Abstract
Introduction:
Diabetic macular edema (DME) is a vision-threatening complication of diabetic retinopathy. The current practice of management is a trial and error method of using intravitreal antivascular endothelial growth factor (VEGF)” or steroids to treat the patient and watch the response. However, if the patient's genetic profile helps us choose appropriate medicine, it would help customize treatment option for each patient. This forms the basis of our study.
Materials and Methods:
A case-control, prospective, observational series, where DME patients were treated with bevacizumab and subclassified as treatment naοve, treatment responders, and treatment nonresponders. Blood samples of 20 subjects were studied, with five patients in each of the groups (nondiabetic- group 1, treatment naοve- group 2, treatment responder- group 3, and treatment nonresponder-group 4). Whole blood RNA extraction followed by labeling, amplification and hybridization was done, and microarray data analyzed. Genes were classified based on functional category and pathways.
Results:
The total number of genes upregulated among all three experimental groups was 5, whereas 105 genes were downregulated. There were no common genes upregulated between the responders and nonresponders. There was only one gene upregulated between the diabetic and diabetic responders posttreatment. There were 19 genes upregulated and 8 genes downregulated in the inflammatory pathway in group 2 versus group 1. There were no downregulated genes detected in vascular angiogenesis and transcription group. There were identical numbers of genes up- and downregulated in the inflammatory pathway. Seventeen genes were upreguated and 11 genes downregulated in receptor activity, which remained the predominant group in the group classification.
Discussion:
In summary, this study would provide an insight into the probable signaling mechanisms for disease pathogenesis as well as progression. This type of study eventually would aid in developing or improvising existing treatment modules with a rational approach towards personalized medicine, in future addressing the differential responses to treatment.
Keywords: Bevacizumab, diabetic macular edema, gene expression profile, microarray analysis
Diabetes is pandemic and according to the international diabetes federation report, the global prevalence of diabetes is 366 million people with 4.6 million deaths in 2011 and by 2030, it is projected to nearly double.[1]
Diabetic macular edema (DME) is a vision-threatening complication of diabetic retinopathy. The incidence of DME is estimated to be 2.3/100 person-years for the overall diabetic population and 4.5 for patients on insulin therapy.[2]
DME is caused by disruption of the blood-retinal barrier. Elevated glucose levels induce increased permeability, cytokine activation; altered blood flow, hypoxia, and inflammation.[3] Hypoxia caused by microvascular disease stimulates the release of vascular endothelial growth factor (VEGF), leading to increased vascular permeability and resultant retinal edema. Higher vitreous VEGF levels were demonstrated in eyes with macular edema compared to eyes without macular edema in diabetic patients, and these high levels correlates to severity of DME (The insufficient amount of anti-VEGF may contribute to the nonresponse of treatment. Therefore, correlates of nonresponse may reflect severity of DME).[1] Multiple studies provide evidence that progression to DME is associated with duration of disease, poor glycemic control, and the need for insulin in type 2 diabetes.[4,5]
Current protocol on management of DME depends on whether there is foveal involvement or whether vision is affected. If there is no foveal involvement, treatment is as per Early Treatment Diabetic Retinopathy Study (ETDRS) guidelines, for example, focal laser. If fovea is involved, it depends if vision is affected or not. If vision is affected, an anti-VEGF injection is considered as monotherapy. If vision is not affected, treatment is as per ETDRS guidelines, for example, focal laser.[6]
Anti-VEGFs form the mainstay of treatment of DME. Current drugs are ranibizumab (humanized antibody fragment directed at all isoforms of VEGF-A) and bevacizumab (full-size, humanized, recombinant monoclonal antibody that inactivates all VEGF isoform), Aflibercept is the latest entrant in this group but yet to be used in DME. The other category of drugs is intravitreal steroids/implants. The current practice is a” trial and error “method to treat the patient and watch the response. However, if the patient's genetic profile helps us choose the appropriate medicine, it would help us individualize our treatment for each and every patient. This will herald the era of pharmacogenomics for titrating individualized treatment.
Materials and Methods
Patient selection
The study was approved by Institutional Review Board/ Institutional Ethics Committee(IRB/IEC)) and was conducted in strict adherence to the tenets of the Declaration of Helsinki. Patients of DME (definition as per ETDRS) who presented to our tertiary eye care institute from June 2012 to January 2013 and followed the inclusion criteria were explained the nature of the study and the informed consent form was obtained. All patients who had best corrected visual acuity <6/9 and thickening on the spectral domain optical coherence tomography (SD-OCT) (criteria above 300 microns; cysts involved or diffuse) with any stage of background diabetic retinopathy, good metabolic control (mean glycosylated hemoglobin HbA1c level <7%) and normal lipid profile were included.
Patients with other ocular pathologies like glaucoma, recent cataract surgery in the last 3 months, SD-OCT suggestive of epiretinal membrane/vitreomacular traction, nephropathy and use of glitazones for diabetic control were excluded. The study was conducted on 20 subjects, with 5 patients per classified group.
All patients underwent vision testing using ETDRS charts, intraocular pressure (Perkin's tonometer), fundus evaluation (indirect ophthalmoscope and slit lamp biomicroscopy), fundus photographs (TRC NW7SF, Topcon), SD-OCT, and a fluorescein angiography (SPECTRALIS® Heidelberg). The systemic parameters evaluated for all were a baseline hemoglobin, serum lipid profile, glycosylated hemoglobin, and serum creatinine.
They were classified into three groups of treatment naive patients, responders, and nonresponders. The definition of a nonresponder was a patient, who received two successive injections of 1.25 mg bevacizumab (Avastin, Genentech/Roche) with stable/worsening/improvement <10% microns thickness on SD-OCT. The responders were those who showed a reduction in thickness >10% central retinal thickness (CRT) on SD-OCT. The control group patients were age and sex-matched nondiabetics.
RNA extraction
Ribonucleic acid (RNA) extraction was done for the whole blood samples using QIAamp RNA Blood Mini kit and then quantified using nanodrop as well as bioanalyzer.
RNA labelling, amplification, and hybridization
The samples were labeled using Agilent Quick Amp labeling Kit (Part number: 5190-0442). 500 ng of total RNA was reverse transcribed using oligodT primer tagged to T7 promoter sequence. Complementary Deoxyribonucleic Acid (cDNA), thus, obtained was converted to double stranded cDNA in the same reaction. Further the cDNA was converted to Complementary Ribonucleic Acid (cRNA) in the in-vitro transcription step using T7 RNA polymerase enzyme and Cy3 dye was added into the reaction mix. During cRNA synthesis Cy3 dye was incorporated into the newly synthesized strands. cRNA obtained was cleaned up using RNeasy columns (Qiagen). Concentration and amount of dye incorporated were determined using nanodrop. Samples that pass the QC for specific activity were taken for hybridization. A total of 600 ng of labeled cRNA were hybridized on the array (AMADID: 27114) using the Gene Expression Hybridization kit in Sure hybridization Chambers at 65°C for 16 h. Hybridized slides were washed using gene expression wash buffers (Part No: 5188-5327). The hybridized, washed microarray slides were then scanned on a G2600D scanner (Agilent Technologies).
Microarray data analysis
Significant genes up and downregulated showing 2.5-fold and above within the group of samples were identified. Differentially, regulated genes were arranged using hierarchical clustering based on Pearson coefficient correlation algorithm to identify significant gene expression patterns. Genes were classified based on functional category and pathways using GeneSpring GX Software version 11.5 and Genotypic Biointerpreter - Biological Analysis Software.
Results
Pathway-specific gene regulation
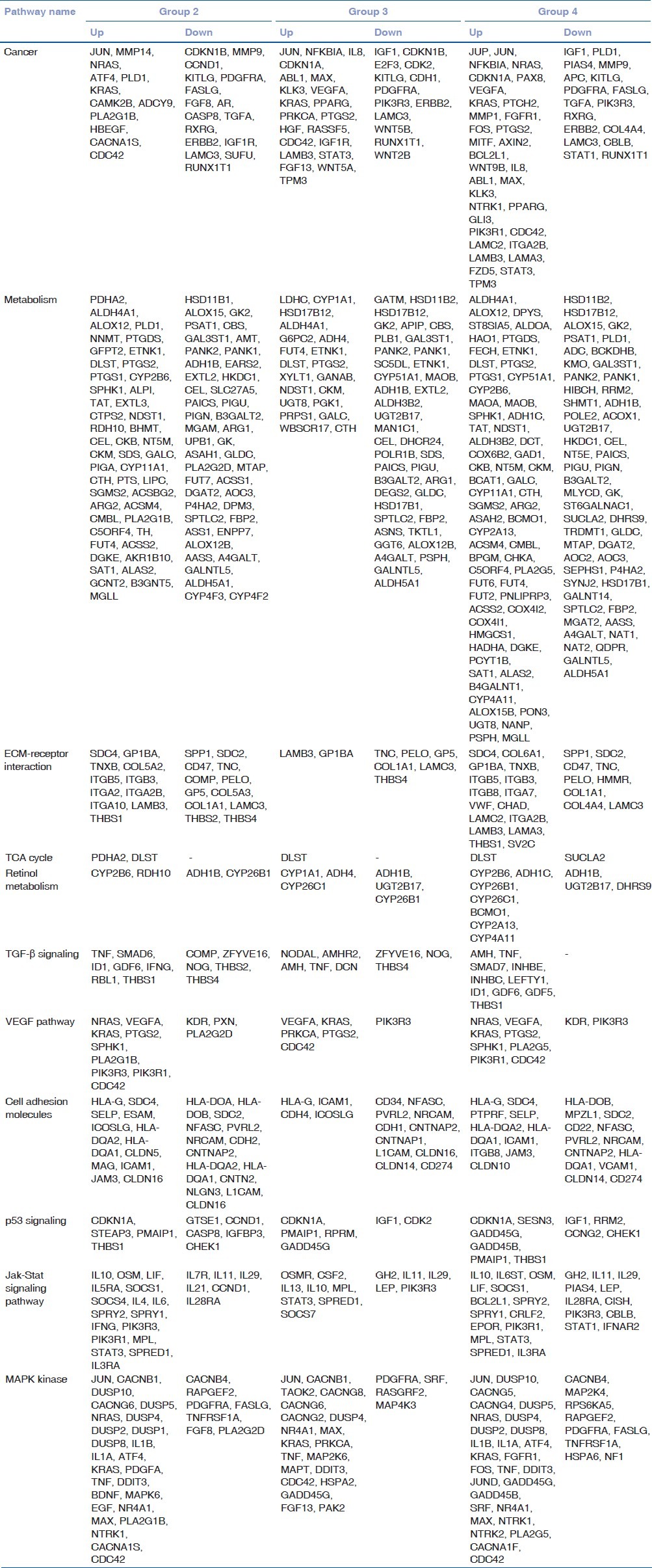
Gene expression analysis was based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. It was done in comparison to the nondiabetic control group [Table 1]. The analysis would provide the probable genes involved in the causation of diabetes mellitus. Broadly genes of cancer, metabolism, extracellular matrix (ECM)-receptor interaction, tricarboxylic acid cycle (TCA cycle), retinol metabolism, transforming growth factor-beta (TGF-β) metabolism, VEGF pathway, cell adhesion molecules, p53 signaling, Jak-Stat signaling pathway and mitogen-activated protein kinases (MAPKs) pathway were analyzed. The genes represented in the metabolic group were predominantly similar to those in the cancer group. The number of ECM receptor genes and cell adhesion molecule genes showed differences between responders and nonresponders.
Table 1.
Representative signaling genes based on KEGG pathway
The total numbers of genes upregulated among all three experimental groups were five, whereas 105 genes were downregulated. There were no common genes upregulated between the responders and nonresponders. However, one gene was detected to be upregulated between the treatment naive and responders group [Fig. 1].
Figure 1.
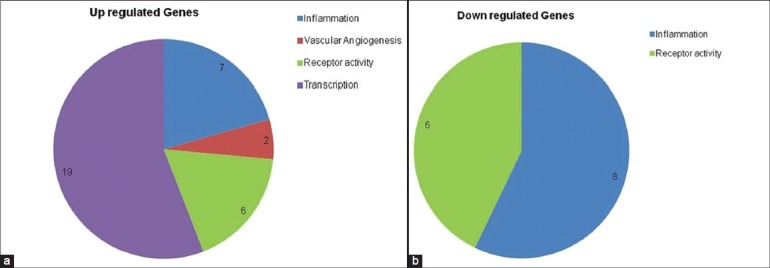
(a) Total number of genes commonly regulated across all the three experimental groups with respect to the control. Upregulated genes are depicted in Figure 3a and the downregulated genes are depicted in Figure 3b. (b) Total number of genes commonly regulated across all the three experimental groups with respect to the control. Upregulated genes are depicted in Fig 3a and the downregulated genes are depicted in Fig 3b
Gene regulation between nondiabetic and diabetic
Gene expression profile was compared between nondiabetic (Group 1) and diabetic (Group 2) patients with a cut-off fold difference of 2.5. We selected a 2.5-fold difference to eliminate detection of the noise created because of population-based variability. The inflammatory molecules, as already reported, were the defining molecules between diabetic and nondiabetic group. Vascular angiogenic genes, though a couple, were also picked in our filtered group. All the transcription factors in the filtered data set were upregulated [Table 2].
Table 2.
Microarray data analysis: Fold increase or decrease of genes between Group 2 versus Group 1 (2.5- fold)
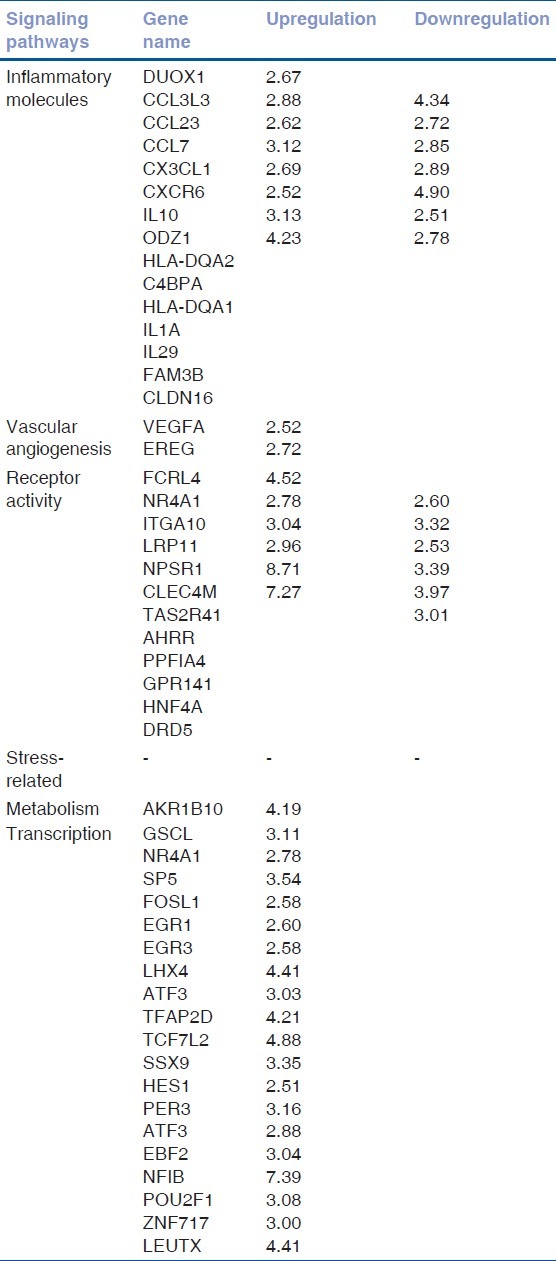
There were 19 genes upregulated and 8 downregulated genes in the inflammatory pathway in group 2 versus group 1. There were no downregulated genes detected in vascular angiogenesis and transcription group. Receptor activity genes were also regulated in group 2 versus group 1 [Fig. 2].
Figure 2.
(a) Depicting number of genes regulated with a 2.5-fold difference across the signaling pathways between diabetic (Group 2) and control group (Group 1). Upregulated genes are shown in Figure 1a and downregulated genes are shown in Figure 1b. (b) Depicting number of genes regulated with a 2.5-fold difference across the signaling pathways between diabetic (Group 2) and control group (Group 1). Upregulated genes are shown in Figure 1a and downregulated genes are shown in Figure 1b
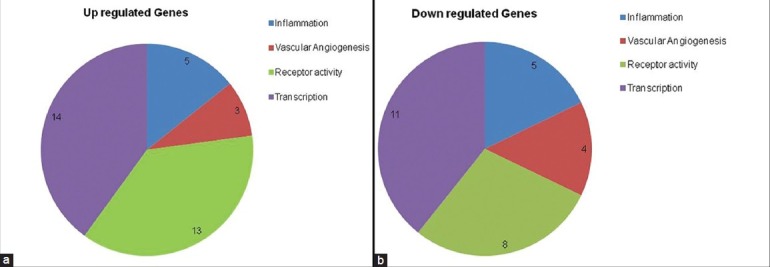
Gene regulation between nonresponder versus responder
Gene expression of the nonresponder (Group 4) was compared with the responders (Group 3) with a cut-off fold difference of 2.5. The transcription factor and receptor activity genes were dominant based on our analysis with a cut-off of 2.5 fold. Since anti-VEGF treatment was not responsive in the nonresponder group, we assumed involvement of a nonvascular pathway in segregating the treatment responders and nonresponders. But surprisingly, we noted higher expression of the vascular angiogenesis pathway genes in anti-VEGF nonresponders compared with responders [Table 3]. Transcription factors as well as stress-related genes were detected in our set filters, suggesting a role in the induction of the nonresponsive treatment parameter. There were identical numbers of genes up- and downregulated in the inflammatory pathway. A total of 17 genes were upregulated and 11 down regulated genes in receptor activity that remained the predominant group [Fig. 3].
Table 3.
Microarray data analysis: Fold increase or decrease of genes between Group 4 versus Group 3 (2.5- fold)
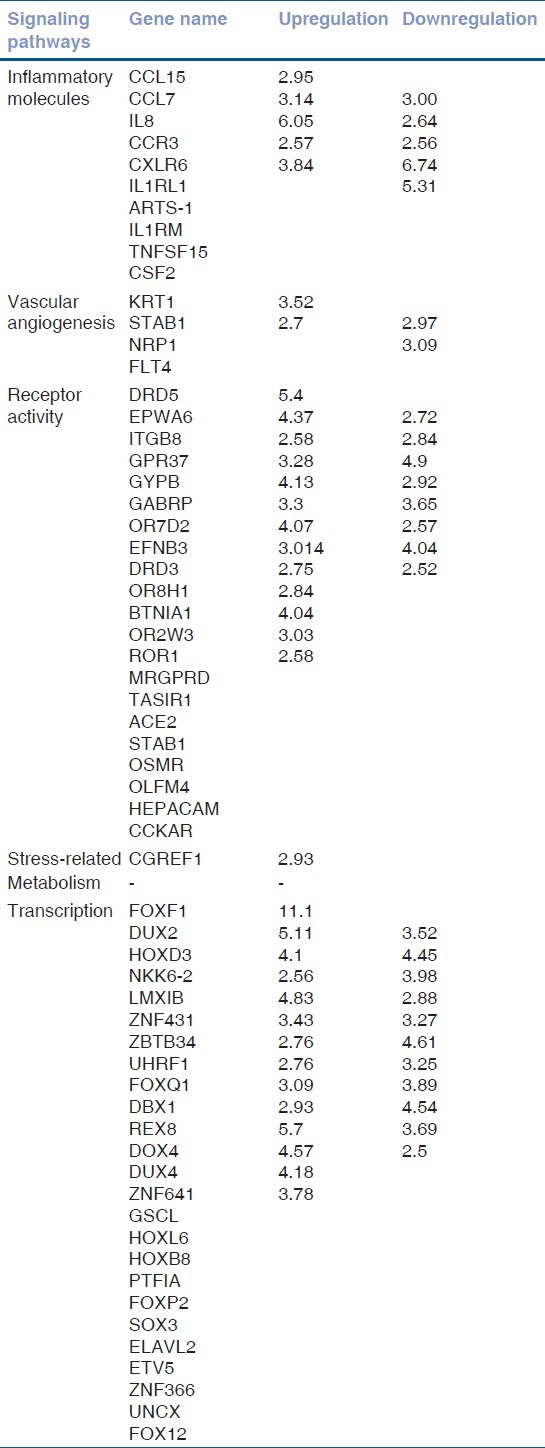
Figure 3.
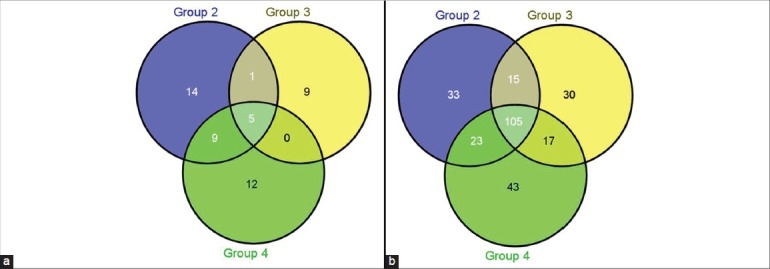
(a) Depicting number of genes regulated with a 2.5-fold difference across the signaling pathways between treatment nonresponder (Group 4) and treatment responder group (Group 3). Upregulated genes are shown in Figure 2a and downregulated genes are shown in Figure 2b. (b) Depicting number of genes regulated with a 2.5-fold difference across the signaling pathways between treatment nonresponder (Group 4) and treatment responder group (Group 3). Upregulated genes are shown in Figure 2a and downregulated genes are shown in Figure 2b
Discussion
The proposed study was to identify specific signaling pathways involved in the development of DME. There has been a distinguishing treatment outcome between DME patients based on their response to anti-VEGF treatment. So far, there is a lack of knowledge in understanding the underlying molecular mechanisms/pathways distinguishing the patient groups. Hence, we carried out a preliminary study to determine the systemic Messenger Ribonucleic Acid (mRNA) expression profile among DME patients classified based on clinical parameters. We conducted a microarray expression profile of DME, treatment naive (Group 2), treatment responders (Group 3), treatment nonresponders (Group 4) and compared all with nondiabetic (Group 1). In this pilot study, we have used whole blood mRNA profiling as it is not clinically feasible to get ocular tissues in all the groups for doing the analysis. Moreover, we hypothesized that diabetes, being a systemic disease; there might be genes and pathways beyond the limitation of eye, which have a role to play in deciphering the underlying distinction between treatment responders and nonresponders. Similar studies have been conducted to understand the pathogenesis of other retinopathies.[7] The advantage of using whole blood over retina samples relies primarily on a plausibility of identifying a biomarker to differentiate the responder and nonresponder groups.[8] In order to minimize variability in our experimental groups, the samples were pooled based on age and sex Samples of each group were in the age range between 50 and 70 years. Each group had two females and three males.
The expression profile/signaling and genes were analyzed based on KEGG pathway database (http://www.genome.jp/kegg/pathway.html). The most predominant number of genes closely related to DME is cancer-related genes. Similar to published reports, we noted an upregulation of oncogenes, cell cycle regulators, growth factor receptors and matrix metalloproteases involved in diabetes.[9,10,11,12] Dysregulation of wnt signalling pathway (WNT)/b-catenin pathway has been implicated in the complications of diabetes such as retinal inflammation, vascular leakage, and neovascularization.[13,14] WNT pathway molecules along with apoptotic regulators are most likely involved in differentiating the treatment responders and nonresponders. In Group 3, 20 genes were upregulated and 40 downregulated, whereas interestingly Group 4 (treatment nonresponders) have 63 genes upregulated and 50 downregulated, indicating probably the pivotal role of metabolic pathway post anti-VEGF treatment.[15,16] A number of proangiogenic, angiogenic, and antiangiogenic factors along with the ECM modulation play a role in diabetic retinopathy. ECM is known to have a definitive role in vascularization.[17,18,19,20] We observed the expression profile and noted that there was a drastic decrease in the number ECM-related genes in the group 3, whereas no change was detected in the number of regulated genes between group 2 and group 4. TGF-β is involved in cellular processes, survival in normal as well as disease state[21] and it was found to be downregulated in group 3 and not in group 4. This seems to indicate the probable role of TGF-b regulation along with VEGF in nonresponder DME. The downregulation of VEGF receptor (kinase insert domain receptor-KDR) in nonresponders suggest most likely the noninvolvement of VEGF pathway, or it could be the low availability of VEGF receptors as it were used up by the endogenous elevated VEGF levels. In the responder group post anti-VEGF treatment, restricted the expression of KDR. The total number of cell adhesion genes regulated in group 3 is 15 compared to 26 genes in group 4. This strongly is suggesting the involvement of higher number of cell adhesion molecules in nonresponder group of patients.[22,23,24] We were unable to detect any differences in the expression levels of mRNA in Jak-Stat and MAPK pathway, although there are reports elucidating the role of Jak-Stat and MAPK pathway in diabetic retinopathy[15] [Table 1]. Other pathways detected by the microarray gene expression profile may indicate other causes that might drive the complications of diabetes.
Inflammation seems to have a major role not only in diabetes but also in distinguishing the responder and nonresponder groups. Recent reports showed the association of several cytokines including interleukin (IL)-6, IL8, and interferon gamma in aqueous humor of DME patients.[24,25,26,27,28] The expression of IL8 in the present study is sixfold higher in nonresponder compared with responder. As mentioned earlier, the vascular angiogenesis molecules are downregulated in nonresponder indicating a canonical-VEGF independent pathway role.[29,30] In nonresponder ephrin receptor signaling and transcription factor gene families such as FOX, HOX were identified to be upregulated compared with the responders [Table 3]. There were few genes in the receptor activity pathway with fold increase as seen in diabetic group compared with nondiabetic group. A list of selective transcription factor genes were also seen upregulated in the diabetic group in comparison with nondiabetic controls[30] [Table 2].
In total of all four groups, five genes are upregulated and 105 genes are downregulated. Interestingly, there are no common genes upregulated between groups 3 and 4, whereas 17 genes were downregulated. Similar to previous reports, our study also shows involvement of inflammatory pathway molecules in diabetics compared with nondiabetic controls[30] [Fig. 1]. But, it is also noted that most of the genes regulated in the nonresponders compared with responders were grouped in receptor activity and transcriptional regulation [Fig. 3]. Inflammatory genes and vascular angiogenesis genes were the minor contributors classifying the responders and nonresponders. Transcriptional regulation is known to have an important role to play in drug response; hence, it might be indicative as an important component for classification. Role of vascular angiogenesis in classifying the responder and nonresponder remains elusive and most likely nonconsequential, as all the patients are treated with anti-VEGF therapy [Fig. 2].
This study limitations are small sample size and microarray analysis approach, as we have addressed the genes with a cut-off of 2.5-fold difference. The choice of the cut-off was based on the implication to eliminate the population-based variable gene expression and not the disease-specific gene expression profile; hence, we might have missed several signaling pathway genes, which might have a definitive role in disease pathogenesis.
To our knowledge this is the first such study, where attempts have been made to understand the signaling pathways and genes playing a definitive role at systemic level to classify the treatment responders and nonresponders. It is conceivable that there has been an overlap of genes in pathways across the analysis, and this is expected as there are a number of genes that are pivotal in cross-talks of signaling pathways rather than a single pathway. Further studies are needed to confirm the list of genes that could classify the treatment responders from nonresponders.
In summary, this study would provide an insight into the underlying mechanisms for disease pathogenesis as well as progression. This eventually would aid in developing or improvising existing treatment modules with a rational approach toward personalized medicine, in future addressing the differential responses to treatment.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
References
- 1.El-Shazly SF, El-Bradey MH, Tameesh MK. Vascular endothelial growth factor gene polymorphism prevalence inpatients with diabetic macular edema and its correlation with anti-VEGF treatment outcomes. Clin Experiment Ophthalmol. 2013 doi: 10.1111/ceo.12182. In Press. [DOI] [PubMed] [Google Scholar]
- 2.Ford JA, Lois N, Royle P, Clar C, Shyangdan D, Waugh N. Current treatments in diabetic macular oedema: Systematic review and meta-analysis. BMJ Open. 2013;3:e002269. doi: 10.1136/bmjopen-2012-002269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhagat N, Grigorian RA, Tutela A, Zarbin MA. Diabetic macular edema: Pathogenesis and treatment. Surv Ophthalmol. 2009;54:1–32. doi: 10.1016/j.survophthal.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Chen E, Looman M, Laouri M, Gallagher M, Van Nuys K, Lakdawalla D, et al. Burden of illness of diabetic macular edema: Literature review. Curr Med Res Opin. 2010;26:1587–97. doi: 10.1185/03007995.2010.482503. [DOI] [PubMed] [Google Scholar]
- 5.Klein R, Klein BE, Moss SE, Cruickshanks KJ. The Wisconsin Epidemiologic Study of Diabetic Retinopathy: XVII. The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology. 1998;105:1801–15. doi: 10.1016/S0161-6420(98)91020-X. [DOI] [PubMed] [Google Scholar]
- 6.American Academy of Ophthalmology. Preferred Practice Pattern: Diabetic Retinopathy. [Last Accessed on 2011 July 20]. http://one.aao.org/CE/PracticeGuidelines/PPP.aspx .
- 7.Pietrzyk JJ, Kwinta P, Bik-Multanowski M, Madetko-Talowska A, Jag³a M, Tomasik T, et al. New insight into the pathogenesis of retinopathy of prematurity: Assessment of whole-genome expression. Pediatr Res. 2013;73:476–83. doi: 10.1038/pr.2012.195. [DOI] [PubMed] [Google Scholar]
- 8.Parrish ML, Wright C, Rivers Y, Argilla D, Collins H, Leeson B, et al. cDNA targets improve whole blood gene expression profiling and enhance detection of pharmocodynamic biomarkers: A quantitative platform analysis. J Transl Med. 2010;8:87. doi: 10.1186/1479-5876-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin M, Kashiwagi K, Iizuka Y, Tanaka Y, Imai M, Tsukahara S. Matrix metalloproteinases in human diabetic and nondiabetic vitreous. Retina. 2001;21:28–33. doi: 10.1097/00006982-200102000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Yildirim N, Sahin A, Erol N, Kara S, Uslu S, Topbas S. The relationship between plasma MMP-9 and TIMP-2 levels and intraocular pressure elevation in diabetic patients after intravitreal triamcinolone injection. J Glaucoma. 2008;17:253–6. doi: 10.1097/IJG.0b013e31815c3a07. [DOI] [PubMed] [Google Scholar]
- 11.Mohammad G, Kowluru RA. The role of Raf-1 kinase in diabetic retinopathy. Expert Opin Ther Targets. 2011;15:357–64. doi: 10.1517/14728222.2011.553604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirwin SJ, Kanaly ST, Hansen CR, Cairns BJ, Ren M, Edelman JL. Retinal gene expression and visually evoked behavior in diabetic long evans rats. Invest Ophthalmol Vis Sci. 2011;52:7654–63. doi: 10.1167/iovs.10-6609. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida S, Shimmura S, Nagoshi N, Fukuda K, Matsuzaki Y, Okano H, et al. Isolation of multipotent neural crest-derived stem cells from the adult mouse cornea. Stem Cells. 2006;24:2714–22. doi: 10.1634/stemcells.2006-0156. [DOI] [PubMed] [Google Scholar]
- 14.Lee K, Hu Y, Ding L, Chen Y, Takahashi Y, Mott R, et al. Therapeutic potential of a monoclonal antibody blocking the Wnt pathway in diabetic retinopathy. Diabetes. 2012;61:2948–57. doi: 10.2337/db11-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speicher MA, Danis RP, Criswell M, Pratt L. Pharmacologic therapy for diabetic retinopathy. Expert Opin Emerg Drugs. 2003;8:239–50. doi: 10.1517/14728214.8.1.239. [DOI] [PubMed] [Google Scholar]
- 16.Khatami M. Regulation of MI transport in retinal pigment epithelium by sugars, amiloride, and pH gradients: Potential impairment of pump-leak balance in diabetic maculopathy. Membr Biochem. 1990;9:279–92. doi: 10.3109/09687689009025847. [DOI] [PubMed] [Google Scholar]
- 17.Costa PZ, Soares R. Neovascularization in diabetes and its complications. Unraveling the angiogenic paradox. Life Sci. 2013;92:1037–45. doi: 10.1016/j.lfs.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Praidou A, Androudi S, Brazitikos P, Karakiulakis G, Papakonstantinou E, Dimitrakos S. Angiogenic growth factors and their inhibitors in diabetic retinopathy. Curr Diabetes Rev. 2010;6:304–12. doi: 10.2174/157339910793360815. [DOI] [PubMed] [Google Scholar]
- 19.Le Goff MM, Lu H, Ugarte M, Henry S, Takanosu M, Mayne R, et al. The vitreous glycoprotein opticin inhibits preretinal neovascularization. Invest Ophthalmol Vis Sci. 2012;53:228–34. doi: 10.1167/iovs.11-8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simunovic F, Steiner D, Pfeifer D, Stark GB, Finkenzeller G, Lampert F. Increased extracellular matrix and proangiogenic factor transcription in endothelial cells after cocultivation with primary human osteoblasts. J Cell Biochem. 2013;114:1584–94. doi: 10.1002/jcb.24500. [DOI] [PubMed] [Google Scholar]
- 21.Lan HY, Chung AC. Transforming growth factor-beta and Smads. Contrib Nephrol. 2011;170:75–82. doi: 10.1159/000324949. [DOI] [PubMed] [Google Scholar]
- 22.Kloeckener-Gruissem B, Barthelmes D, Labs S, Schindler C, Kurz-Levin M, Michels S, et al. Genetic association with response to intravitreal ranibizumab in patients with neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52:4694–702. doi: 10.1167/iovs.10-6080. [DOI] [PubMed] [Google Scholar]
- 23.Knudsen ST. Ambulatory blood pressure, endothelial perturbation, and microvascular complications in type 2 diabetes. Dan Med Bull. 2010;57:B4145. [PubMed] [Google Scholar]
- 24.Meleth AD, Agrón E, Chan CC, Reed GF, Arora K, Byrnes G, et al. Serum inflammatory markers in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2005;46:4295–301. doi: 10.1167/iovs.04-1057. [DOI] [PubMed] [Google Scholar]
- 25.Lee WJ, Kang MH, Seong M, Cho HY. Comparison of aqueous concentrations of angiogenic and inflammatory cytokines in diabetic macular oedema and macular oedema due to branch retinal vein occlusion. Br J Ophthalmol. 2012;96:1426–30. doi: 10.1136/bjophthalmol-2012-301913. [DOI] [PubMed] [Google Scholar]
- 26.Jonas JB, Jonas RA, Neumaier M, Findeisen P. Cytokine concentration in aqueous humor of eyes with diabetic macular edema. Retina. 2012;32:2150–7. doi: 10.1097/IAE.0b013e3182576d07. [DOI] [PubMed] [Google Scholar]
- 27.Rangasamy S, McGuire PG, Das A. Diabetic retinopathy and inflammation: Novel therapeutic targets. Middle East Afr J Ophthalmol. 2012;19:52–9. doi: 10.4103/0974-9233.92116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koskela UE, Kuusisto SM, Nissinen AE, Savolainen MJ, Liinamaa MJ. High vitreous concentration of IL-6 and IL-8, but not of adhesion molecules in relation to plasma concentrations in proliferative diabetic retinopathy. Ophthalmic Res. 2013;49:108–14. doi: 10.1159/000342977. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimura T, Sonoda KH, Sugahara M, Mochizuki Y, Enaida H, Oshima Y, et al. Comprehensive analysis of inflammatory immune mediators in vitreoretinal diseases. PLoS One. 2009;4:e8158. doi: 10.1371/journal.pone.0008158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tremolada G, Lattanzio R, Mazzolari G, Zerbini G. The therapeutic potential of VEGF inhibition in diabetic microvascular complications. Am J Cardiovasc Drugs. 2007;7:393–8. doi: 10.2165/00129784-200707060-00002. [DOI] [PubMed] [Google Scholar]


