there is a specific interaction between BRI2 and BACE1. To investigate whether the endogenous proteins interact, we immunoprecipitated endogenous BACE1 (Fig. 2C, panel a, lane 2) from extracts of the human neuroblastoma cell line SH-SY5Y, and showed that this resulted in co-precipitation of BRI2 (Fig. 2C, panel b, lane 2). No interaction was observed when the IP antibody was pre-absorbed with its antigenic peptide (Fig. 2C, panel b, lane 1). Therefore, endogenous BRI2 and BACE1 interact specifically. In support, BRI3 has also been found to interact with BACE1 [22].
Fig. (2). BRI2 interacts specifically with BACE1.
(A) HEK293 cells were co-transfected with the cDNA encoding for BACE1 and either the myc-tag bearing vector, pRK5 (lane 1) or the cDNA encoding for myc-tagged BRI2 (mycBRI2) (lanes 2–4). Immunoprecipitates (IPs) with the anti-myc antibody 9B11 (to detect mycBRI2), 9B11 preabsorbed with its antigenic peptide or anti-transferrin receptor antibody were analyzed by western blot using the anti-myc antibody 9E10 (panel a), anti-BACE1 antibody 7523 (panel b), or the anti-transferrin receptor antibody (panel c). IgG(h) denotes the heavy chain of IgG. Extracts of cells expressing BACE1 or BACE1 and mycBRI2 were analyzed by western blot using the antibodies 9E10 (panel e) or 7523 (panel f). (B) HEK293 cells were co-transfected with cDNAs encoding for mycBRI2 and BACE1. IPs with the BACE1 specific antibody 7520 or 7520 preabsorbed with its antigenic peptide were analyzed by western blot using the anti-BACE1 antibody 7523 (panel a) or the anti-myc antibody 9E10 (panel b). Successful BACE1 and mycBRI2 expression can be seen in western blots in panels c and d, respectively. C. Extracts of SH-SY5Y cells were incubated with the BACE1 specific antibody 7520 or 7520 preabsorbed with its antigenic peptide and IPs and cell extracts were analyzed by western blot using the anti-BACE1 antibody 7523 (panel a) or the anti-BRI2 antibody T-1515 (panel b).
BRI2 Co-Localizes with BACE1 in Intracellular Compartments
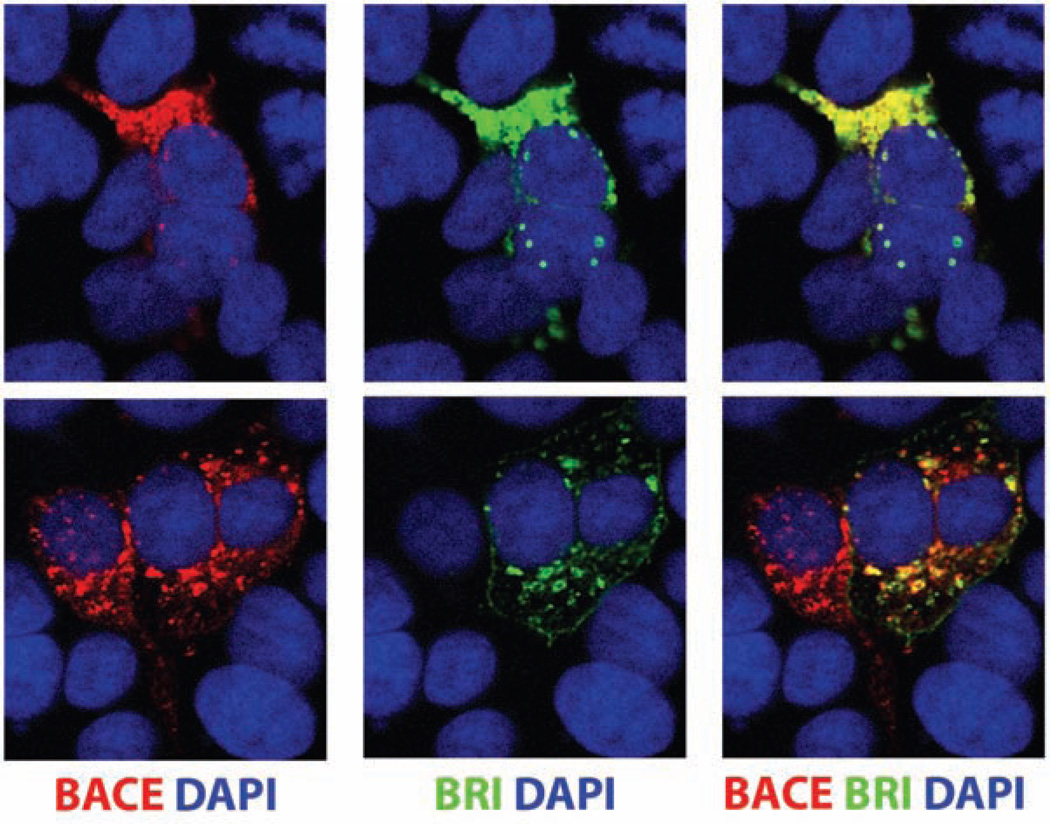
To further investigate the interaction between BRI2 and BACE1, we examined whether they co-localize in certain cellular compartments (Fig. 3). HEK293 cells co-expressing mycBRI2 and BACE1 were incubated with primary antibodies against the myc epitope of BRI2 (9E10) or BACE1 (7523) and were then labeled with the goat anti-mouse Alexa 488 antibody (for BRI2 detection, green) as wells as the goat anti-rabbit Alexa 546 antibody (for BACE1 detection, red). Using a confocal microscope analysis, we observed a substantial co-localization of both proteins (yellow merged signal) in intracellular compartments adjacent to the cell nucleus, that most likely represent the ER and Golgi compartments. Colocalization was also observed in long cellular processes (not shown). When BACE1 was transfected in the absence of mycBRI2 there was no green signal detected and when mycBRI2 was transfected in the absence of BACE1 there was no red signal detected (data not shown). BRI3 has also been shown to co-localize with BACE1 [22].
Fig. (3). BRI2 and BACE1 co-localize in intracellular compartments.
HEK293 cells were transfected with the cDNAs that encode for mycBRI2 and BACE1. 24 hours post-transfection the cells were stained with the 9E10 anti-myc antibody (green) and 7520 anti-BACE1 antibody (red). The cell nuclei are stained with DAPI (red). Staining was followed by confocal microscope analysis.
BRI2 Reduces the Cellular Levels of BACE1
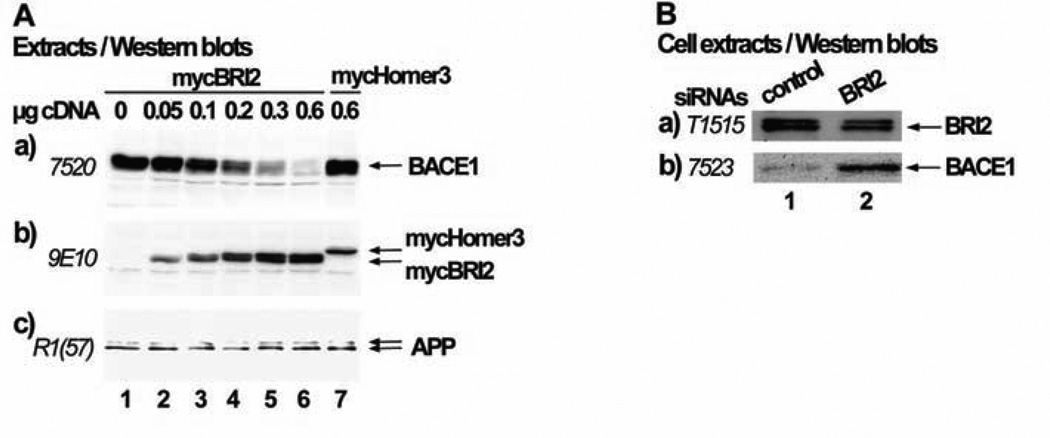
The interaction between BRI2 and BACE1 could be responsible for the reduced processing of APP by BACE1. One possibility would be that BRI2 alters the subcellular localization of BACE1, preventing it from reaching the compartments in which it cleaves APP. However, our confocal microscope experiments did not show any effect of BRI2 on the intracellular distribution of BACE1. The possibility that BRI2 affects BACE1 transcription is excluded, since the plasmid in which BACE1 is subcloned contains the constitutive CMV promoter. Another option would be that BRI2 affects the cellular levels of BACE1 protein. In the initial immunoprecipitation experiments described above, we transfected the cDNAs of mycBRI2 and BACE1 in a ratio of 0,25:1, since transfecting equal cDNA amounts resulted in very low levels of BACE1 expression. This observation led us to test the possibility that BRI2 reduces the protein levels of BACE1. For that purpose, we co-transfected cells with increasing amounts of mycBRI2 encoding cDNA (0.05µg to 0.6µg) or 0.6 µg of mycHomer3 protein encoding cDNA and 0,3µg of BACE1 cDNA (Fig. 4A). Western blot analysis of the cell extracts revealed that the levels of BACE1 protein decrease (panel a, lanes 1–6) as the levels of BRI2 protein increase (panel b, lanes 1–6). Overexpression of mycHomer3 protein (panel b, lane 7) had no effect on the levels of BACE1 (panel a, lane 7). Furthermore, overexpression of mycBRI2 did not reduce the cellular levels of APP (panel c). Importantly, knocking down the expression of endogenous BRI2 in cells using siRNAs (Fig. 4B, panel a, line 2) led to enhanced levels of endogenous BACE1 (Fig. 4B, panel b, line 2).
Fig. (4). Expression of BRI2 reduces the cellular levels of BACE1.
(A) HEK293 cells were co-transfected with 0.3µg BACE1 cDNA and either the myc-tag bearing vector, pRK5 (lane 1), increasing amounts of the cDNA encoding for BRI2 (0.05µg, 0.1µg, 0.2µg, 0.4µg and 0.6µg respectively for lanes 2–6) or 0.6 µg cDNA encoding for mycHomer3 protein (lane 7) (pRK5 vector was added when needed in order to have 1µg total DNA for each transfection). Cell extracts were analyzed by western blot using the antibodies 7520 against BACE1 (panel a), 9E10 against the myc tag (panel b) or R1(57) against APP (panel c). (B) HEK293 cells were transfected with control siRNA (lane 1) or BRI2 siRNA (lane 2). Cell extracts were analyzed by western blot using the anti-BRI2 antibody T-1515 (panel a) or the anti-BACE1 antibody 7523 (panel b).
Regions of BRI2 Involved in its Effect on the Cellular Levels of BACE1 and its Interaction with BACE1
Next, we aimed to identifying the BRI2 regions mediating its effect on the cellular levels of BACE1. To address this issue, we used several deletion constructs of mycBRI2 including mycBRI2Δ1-46, mycBRI2Δ107-266, mycBRI2Δ147-266 and mycBRI2Δ187-266, which lack the corresponding sequences of BRI2 [8]. We observed that similar to wild type mycBRI2 all individual deletion constructs of BRI2 reduced the cellular levels of BACE1 (data not shown). To examine the mechanism by which expression of BRI2 results in reduced levels of BACE1, we examined whether the above mentioned deletion mutants of mycBRI2 interact with BACE1. Immunoprecipitation of BACE1 from the cell extracts with the antibody 7520 resulted in coimmunoprecipitation of wild type mycBRI2 or of the deletion mutant mycBRI2 proteins (Data not shown).
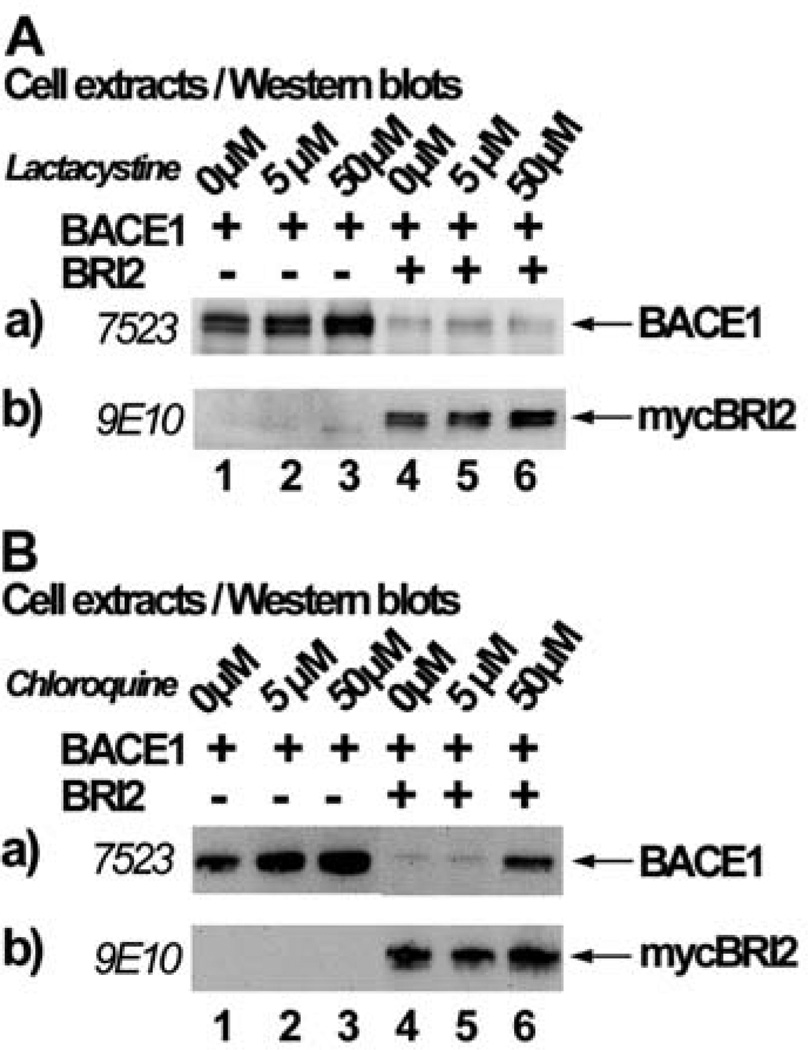
BRI2 Induces Degradation of BACE1 Through the Lysosomal Pathway
To unravel the cellular pathway through which BRI2 reduces BACE1 levels, we investigated if this happens through the two pathways previously described to be responsible for the degradation of BACE1 protein. According to one report, BACE1 is degraded through the proteasomal pathway [23]. To examine if this degradation takes place in our cellular system, we used lactacystine, which is a highly specific inhibitor of the 20S proteasomal subunit (Fig. 5A). We transfected HEK293 cells with the cDNA encoding BACE1 and pRK5 vector (lanes 1, 2 and 3) or BACE1 and mycBRI2 (lanes 4, 5 and 6) and 24 h post-transfection cells were incubated with 0, 5 or 50 µM of lactacystin for 24 h. Western blot analysis of extracts of cells transfected with BACE1 and vector showed an accumulation of BACE1 in the presence of lactacystin (lanes 1, 2 and 3), verifying that BACE1 can be degraded through the proteasomal pathway in our system. When BACE1 was co-transfected with mycBRI2, there was a reduction of BACE1 levels either in the absence or presence of lactacystin (lanes 4, 5 and 6). This shows that blocking this degradation pathway does not prevent BRI2 from exerting its effect on BACE1 protein levels. Identical results were obtained using 10 or 50 µM MG132 proteasomal inhibitor (not shown).
Fig. (5). BRI2 partially reduces the levels of BACE1 by triggering its degradation through the lysosomal pathway.
HEK293 cells transfected with the cDNA encoding for BACE1 and the myctag bearing vector pRK5 or the cDNA encoding for mycBRI2 were incubated with the indicated concentrations of the proteasomal inhibitor lactacystin (A) or the lysosomal inhibitor chloroquine (B). Cell extracts were analyzed by western blot using the anti-BACE1 antibody 7523 (panels a) or the anti-myc antibody 9E10 (panels b).
We then examined whether BRI2 causes BACE1 degradation through the lysosomal pathway, which degrades BACE1 after its recycling from the cell surface [24]. BACE1 and pRK5 vector (Fig. 5B, lanes 1–3) or BACE1 and mycBRI2 (lanes 4–6) were transfected in HEK293 cells and 24h post-transfection cells were treated for 24 h with 0, 5 or 50 µM of the lysosomal inhibitor chloroquine. We observed that in the absence of mycBRI2, chloroquine causes an increase in the protein levels of BACE1 (panel a, lanes 1–3), verifying that BACE1 is indeed degraded through the lysosomal pathway. mycBRI2 expression in the absence of chloroquine or in the presence of 5 µM of chloroquine reduced the cellular levels of BACE1 (panel a, lanes 4 and 5). However, in the presence of 50µM of chloroquine BACE1 levels were elevated (panel a, lane 6), although they never reached the control levels. This result suggests that BRI2, at least partially, affects BACE1 levels by triggering its degradation through the lysosomal pathway.
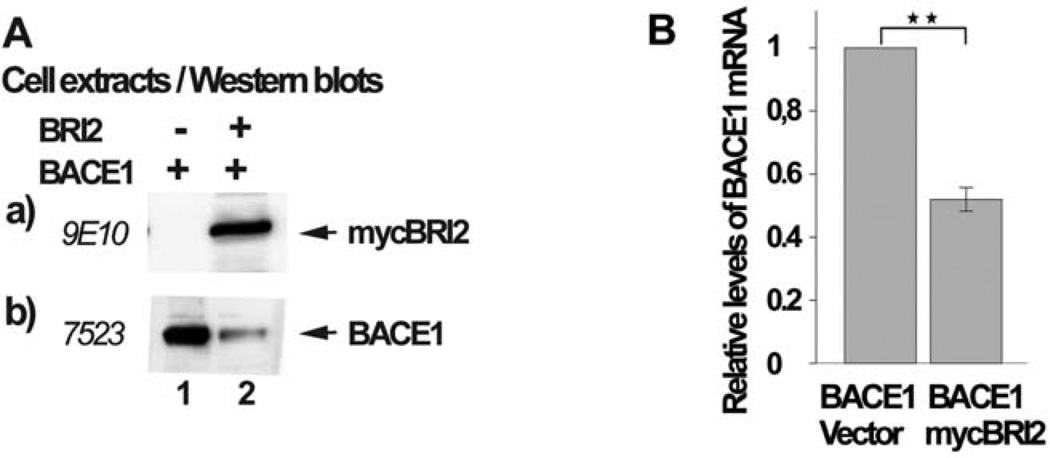
BRI2 Reduces the Levels of BACE1 mRNA
The data described above indicates that BRI2 reduces the levels of BACE1 protein by inducing its enzymatic degradation through the lysosomal pathway and by additional mechanisms. Therefore, we examined the possibility that BRI2 also reduces the levels of BACE1 mRNA. We expressed in HEK293 cells BACE1 or BACE1 and mycBRI2. A fraction of the cells was used to prepare cell extracts (Fig. 6A) or total mRNA (Fig. 6B). Fig. 6A shows that mycBRI2 was successfully expressed (Fig. 6A, panel a, lane 2) and its expression resulted in reduced levels of BACE1 protein (Fig. 6A, panel b, lane 2). Following cDNA synthesis, we performed quantitative real time PCR for both BACE1 mRNA and GAPDH mRNA and we compared the ratio of the levels of the BACE1 mRNA versus the levels of GAPDH mRNA in cells expressing BACE1 with the corresponding ratio in cells expressing both BACE1 and mycBRI2. Our analysis showed that BRI2 expression caused a 50% reduction in BACE1 mRNA levels (Fig. 6B).
Fig. (6). BRI2 reduces the mRNA levels of BACE1.
HEK293 cells transfected with the cDNA encoding for BACE1 and the myc-tag bearing vector pRK5 (lane 1) or the cDNA encoding for mycBRI2 (lane 2). Part of the cells was used to prepare cell extracts and part to prepare mRNA and cDNA. (A) Cell extracts were analyzed by western blot using the anti-myc antibody 9E10 (panel a) or anti-BACE1 antibody 7523 (panel b). (B) The cDNA was used in Real-Time PCR reactions in order to calculate the absolute concentration of the mRNA of BACE1 and GAPDH and their ratios, which were plotted in the barogram. The experiment was repeated three times and the values are the average. The error bars represent the standard deviation (SD), + S.D. 0,074, p < 0,01.
DISCUSSION
BRI2 is a protein mutated in two neurodegenerative diseases, FBD and FDD, which are characterized by the deposition of the amyloidogenic peptides ABri and ADan in the brains of patients. These peptides derive from the proteolytic processing of the mutated forms of BRI2 in FBD and FDD, respectively [1, 2]. Although the above conditions are rare, interest in their study lies on the fact that they present striking neuropathological similarities with the most prevalent form of dementia, Alzheimer’s disease (AD). AD is characterized by amyloid deposits of Aβ peptide in the brain. This peptide derives from the sequential processing of the APP by β-secretase (BACE1) and γ-secretase. APP is cleaved by BACE1 to release a secreted fragment (sAPPβ) and a membrane bound C-terminal fragment (APP/CTFβ). CTFβ is then cleaved intramembraneously by the complex of γ-secretase, which results in the release of Aβ peptide [12]. An important line of evidence indicates that Aβ deposition is a central event in AD pathology, since numerous studies indicate that oligomeric species of this peptide are neurotoxic [25, 26]. Apart from a direct role in neuronal toxicity, Aβ species have been implicated in both the inflammatory processes and the oxidative stress observed in the AD brain. Therefore, the study of the mechanisms that regulate the production of Aβ is of undoubted significance. Amyloid deposition is not the only similarity of FBD/FDD and AD. Other resemblances include the presence of neurofibrillary tangles, indications of local inflammatory response in the brain and extensive loss of neurons [27].
The above similarities raise the intriguing possibility that common cellular pathways are disturbed in all three conditions and result in similar neuropathological outcomes. In support, it has been shown that the two main proteins involved in FBD/FDD and AD, namely BRI2 and APP, physically interact and that expression of BRI2 results in decreased processing of APP by α-secretase and decreased levels of Aβ [8, 9]. However, the literature does not provide a clear picture regarding the effects of BRI2 on the processing of APP by BACE1. Initially, Matsuda et al. suggested that BRI2 increases processing of APP by BACE1, based on indirect data [9]. In disagreement with this interpretation, the same group presented data indicating that first, downregulation of BRI2 results in increased processing of APP by both α-secretase and BACE1 [13] and second, BRI3 inhibits processing of APP by BACE1 [21]. To clarify this issue, we examined the effect of BRI2 overexpression on the levels of sAPPβ, using an antibody that specifically recognizes sAPPβ (Fig. 1). Our data clearly indicates that BRI2 inhibits processing of APP by BACE1 and reduces the levels of sAPPβ. Down-regulation of BRI2 reverses its effect on APP processing and also Aβ deposition is reduced in mice models expressing both BRI2 and an APP mutant form that normally leads to enhanced amyloid deposition [13, 14]. Furthermore, transgenic mice expressing one Danish mutant and one wild type allele of Bri2 show increased levels of Aβ and sAPPβ suggesting that the mutant allele causes loss of function of BRI2 [16–18]. These experimental evidence points towards a significant physiological role of BRI2 as a regulator of APP processing.
As mentioned above, BRI2 reduces APP cleavage by BACE 1 and leads to reduced Aβ production. BRI2 could exert this effect through its interaction with APP by blocking the access of the secretases to their substrate. One group has provided evidence, based on domain swapping studies, that this is a possible option [13]. Another possibility is that BRI2 directly affects the secretases that produce Aβ, and more specifically BACE1, since it reduces sAPPβ production from APP. This, of course, does not exclude the possibility that BRI2 could also compete with the secretases for APP binding. To examine whether BRI2 physically interacts with BACE1, we co-expressed the proteins in cells and showed that immunoprecipitation of BRI2 results in specific coprecipitation of BACE1 and vice versa (Fig. 2A and 2B). Most importantly, we demonstrated that this interaction exists between the endogenous proteins in the human neuroblastoma cell line SH-SY5Y (Fig. 2C). These results indicate that BRI2 and BACE1 interact specifically. In addition, there is substantial co-localization of BRI2 and BACE1 in intracellular compartments as shown by confocal microscopy experiments (Fig. 3). In support of our results, Wickham et al. have shown that BRI3 also interacts and co-localizes with BACE1 [22].
After confirming that these two proteins interact, the question remained as to how BRI2 affects BACE1 and leads to reduced APP processing. In our immunoprecipitation experiments, when we co-transfected the cDNAs encoding for BRI2 and BACE1 in equal levels, we faced problems pertaining to low expression levels of BACE1. This observation led to the hypothesis that BRI2 could affect the cellular levels of BACE1. This scenario would also explain the reduced APP cleavage by BACE1 upon BRI2 expression. To explore this possibility, we co-expressed increasing amounts of BRI2 and a certain amount of BACE1 cDNA. We observed that as the levels of BRI2 protein increased the levels of BACE1 decreased whilst the levels of endogenous APP remained unchanged (Fig. 4A). However, overexpression of mycHomer3 protein had no effect on BACE1 levels. Furthermore, knock-down of endogenous BRI2 in SH-SY5Y cells using siRNAs, resulted in increased levels of BACE1 (Fig. 4B). Taken together, the above results indicate that expression of BRI2 results in reduced levels of BACE1.
Next, we sought to identify the BRI2 domain that mediates this effect. We obtained data indicating that deletion of the BRI2 N-terminal cytoplasmic domain (mycBRI2Δ1-45) or most of the C-terminal extracellular domain (mycBRI2 Δ107-266) did not affect interaction of BRI2 with BACE1 or the effect of the expression of BRI2 on the levels of BACE1 (data not shown). This data shows that the BRI2 sequence between aminoacids 45–107 is sufficient for the interaction with BACE1 and the effect of BRI2 on the protein levels of BACE1. It is interesting to note that the same sequence was found to be involved in the interaction of BRI2 with APP [8].
Our attempts then aimed at elucidating the exact mechanism through which BRI2 reduces the cellular levels of BACE1 protein. The levels of BACE1 are regulated through a variety of ways. For example, the gene that encodes for BACE1 is subjected to a tight transcriptional control and the same is true for the translation of the mRNA of the protein [28]. The possibility that BRI2 affects BACE1 transcription is excluded, since the plasmid in which BACE1 is subcloned contains a constitutive promoter. Based on our findings, we hypothesized that BRI2 reduces the levels of BACE1, at least partially, through their physical interaction. We reasoned that this could occur if BRI2 promotes BACE1 protein degradation. An initial study showed that BACE1 is degraded through the ubiquitin-proteasome pathway [23], which was then challenged by another study showing that BACE1 is degraded through the lysosomes and not the proteasome [24]. Using specific inhibitors against lysosomal (chloroquin) and proteasomal (lactacystin) degradation, we showed that in our system BACE1 is degraded through both pathways. However, only blocking the lysosomal degradation partially reversed the effect of BRI2 on BACE1 stability (Fig. 5B), implying that BRI2 could trigger BACE1 degradation through this pathway. This result is corroborated by the fact that our confocal analysis showed co-localization of the proteins in doughnut-like structures resembling the lysosomes (Fig. 3). The co-localization in other compartments implies that BRI2 could additionally affect BACE1 levels by promoting its degradation through other cellular pathways. For example, the proteins strongly co-localize in intracellular compartments adjacent to the nucleus that most likely represent the ER, making it possible that BRI2 could also have an effect on BACE1 earlier in the biosynthetic pathway.
Given that BRI2-induced degradation of BACE1 does not fully explain the effect of BRI2 on the cellular levels of the protease, we examined whether BRI2 affects the levels of BACE1 mRNA. Our results indicate that expression of BRI2 results in reduction of the mRNA levels of BACE1 by about 50% (Fig. 6). The exciting mechanism by which BRI2 affects the cellular levels of BACE1 deserves more detailed studies. To our knowledge, BRI2 is the first protein reported to have a direct impact on the levels of BACE1 protein and mRNA. At this point it is interesting to note that BRI2 has been found to increase secretion of the insulin degrading enzyme (IDE) and to promote Aβ degradation [15]. The mechanism by which BRI2 increases secretion of IDE is not known. An interesting possibility is that BRI2 could affect the levels and/or the maturation of IDE mRNA.
A previous report showed that BRI2 reduces the levels of sAPPβ, and concomitantly of Aβ, through its physical interaction with APP, by blocking the access of the secretases to their substrate [13]. Our previous data indicating that wild type BRI2 and the deletion mutants used here interact with APP [8] supports the work by Matsuda et al. [9]. Our current data does not exclude the possibility that the effect of BRI2 on APP cleavage could be partly mediated through the BRI2-APP binding.
Concerning AD therapy, the secretases that cleave APP have long been the center of attempts to control Aβ production. With regard to γ-secretase it is known that this complex has an array of physiological targets, including Notch1 receptor, and its ablation in mice leads to severe or lethal phenotypes. Even its partial ablation leads in strong autoimmune phenotypes in adulthood [29]. Before an approach to specifically inhibit γ-secretase cleavage of APP and not of other substrates is found, the possibility to use γ-secretase as a therapeutic target is distant. This statement is supported by the failure of γ-secretase inhibitors in clinical trials [30]. On the other hand, BACE1-deficient mice have only been reported to exhibit some abnormalities in specific behavioral memory tests [31–33]. However, complete inhibition of BACE1 is risky, given that this enzyme has been found to have also many putative substrates, including the LRP [34], the P-selectin glycoprotein ligand-1 (PSGL-1), [35] and the sialyl-transferase (ST6Gal) [36]. Partial inhibition of BACE1, though, could be a viable therapeutic target, since heterozygous BACE −/+ mice exhibit almost heterozygous BACE −/+ mice exhibit almost complete restoration of the memory deficits of homozygous −/− mice. Also, knockout of BACE1 in APP Tg2576 mice ameliorated Aβ pathology and restored the memory deficits observed in these mice.
BACE 1 levels and activity appear to be elevated in the AD compared to the normal brain. Although the reasons for this elevation seem complex it would be interesting to investigate whether BRI2, through its effect on BACE1 protein levels, participates in that phenomenon. On that context, examination of the protein levels of BRI2 in the brain of AD patients or Tg AD mice is vital. Surprisingly, preliminary results from our group show that BRI2 levels are indeed reduced in AD Tg mice versus their non-transgenic littermates. In conclusion, our study expands the understanding of BRI2 and BACE1 cell biology and paves the way for further examination of BACE1 and BRI2 interaction as a novel cellular target for AD therapy.
ACKNOWLEDGEMENT
This research was funded by the Alzheimer’s Association grant IIRG-09-133340 and by the NIH grant AG030539.
We thank Prof. Christian Haass for kindly providing us with the anti-BACE1 antibodies 7523 and 7520. We also thank Dr. P. Mehta for kindly providing the R1(57) anti-APP antibody.
ABBREVIATIONS
- FBD
Familial British Dementia
- FDD
Familial Danish Dementia
- ABriPP
Amyloid Bri Precursor Protein
- ADanPP
Amyloid Dan Precursor Protein
- AD
Alzheimer’s disease
- APP
Amyloid Precursor Protein
- APP/CTFs
APP C-terminal fragments
- APP/CTFα
α-secretase derived APP/CTF
- APP/CTFβ
β-secretase derived APP/CTF
- sAPPα
α-secretase secreted APP
- sAPPβ
β-secretase secreted APP
- BACE1
Beta-Site Amyloid Beta A4 Precursor Protein-cleaving Enzyme1 or β-secretase
- AICD
APP IntraCellular Domain
- SPPLs
Signal Peptide Peptidase-like proteases
Footnotes
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Vidal R, Frangione B, Rostagno A, Mead S, Revesz T, Plant G, et al. A stop-codon mutation in the BRI gene associated with familial British dementia. Nature. 1999;399:776–781. doi: 10.1038/21637. [DOI] [PubMed] [Google Scholar]
- 2.Vidal R, Revesz T, Rostagno A, Kim E, Holton JL, Bek T, et al. A decamer duplication in the 3' region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred. Proc Natl Acad Sci U S A. 2000;97:4920–4925. doi: 10.1073/pnas.080076097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SH, Creemers JW, Chu S, Thinakaran G, Sisodia SS. Proteolytic processing of familial British dementia-associated BRI variants: evidence for enhanced intracellular accumulation of amyloidogenic peptides. J Biol Chem. 2002;277:1872–1877. doi: 10.1074/jbc.M108739200. [DOI] [PubMed] [Google Scholar]
- 4.Kim SH, Wang R, Gordon DJ, Bass J, Steiner DF, Lynn DG, et al. Furin mediates enhanced production of fibrillogenic ABri peptides in familial British dementia. Nat Neurosci. 1999;2:984–988. doi: 10.1038/14783. [DOI] [PubMed] [Google Scholar]
- 5.Kim SH, Wang R, Gordon DJ, Bass J, Steiner DF, Thinakaran G, et al. Familial British dementia: expression and metabolism of BRI. Ann N Y Acad Sci. 2000;920:93–99. doi: 10.1111/j.1749-6632.2000.tb06909.x. [DOI] [PubMed] [Google Scholar]
- 6.Delacourte A. Tauopathies: recent insights into old diseases. Folia Neuropathol. 2005;43:244–257. [PubMed] [Google Scholar]
- 7.Holton JL, Lashley T, Ghiso J, Braendgaard H, Vidal R, Guerin CJ, et al. Familial Danish dementia: a novel form of cerebral amyloidosis associated with deposition of both amyloid-Dan and amyloid-beta. J Neuropathol Exp Neurol. 2002;61:254–267. doi: 10.1093/jnen/61.3.254. [DOI] [PubMed] [Google Scholar]
- 8.Fotinopoulou A, Tsachaki M, Vlavaki M, Poulopoulos A, Rostagno A, Frangione B, et al. BRI2 interacts with amyloid precursor protein (APP) and regulates amyloid beta (Abeta) production. J Biol Chem. 2005;280:30768–30772. doi: 10.1074/jbc.C500231200. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda S, Giliberto L, Matsuda Y, Davies P, McGowan E, Pickford F, et al. The familial dementia BRI2 gene binds the Alzheimer gene amyloid-beta precursor protein and inhibits amyloid-beta production. J Biol Chem. 2005;280:28912–28916. doi: 10.1074/jbc.C500217200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang YW, Thompson R, Zhang H, Xu H. APP processing in Alzheimer's disease. Mol Brain. 2011;4:3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barthet G, Georgakopoulos A, Robakis NK. Cellular mechanisms of γ-secretase substrate selection, processing and toxicity. Prog Neurobiol. 2012 Aug;98(2):166–175. doi: 10.1016/j.pneurobio.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow VW, Mattson MP, Wong PC, Gleichmann M. An overview of APP processing enzymes and products. Neuromolecular Med. 2010;12:1–12. doi: 10.1007/s12017-009-8104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda S, Giliberto L, Matsuda Y, McGowan EM, D'Adamio L. BRI2 inhibits amyloid beta-peptide precursor protein processing by interfering with the docking of secretases to the substrate. J Neurosci. 2008;28:8668–8676. doi: 10.1523/JNEUROSCI.2094-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Miller VM, Levites Y, West KJ, Zwizinski CW, Moore BD, et al. BRI2 (ITM2b) inhibits Abeta deposition in vivo. J Neurosci. 2008;28:6030–6036. doi: 10.1523/JNEUROSCI.0891-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilger E, Buehler A, Woelfing H, Kumar S, Kaeser SA, Nagarathinam A, et al. BRI2 Protein Regulates {beta}-Amyloid Degradation by Increasing Levels of Secreted Insulin-degrading Enzyme (IDE) J Biol Chem. 2011;286:37446–37457. doi: 10.1074/jbc.M111.288373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda S, Tamayev R, D'Adamio L. Increased AbetaPP Processing in Familial Danish Dementia Patients. J Alzheimers Dis. 2011 doi: 10.3233/JAD-2011-110785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giliberto L, Matsuda S, Vidal R, D'Adamio L. Generation and initial characterization of FDD knock in mice. PLoS One. 2009;4:e7900. doi: 10.1371/journal.pone.0007900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamayev R, Matsuda S, Giliberto L, Arancio O, D'Adamio L. APP heterozygosity averts memory deficit in knockin mice expressing the Danish dementia BRI2 mutant. Embo J. 2011;30:2501–2509. doi: 10.1038/emboj.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parisiadou L, Bethani I, Michaki V, Krousti K, Rapti G, Efthimiopoulos S. Homer2 and Homer3 interact with amyloid precursor protein and inhibit Abeta production. Neurobiol Dis. 2008;30:353–364. doi: 10.1016/j.nbd.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Gatta LB, Albertini A, Ravid R, Finazzi D. Levels of beta-secretase BACE and alpha-secretase ADAM10 mRNAs in Alzheimer hippocampus. Neuroreport. 2002;13:2031–2033. doi: 10.1097/00001756-200211150-00008. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda S, Matsuda Y, D'Adamio L. BRI3 inhibits amyloid precursor protein processing in a mechanistically distinct manner from its homologue dementia gene BRI2. J Biol Chem. 2009;284:15815–15825. doi: 10.1074/jbc.M109.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickham L, Benjannet S, Marcinkiewicz E, Chretien M, Seidah NG. Beta-amyloid protein converting enzyme 1 and brain-specific type II membrane protein BRI3: binding partners processed by furin. J Neurochem. 2005;92:93–102. doi: 10.1111/j.1471-4159.2004.02840.x. [DOI] [PubMed] [Google Scholar]
- 23.Qing H, Zhou W, Christensen MA, Sun X, Tong Y, Song W. Degradation of BACE by the ubiquitin-proteasome pathway. Faseb J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- 24.Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G. BACE is degraded via the lysosomal pathway. J Biol Chem. 2005;280:32499–32504. doi: 10.1074/jbc.M506199200. [DOI] [PubMed] [Google Scholar]
- 25.Lublin AL, Gandy S. Amyloid-beta oligomers: possible roles as key neurotoxins in Alzheimer's Disease. Mt Sinai J Med. 2010;77:43–49. doi: 10.1002/msj.20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakono M, Zako T. Amyloid oligomers: formation and toxicity of Abeta oligomers. Febs J. 2010;277:1348–1358. doi: 10.1111/j.1742-4658.2010.07568.x. [DOI] [PubMed] [Google Scholar]
- 27.Tsachaki M, Ghiso J, Efthimiopoulos S. BRI2 as a central protein involved in neurodegeneration. Biotechnol J. 2008;3:1548–1554. doi: 10.1002/biot.200800247. [DOI] [PubMed] [Google Scholar]
- 28.Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer's disease. Prog Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Tournoy J, Bossuyt X, Snellinx A, Regent M, Garmyn M, Serneels L, et al. Partial loss of presenilins causes seborrheic keratosis and autoimmune disease in mice. Hum Mol Genet. 2004;13:1321–1331. doi: 10.1093/hmg/ddh151. [DOI] [PubMed] [Google Scholar]
- 30.Imbimbo BP, Panza F, Frisardi V, Solfrizzi V, D'Onofrio G, Logroscino G, et al. Therapeutic intervention for Alzheimer's disease with gamma-secretase inhibitors: still a viable option? Expert Opin Investig Drugs. 2011;20:325–341. doi: 10.1517/13543784.2011.550572. [DOI] [PubMed] [Google Scholar]
- 31.Olsen MK, Roberds SL, Ellerbrock BR, Fleck TJ, McKinley DK, Gurney ME. Disease mechanisms revealed by transcription profiling in SOD1-G93A transgenic mouse spinal cord. Ann Neurol. 2001;50:730–740. doi: 10.1002/ana.1252. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, et al. Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- 33.Harrison SM, Harper AJ, Hawkins J, Duddy G, Grau E, Pugh PL, et al. BACE1 (beta-secretase) transgenic and knockout mice: identification of neurochemical deficits and behavioral changes. Mol Cell Neurosci. 2003;24:646–655. doi: 10.1016/s1044-7431(03)00227-6. [DOI] [PubMed] [Google Scholar]
- 34.von Arnim CA, Kinoshita A, Peltan ID, Tangredi MM, Herl L, Lee BM, et al. The low density lipoprotein receptor-related protein (LRP) is a novel beta-secretase (BACE1) substrate. J Biol Chem. 2005;280:17777–17785. doi: 10.1074/jbc.M414248200. [DOI] [PubMed] [Google Scholar]
- 35.Lichtenthaler SF. Ectodomain shedding of the amyloid precursor protein: cellular control mechanisms and novel modifiers. Neurodegener Dis. 2006;3:262–269. doi: 10.1159/000095265. [DOI] [PubMed] [Google Scholar]
- 36.Kitazume S, Tachida Y, Oka R, Shirotani K, Saido TC, Hashimoto Y. Alzheimer's beta-secretase, beta-site amyloid precursor proteincleaving enzyme, is responsible for cleavage secretion of a Golgiresident sialyltransferase. Proc Natl Acad Sci U S A. 2001;98:13554–13559. doi: 10.1073/pnas.241509198. [DOI] [PMC free article] [PubMed] [Google Scholar]