Abstract
While the human genome sequence is relatively uniform between the cells of an individual, the DNA methylation of the genome (methylome) has unique features in different cells, tissues and stages of development. Recent genome-wide sequencing of the methylome has revealed large partially methylated domains (PMDs) in the human placenta. Unlike CpG islands and Polycomb-regulated regions, which can also have low levels of methylation, placental PMDs cover approximately 37% of the human genome and are associated with inaccessible chromatin and the repression of tissue-specific genes. Here, we summarize the interesting biological questions that have arisen as a result of finding PMDs in the human placenta, including how PMDs form, what they do, how they evolved and how they might be relevant to human disease.
Keywords: cancer, DNA methylation, epigenetics, epigenomics, mammalian evolution, methylome, placenta, trophoblast
Short history of partially methylated domains
DNA methylation of CpG dinucleotides has been studied for decades; however, recent new techniques and discoveries have caused a major re-examination of the role of DNA methylation in gene regulation. In mammals, it was classically thought that a gene’s promoter and nearby CpG islands are methylated when the gene is transcriptionally repressed and unmethylated when the gene is active (FIGURE 1A). This was certainly true for the classic epigenetically regulated genes in which the methylated promoter correlated with the silent allele for imprinted genes or X-inactivated genes in females [1-4]. However, this led to the generally held belief that DNA methylation was an epigenetic marker of gene repression [5], a limited view of the methylome, which was due to the limitations of available technology. The gold standard for analyzing DNA methylation, Sanger bisulfite sequencing, involved labor-intensive cloning and sequencing of individual 1-kb PCR fragments (Box 1). Therefore, analyses focused on short sequences where potential methylation sites were clustered, such as promoters and CpG islands, which were most likely to have regulatory functions. However, recent advances in genome-wide sequencing technology and chemical biology have shed light on new epigenetic mysteries: hydroxymethylcytosine [6,7], non-CpG methylation [8], R loops in promoter CpG islands [9], and the diversity of DNA methylation patterns and functions in various species [10-12]. Another of these is the discovery of partially methylated domains (PMDs).
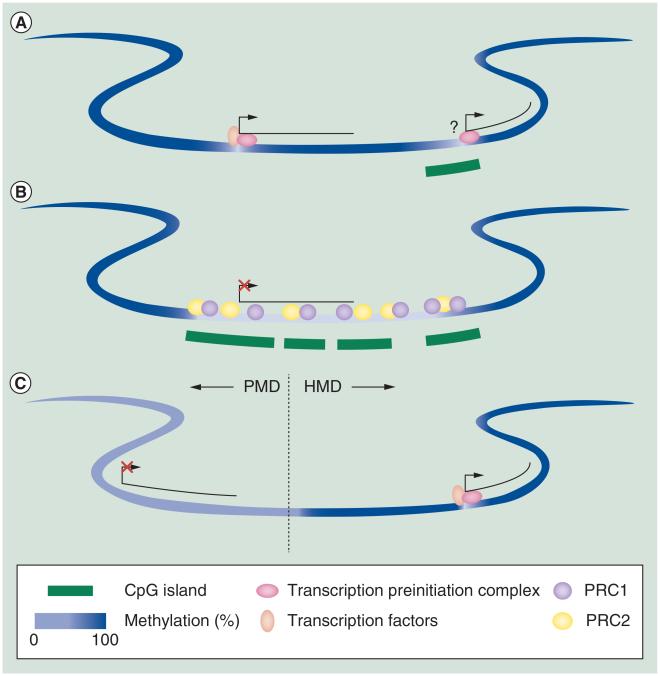
Figure 1. DNA methylation patterns in mammals.
(A) Low methylation over active gene promoters and promoter CpG islands. It is still uncertain what causes low methylation over CpG islands; (B) low methylation over Polycomb-regulated genes in tissues where the gene is repressed; (C) low methylation in PMDs where genes are typically repressed.
HMD: Highly methylated domain; PMD: Partially methylated domain.
Box 1. Why partially methylated domains were not discovered sooner: DNA methylation analysis technologies and their limitations for partially methylated domain analysis.
For decades, the limited scope of the available technology was the major hindrance to studying genome-wide DNA methylation. With the advent of DNA methylation arrays and sequencing, however, the major considerations became coverage, cost and the amount of DNA available for analysis [63,64]. The most straightforward method to expand the genome-wide gold standard Sanger bisulfite sequencing protocol is to create a sequencing library, bisulfite convert and perform whole genome sequencing (called MethylC-seq, or sometimes BS-seq, bisulfite-seq or WGSBS) [63,65,66]. However, since traditional Sanger bisulfite sequencing data usually required coverage of ten or more clones to obtain a good estimate of the percentage of methylation at individual CpG sites [8], researchers realized that it would be prohibitively expensive to get that level of coverage genome-wide using early high-throughput sequencing technologies. Instead, researchers relied on chromatin immunoprecipitation using antibodies to methylated cytosine (MeDIP-chip or MeDIP-seq), methylation-sensitive restriction enzymes (Methyl-seq and MRE-seq), restriction enzymes combined with bisulfite sequencing (reduced representation bisulfite sequencing), pull-downs using protein domains that bind to methylated DNA (MBD-seq) and methylation-specific microarrays [63,65-67]. Unfortunately, many of these technologies, such as Methyl-seq, reduced representation of bisulfite sequencing and early versions of the Illumina (CA, USA) Infinium® Methylation BeadChips, were either biased toward closely spaced CpG sites or were specifically designed with a biased targeting of CpG islands and promoters. Thus, little data were produced over CpG-poor genes and gene deserts where PMDs localize [28]. Methods such as MBD-seq and MeDIP-seq, on the other hand, were poor at detecting and discriminating intermediate levels of methylation that are observed over PMDs [68]. Although large genomic regions of low methylation (PMDs) had been detected using technologies such as padlock probes [15] and the Infinium 450k Methylation platform (Illumina) [28], it has taken the full genomic landscape coverage of MethylC-seq to uncover the significance and scope of PMDs [8,13,17,19,28]. Fortunately, given sufficient methylation differences between PMDs and HMDs in the genome, PMD locations can be detected with fairly low coverage (1–2×) using MethylC-seq, making PMD analysis experiments more economically feasible for multiple sample comparisons [13]. However, as technologies such as Illumina Methylation BeadChips platforms increase the coverage of probes over intergenic regions and gene deserts, array-based methodologies may become more useful for finding and comparing PMDs.
HMD: Highly methylated domain; PMD: Partially methylated domain.
PMDs are large domains of DNA (often greater than 100 kb) that have lower levels of DNA methylation than the rest of the genome (FIGURE 1B). A genome with PMDs can thus be divided into PMD regions and highly methylated domains (HMDs), with PMDs covering approximately 40% of the genome in human IMR90 fetal lung fibroblasts [8,13]. However, most adult human tissues do not have PMDs, instead showing a highly methylated genome of 75–80% methylation and only relatively small hypomethylated regions within CpG islands and Polycomb-regulated genes [14]. PMDs were perhaps first discovered in a partial genomic sampling of ENCODE pilot regions using bisulfite padlock probes [15]. The first definitive whole-genome discovery of PMDs came in 2009 when Lister et al. used MethylC-seq to survey the entire methylome of the human IMR90 fetal lung fibroblast cell line, demonstrating that IMR90 cells, but not the H1 embryonic stem cell (ESC) line, showed the methylomic landscape feature of PMDs [8]. PMDs were later described in human SH-SY5Y neuroblastoma cells [13], breast cancer cells [16,17] and colorectal cancer cells [18,19].
One of the difficulties of studying PMDs is the lack of definitions and consistent nomenclature within the epigenetics community (Box 2). Genomic regions of low methylation can represent promoters, CpG islands, enhancers [20-22], Polycomb-regulated regions [23], PMDs or other previously uncharacterized regulatory regions. Unfortunately, it has become common practice in the recent literature to give novel names to differentially methylated regions, leaving it up to the reader to reanalyze the data themselves to glean what type of regulatory elements/regions might be involved. Although authors often break down whether the regions are in promoters or CpG islands, classifications such as ‘genic’ and ‘intergenic’ are less useful. Another difficulty of studying PMDs is the surprising diversity of large-scale genome methylation patterns in human tissues, cell lines and cancers. In addition to the globally high methylation seen in most adult human tissues and the handful of cell lines and tissues showing PMD/HMD distinctions, there are also tissues and cell lines showing consistently lower methylation throughout their genomes, such as developing red blood cells and adipose stem cells grown in culture [24,25]. In addition, careful examination of some genomes, such as neural cells derived from human ESCs show large domains of slightly lower methylation that cover only a few genes, often those in gene family clusters [26]. It is therefore an open question as to how much of the genome may be covered by PMDs, how long PMDs should be and how great the difference in methylation must be between PMDs and HMDs for a genome to be considered to have true PMDs.
Box 2. The alphabet soup of DNA methylation: differences between differentially methylated regions, partially methylated domains, CpG islands, CpG island shores and Polycomb regions.
DMR
-
■
A term widely used in the literature to denote regions with different methylation between alleles, tissues, diseased and normal samples, treatment and control samples and individuals or organisms. There is no implied common biological mechanism or common algorithm/parameters used to define DMRs.
CpG island
-
■
Although CpG islands have been defined using a variety of algorithms and thresholds [69-72], in general they are genomic regions greater than 200 bp in length, with an unusually high CpG content. In humans, CpG islands in gene promoters tend to have low methylation, except in some cases when the gene is repressed. CpG islands in gene bodies tend to have higher methylation. Using the Gardiner–Garden and Frommer algorithm the mean CpG island length in humans is 764 bp, but they can be as long as 54,000 bp. There is probably one or more biological mechanisms that cause CpG islands to be protected at gene promoters during mammalian evolution [73-75].
CpG island shore
-
■
Often defined as the 1–2 kbs flanking both sides of a CpG island. They can have methylation levels similar to the CpG island, the surrounding genomic DNA or in between. Differential methylation within CpG island shores has been associated with colon cancer [76].
PMD and HMD
-
■
Found in genomes where large portions of the genome have HMDs interspersed with large domains of PMDs (usually greater than 100 kb in size). These domains are much larger than typical CpG islands or Polycomb-regulated regions and probably represent a unique biological phenomenon [13,28]. To date, only a handful of human cells and tissues have been found to have PMDs and HMDs.
DMV
-
■
Regions of low methylation that are at least 5 kb in length, some of which were found in all cell types examined, including human cerebral cortex, kidney, placenta, natural killer and embryonic stem cells [33].
UMR, LMR and FMR
-
■
In the study by Stadler et al. in mouse ESCs and neuronal progenitors, UMRs were found to correspond to CpG islands. LMRs were smaller than CpG islands and had properties indicative of enhancers. FMRs covered the vast majority of the genome [21].
DMR: Differentially methylated region; DMV: DNA methylation valley; FMR: Fully methylated region; HMD: Highly methylated domain; LMR: Low-methylated region; PMD: Partially methylated domain; UMR: Unmethylated region.
Much, if not all, of the above difficulties in PMD and DNA methylation definitions and nomenclature will be addressed once we have a fuller understanding of the mechanisms causing the PMD/HMD landscape and the possible role of PMDs in gene regulation. Unfortunately, to date, very little is known about what causes a cell type to form PMDs and how or why the PMDs become highly methylated in mature tissues. Furthermore, since single-cell analyses of PMDs have not been performed, it is not clear what PMDs would look like at the cellular level.
PMDs contain genes that are transcriptionally repressed [8] and often overlap H3K9me3 and H3K27me3 [12], but these chromatin marks are by no means predictive of PMD locations. PMDs do not, however, appear to be a signature of total heterochromatin since there are many genes within HMDs that are also not transcriptionally active [13]. Instead, PMDs may mark the locations of a developmentally important chromatin program for repressing tissue-specific genes in the inappropriate cell type [13]. For example, SH-SY5Y cells have PMDs covering genes involved in respiratory tube development, defense response and keratinization, among others [13]. By contrast, the IMR90 cells have PMDs covering genes involved in neuron differentiation, defense response and keratinization. Thus, the developmentally important genes in a cell are those that are uniquely not in a PMD in that cell type. For instance, many neurotransmitter receptors and neuronal adhesion molecules are in PMDs in fetal fibroblasts, but not neuroblastoma cells that express these molecules [13]. Genes within PMDs have significantly higher promoter and promoter CpG island methylation than genes within HMDs, consistent with the known association between increased promoter methylation and reduced gene expression [27].
PMDs are also distinct from Polycomb-regulated regions, which have low DNA methylation levels [23,28]. Genes regulated by the Polycomb complex encode developmentally regulated transcription factors marked in ESCs by the histone methyltransferase EZH2 and bivalency of both repressive (H3K27me3) and active H3K4me3 histone marks [29]. Although PMDs often encompass Polycomb-regulated genes, PMDs tend to be much larger and are not particularly enriched for transcription factor genes [28]. In addition, PMDs exhibit tissue-specific hypomethylation, while the much smaller hypomethylated domains over Polycomb-regulated genes are hypomethylated in most tissues and are characterized by closely spaced clusters of CpG islands. However, the mechanisms involved in creating and maintaining PMDs and how they interact with Polycomb gene repression is entirely unknown.
PMDs in the placenta: more questions than answers
Until recently it was uncertain whether PMDs were mostly a phenomenon in cancers and an artifact of the rapid cell divisions observed in cell tissue culture conditions. However, the discovery that the human placenta has PMDs, and that the PMDs are maintained from the first to third trimester [28,30], opens up whole new questions for a potential role of PMDs in normal human development.
Are PMDs only found in the placenta or are there other tissues with PMDs?
Although normal human breast and colon tissue have some indications of PMDs, recognized as slightly lower methylation levels over the regions defined as PMDs in their corresponding cancers [17,19], thus far no other human tissues have been found with evidence of clear-cut boundaries between PMDs and HMDs. This may change as more and more human tissues are analyzed by MethylC-seq. Since genes in PMDs in one tissue may be in a HMD and active in another, more functionally relevant tissue, knowing the tissue specificity of the PMD genes can give clues to other human tissues that could potentially have PMDs. Given that the genes in PMDs are involved in embryonic functions in a variety of tissues, the question becomes not where PMDs may be found, but when during development, they occur. It could be that PMDs are found only in a transient state during cellular differentiation when cell fate or maturational decisions are being made. This could make finding PMDs in other tissues more difficult, highlighting the need to discover the mechanisms surrounding PMDs to help identify other candidate cell types for MethylC-seq analysis.
However, another possibility is that the placenta is the only human tissue with PMDs. Unlike other human tissues, most of the fetal side of the human placenta never originates directly from ESCs [31]. In vivo, trophoblast cells arise from the trophectoderm, not the inner cell mass, of the blastocyst. Following fertilization, the zygote undergoes waves of demethylation, first of the paternal and then the maternal chromosomes. By the morula stage, most of the genome has low methylation. By the blastocyst stage, the inner cell mass starts to become remethylated, whereas the trophectoderm maintains low methylation [32]. This does not answer how distinct PMD and HMD regions form in the placenta, but the mechanism raises interesting questions about how PMDs form in cancer tissues and whether such a mechanism could possibly occur in other normally developing tissues.
Interestingly, two studies examined the DNA methylation patterns in trophoblast-like cells differentiated in vitro from human ESCs [25,33]. Although such cells had trophoblast characteristics [34,35], they did not have PMDs. Likewise in mouse cells, differentiation of trophoblast cells from mouse ESCs did not fully recapitulate the low levels of methylation seen in E9.5 trophectoderm tissue [36]. This could be due to either the sensitivity of trophoblast differentiation to cell culture conditions [37] or because sufficiently low levels of methylation are difficult to achieve from ESCs, which have already partially or completely undergone remethylation in the embryo.
What causes PMDs?
Some clues to the cause of the hypomethylated state of PMDs come from observations of the chromatin states within PMDs. PMDs have been found to associate with late-replicating regions in dividing cells [38]: nuclear-lamin associated domains at the nuclear periphery [19] and the repressive chromatin histone marks, H3K27me3 and H3K9me3 [20]. Thus, PMDs could mark a tissue-specific, transcriptionally repressive heterochromatic environment. Combined MethylC-seq and RNA-sequencing analyses of the placenta identified transcriptionally repressed domains that consistently overlapped with PMDs [28]. These results suggest that domain-specific transcriptional repression is deeply intertwined with heterochromatin and partial methylation, but the question of whether the heterochromatin state of gene repression is the cause or consequence of PMD formation remains.
PMDs might simply be a consequence of the heterochromatin environment and/or nuclear localization. Intriguingly, all of the cells and tissues found to contain PMDs are also in a state of rapid growth. This observation led to one hypothesis that, due to the rapid DNA replication, the maintenance DNA methyltransferase (DNMT) enzyme, DNMT1, may not have sufficient time or activity to fully methylate the DNA in the late-replicating heterochromatic regions [17]. An alternative hypothesis, suggested by the overlap with nuclear lamins, is that PMDs are hypomethylated simply because they are inaccessible to DNMT1 and/or the de novo DNMTs. Such compacted heterochromatin might also be inaccessible to transcription factors and other transcriptional activators, making the methylation state of the genes and their promoters irrelevant.
PMDs might instead be an important part of gene regulation, being an identifying mark of a unique developmental mechanism to repress unnecessary genes during cell differentiation and/or migration. In this scenario, PMDs would later be replaced by more permanent heterochromatin and histone marks in the fully mature cell, and gene repression would become independent of gene-body DNA methylation levels. However, this hypothesis opens up major questions about the causality of repression of PMD genes. Do PMDs cause gene repression or are they simply a marker of transcriptional repression in uncommitted cell types? Are PMDs hypomethylated because they are simply protected from DNMTs during development or are they actively demethylated? Alternatively, are there mechanisms that specifically methylate genomic domains to convert from a PMD to HMD state in a hypomethylated genome?
What is the significance of PMDs for placental function?
The major function of the placenta is to provide a feto–maternal interface for nutrient, oxygen and waste exchange, as well as regulating fetal growth and development through endocrine and growth factors [39]. In addition, both immune and trophoblastic cells serve as defense responders to a wide range of foreign microbes, as well as to the mother’s own immune response to fetal antigens [40]. Major problems in placental function result in early miscarriage and reproductive failure, while more subtle pathologies in placental development are observed in pregnancy complications such as preterm birth, pre-eclampsia or fetal interuterine growth restriction [41].
The discovery of PMDs in the human placenta begs the question ‘why are they in the placenta?’ Some clues can be gleaned from the function of the genes that reside in placental HMDs that are PMDs (thus repressed) in other cells and tissues. These placental HMD genes, which are specifically not repressed in placenta, could be important for placental function. Placental HMDs are enriched for genes involved in the defense response [28] and include α-defensins, α-interferons, selectins-E and -P, chemokine receptors, chemokine ligands and interleukins. Many of the genes in placental HMDs are important for normal pregnancy: low levels of IR1RN are associated with pre-eclampsia, high levels of THBD are associated with preterm birth [42,43], and CRH is a marker for the length of gestation and the timing of parturition [44]. Other genes are important for placental development and structure, including CCR1, CCR2, CCR3 and CCL14, which are thought to be involved in trophoblast migration [45] and the desmosome genes, which are involved in cell–cell adhesion in the placenta [46]. Thus, many of these genes are involved in placental development, pregnancy and immune response and their presence in the placenta-specific HMDs suggests a unique method of tissue-specific repression.
Many interesting questions remain about PMD function in the placenta. For example, an important question is which cell types in the placenta have the PMDs? Cells within the placenta are heterogeneous, including fetal trophoblasts that further differentiate into syncytiotrophoblasts, cytotrophoblasts and extravillous trophoblasts. In addition, cells of the maternal immune system are generated and reside within the placenta, creating a tolerogenic state [40]. A look at the cell specificity of placental HMD genes, discussed above, gives a confusing mix of potential cell types. For example, KLRC genes are specifically expressed in natural killer cells, α-defensins are most commonly found in neutrophils and many of the chemokine receptors are expressed by syncytiotrophoblasts [47]. It is not entirely implausible that all of these cells would have PMDs, since many immune cells are found in the placenta and some, such as decidual natural killer cells, have a specialized contingent that stays in the placenta [48]. However, it should be noted that just because a gene is not in a PMD does not mean that it is being actively expressed [13,28]. Future studies will need to isolate individual cell types from the placenta to determine their DNA methylation patterns and their potential for specific roles in placental health and fetal growth.
How did placental PMDs evolve?
Coupled with the question of PMD function in the placenta is the question of how they evolved in relation to the evolution of the mammalian placenta. Although it has long been known that the mouse placenta also has lower levels of methylation compared with other mouse tissues [49,50], it is still unknown whether the mouse placenta has PMDs. If it does have PMDs, important information could be gleaned from studies in mice. For example, it has been recently shown in mice that certain endogenous retroviruses can act as enhancers when in hypomethylated tissues, such as the placenta [51]. Another study showed that although Dnmt3a2 and Dnmt3b levels drop along with DNA methylation as mouse ESCs differentiate into trophoblasts, individual overexpression of Dnmts and Np95/Uhrf1 did not cause an increase in methylation levels in the trophoblasts, suggesting that other mechanisms may be involved that prevent remethylation [36].
It is plausible, however, that DNA methylation and regulatory mechanisms could be entirely different between mice and humans, especially considering the evolution of the placenta itself. Among placental mammals, the diversity of gross morphology and cellular organization of the placenta is remarkable, with morphology ranging from discoid/bidiscoid, zonary, cotyledonary and diffuse [52,53]. Compared with humans, mice have a different mode of implantation and trophoblast invasion is much more limited [54], although they share a discoid, hemochorial placenta type. As expected, species more closely related to humans have more similar morphology. Although prosimians (early primates) have a noninvasive epitheliochorial-type placenta, the rest of the primates have an invasive hemochorial placenta. Great apes have a similar, highly invasive placenta to humans, and pre-eclampsia has been documented in gorillas and chimpanzees, showing that they can have similar pregnancy complications [53]. It will be interesting to determine whether all mammalian placentas have low methylation and/or PMDs and, if not, how they correlate with placental morphology, maternal–fetal interactions, and species-specific placental gene regulation.
What is the significance of placental PMDs to cancer epigenetics?
Improved understanding of the placental methylome and PMDs is expected to be beneficial to diseases outside of those directly involving complications of pregnancy. In the first 10–12 weeks of human pregnancy, fetal trophoblast cells of the placenta invade the maternal uterine lining and spiral arteries, making the action of these placental cells similar to the invasive properties of cancer cells [55]. Human tumors aberrantly express placenta-specific genes, and together with germline-specific genes, placental genes can be biomarkers of aggressive metastasis in cancer [56].
Many human cancers also have a characteristic epigenomic profile, with promoter hyper-methylation of tumor suppressor genes and/or genome-wide global hypomethylation [57]. The placental methylome showed a similar epigenomic landscape to that observed in many cancers in its global hypomethylation combined with promoter hypermethylation [28]. However, the observation of PMDs through MethylC-seq in both placental and cancer methylomes may help to explain the decades-long conundrum of both hypermethylation and hypomethylation in cancer. For instance, the genomic regions of large-scale hypomethylation and focal hyper-methylation in colorectal cancers were found to coincide with nuclear lamina-associated domains, as well as PMDs [19]. Human breast cancer was also found to have large hypomethylated domains corresponding to PMDs and repressed chromatin [16,17]. However, despite the overall partial methylation of the large-scale domain-wide methylation patterns, promoter CpG island methylation was actually higher when a gene was within a PMD compared with HMD in the placenta [45]. This observation is similar to that observed in cancer, where focal areas of hypermethylation were contained within large hypomethylated domains [19].
This new genome-wide view of DNA hypoand hyper-methylation in the placenta and cancer compared with other human tissues suggests that a new perspective on methylation data in cancer is warranted. For instance, in interpreting the hypermethylation observed in the CpG island promoter of a gene of interest, it may be necessary to also investigate regions just outside of the gene to determine whether the change in the methylation state of the gene’s CpG island promoter is due to active hypermethylation of that specific gene or, alternatively, part of a more global phenomenon due to the gene being in a PMD. In addition, since PMDs are observed in the early developmental tissue of the placenta [28], could the epigenomic alterations seen in cancer possibly be due to the transformed cells becoming arrested in an earlier developmental stage? If so, methylation changes in cancer that have been assumed to be something gained in the tumor, could instead be reinterpreted as an earlier epigenetic state that is simply not lost. These genome-wide views of the methylome should be important in understanding the response of human cancers to epigenetic therapies for globally inhibiting DNA methylation [58].
Conclusion & future perspective
Our understanding of the DNA methylome, including the dynamic changes that occur in development, lineage commitment and disease states, is still in its infancy. Clearly, the advent of genome-wide sequencing approaches to DNA methylation analyses has revolutionized the field, allowing views of the methylome that had not been seen previously. PMDs have recently emerged as an unexpected landscape feature of the human methylome that may shed light on epigenetic mysteries that have long stymied the cancer field. This genome-wide view of the methylome has suggested that investigations of DNA methylation differences in human disease states needs to move beyond the low hanging fruit of CpG islands to the examination of all CpG sites in the genome. Future genome-wide investigations, including single-cell analyses, are expected to continue to improve our understanding of the role of DNA methylation patterns in development and disease.
The study of PMDs in the human placenta has raised many questions that are pertinent not only to human reproductive biology, but also tissue development, molecular mechanisms of transcriptional repression and cancer. Future investigations of genome-wide DNA methylation in additional human tissues and early cell lineages will be important to determine whether tissues other than the placenta contain PMDs and which specific cell types within the heterogeneous placental tissue contain PMDs. Clearly, understanding the mechanism of how PMDs are gained and lost in specific tissues and developmental stages will be important in interpreting their functional importance. The development of cell culture and mutant mouse models that recapitulate the gain and/or loss of PMDs will therefore be critical to dissecting the molecular pathways regulating PMDs and their functional consequences. In addition, evolutionary studies with placentas from different mammalian species, particularly our closest primate relatives, will provide important clues about when placental PMDs were gained during evolution and how they are relevant to placental function.
With a greater understanding of the function and importance of the placental epigenetic landscape, the placenta might one day be seen as a rich source of biomarkers to be collected from all births, rather than a tissue to be quickly discarded. While transient in its existence, the human placenta is a large tissue that can be stored, frozen and shared for research and diagnosis by multiple investigators. With an emerging acceptance of the ‘developmental origins of adult disease’ hypothesis [59,60], the study of placental tissues may provide critical clues to multiple common and rare diseases. Since placental tissue is like a time capsule of environmental exposures in utero, investigations of toxicologic agents, such as heavy metals, organic pollutants, pesticides and other exposures, could be performed and integrated with genetic and epigenetic investigations in prospective human studies. Placental tissue can also reveal early genetic abnormalities that were rescued in the fetus, such as confined placental mosaicism [61], which may be impacting methylation levels and patterns. Furthermore, since placental tissue is the major source of fetal DNA identified in maternal blood, assaying the placental methylome for fetal health may be important in noninvasive prenatal diagnosis [62]. Perhaps in the future, both the genome and methylome of human placentas will be sequenced at birth, providing a combined predictive roadmap of disease risks throughout the individual’s lifespan.
Executive summary.
The short history of partially methylated domains
-
■
Human placenta, fetal lung fibroblasts and some cancer tissues and cell lines have a unique methylomic feature: large domains of low methylation (partially methylated domains; PMDs) interspersed with large domains of high methylation (highly methylated domains; HMDs).
-
■
PMDs cover regions of the human genome that contain fewer genes and CpG islands and have therefore been overlooked in genome-wide studies focused on CpG islands and gene promoters.
-
■
PMDs contain tissue-specific and developmentally regulated genes that are in a repressed chromatin state. Genes in PMDs in one tissue may be in an HMD and expressed in another tissue.
PMDs in placenta: more questions than answers
-
■
PMDs may be unique to the placenta because of the distinct cell lineage of trophoblast cells from early embryonic tissues. Alternatively, PMDs could be an undiscovered transient state in many developing human tissues.
-
■
It is still an open question whether PMDs are a side effect of chromatin inaccessibility or are an active part of a mechanism for gene repression.
-
■
Genes that are in placenta-specific HMDs are involved in placenta development, pregnancy and immune response. More studies will be needed to determine which of the many cell types in the placenta contain PMDs.
-
■
While hypomethylation of placenta is observed in mice, it is not currently clear whether the landscape of PMDs and HMDs is conserved across placental mammals given the diversity of placental types.
-
■
Since trophoblast cells in the placenta are tumor-like in their invasive properties, it may not be surprising that the epigenomic landscape of cancer resembles that of the placenta, including promoter hypermethylation and global hypomethylation.
Future perspective
-
■
PMDs in human placental tissue could be useful for identifying epigenetic biomarkers relevant for human disease.
-
■
Future studies will be required to address many outstanding questions that are raised by the presence of PMDs in the placenta, including developmental, evolutionary and mechanistic investigations.
Acknowledgments
The authors thank the NIH (R01HD041462 and R01ES021707) and the Department of Defense (1210491) for research support in this area.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Razin A, Shemer R. DNA methylation in early development. Hum. Mol. Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 2.Lalande M. Parental imprinting and human disease. Ann. Rev. Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- 3.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 1992;51(6):1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 4.Norris DP, Patel D, Kay GF, et al. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77(1):41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 5.Bird AP, Wolffe AP. Methylation-induced repression – belts, braces, and chromatin. Cell. 1999;99(5):451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 6.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ■■.First paper to describe the human methylome at base-pair resolution. The authors discovered partially methylated domains (PMDs) in human fetal lung fibroblast (IMR90) cells and coined the name PMD.
- 9.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012;45(6):814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 11.Feng S, Cokus SJ, Zhang X, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veluchamy A, Lin X, Maumus F, et al. Insights into the role of DNA methylation in diatoms by genome-wide profiling in Phaeodactylum tricornutum. Nat. Commun. 2013;4:2091. doi: 10.1038/ncomms3091. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder DI, Lott P, Korf I, LaSalle JM. Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res. 2011;21(10):1583–1591. doi: 10.1101/gr.119131.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orlando DA, Guenther MG, Frampton GM, Young RA. CpG island structure and trithorax/Polycomb chromatin domains in human cells. Genomics. 2012;100(5):320–326. doi: 10.1016/j.ygeno.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27(4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shann YJ, Cheng C, Chiao CH, Chen DT, Li PH, Hsu MT. Genome-wide mapping and characterization of hypomethylated sites in human tissues and breast cancer cell lines. Genome Res. 2008;18(5):791–801. doi: 10.1101/gr.070961.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hon GC, Hawkins RD, Caballero OL, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22(2):246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ■.This study found PMDs in low-passage breast cancer cells and showed that the genes within them are transcriptionally repressed.
- 18.Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2012;44(1):40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ■.The study found PMDs in colon cancer tissue. PMDs correlated with regions of late DNA replication and attachment to the nuclear lamina.
- 20.Hawkins RD, Hon GC, Lee LK, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6(5):479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadler MB, Murr R, Burger L, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 22.Ong CT, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13(5):423–430. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch MD, Smith AJ, De Gobbi M, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31(2):317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334(6057):799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins-Taylor K, Schroeder DI, LaSalle JM, Lalande M, Xu RH. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics. 2012;7(1):71–82. doi: 10.4161/epi.7.1.18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr. Top. Microbiol. Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 28.Schroeder DI, Blair JD, Lott P, et al. The human placenta methylome. Proc. Natl Acad. Sci. USA. 2013;110(15):6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ■■.First description of PMDs in a normal human tissue, the placenta.
- 29.Mendenhall EM, Bernstein BE. Chromatin state maps: new technologies, new insights. Curr. Opin. Genet. Dev. 2008;18(2):109–115. doi: 10.1016/j.gde.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu T, Handley D, Bunce K, Surti U, Hogge WA, Peters DG. Structural and regulatory characterization of the placental epigenome at its maternal interface. PLoS ONE. 2011;6(2):el4723. doi: 10.1371/journal.pone.0014723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rugg-Gunn PJ. Epigenetic features of the mouse trophoblast. Reprod. Biomed. Online. 2012;25(1):21–30. doi: 10.1016/j.rbmo.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013;368(1609):20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie W, Schultz MD, Lister R, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu RH, Chen X, Li DS, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 2002;20(12):1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 35.Amita M, Adachi K, Alexenko AP, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc. Natl Acad. Sci. USA. 2013;110(13):e1212–e1221. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oda M, Oxley D, Dean W, Reik W. Regulation of lineage specific DNA hypomethylation in mouse trophectoderm. PLoS ONE. 2013;8(6):e68846. doi: 10.1371/journal.pone.0068846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giakoumopoulos M, Golos TG. Embryonic stem cell-derived trophoblast differentiation: a comparative review of the biology, function, and signaling mechanisms. J. Endocrinol. 2013;216(3):R33–R45. doi: 10.1530/JOE-12-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2011;20(4):670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 39.Koukoura O, Sifakis S, Spandidos DA. DNA methylation in the human placenta and fetal growth (review) Mol. Med. Rep. 2012;5(4):883–889. doi: 10.3892/mmr.2012.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat. Med. 2013;19(5):548–556. doi: 10.1038/nm.3160. [DOI] [PubMed] [Google Scholar]
- 41.Sterner K, Jameson N, Wildman D. Placental development, evolution, and epigenetics of primate pregnancies. In: Clancy K, editor. Building Babies: Primate Development in Proximate and Ultimate Perspective. Springer; NY, USA: 2013. pp. 55–83. [Google Scholar]
- 42.Faupel-Badger JM, Wang Y, Karumanchi SA, et al. Associations of pregnancy characteristics with maternal and cord steroid hormones, angiogenic factors, and insulin-like growth factor axis. Cancer Causes Control. 2011;22(11):1587–1595. doi: 10.1007/s10552-011-9835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faupel-Badger JM, Fichorova RN, Allred EN, et al. Cluster analysis of placental inflammatory proteins can distinguish preeclampsia from preterm labor and premature membrane rupture in singleton deliveries less than 28 weeks of gestation. Am. J. Reprod. Immunol. 2011;66(6):488–494. doi: 10.1111/j.1600-0897.2011.01023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abou-Seif C, Shipman KL, Allars M, et al. Tissue specific epigenetic differences in CRH gene expression. Front. Biosci. 2012;17:713–725. doi: 10.2741/3953. [DOI] [PubMed] [Google Scholar]
- 45.Hannan NJ, Jones RL, White CA, Salamonsen LA. The chemokines, CX3CL1, CCL14, and CCL4, promote human trophoblast migration at the feto–maternal interface. Biol. Reprod. 2006;74(5):896–904. doi: 10.1095/biolreprod.105.045518. [DOI] [PubMed] [Google Scholar]
- 46.Aplin JD, Jones CJ, Harris LK. Adhesion molecules in human trophoblast – a review. I. Villous trophoblast. Placenta. 2009;30(4):293–298. doi: 10.1016/j.placenta.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Douglas GC, Thirkill TL, Sideris V, Rabieh M, Trollinger D, Nuccitelli R. Chemokine receptor expression by human syncytiotrophoblast. J. Reprod. Immunol. 2001;49(2):97–114. doi: 10.1016/s0165-0378(00)00083-8. [DOI] [PubMed] [Google Scholar]
- 48.Hanna J, Goldman-Wohl D, Hamani Y, et al. Decidual NK cells regulate key developmental processes at the human fetal–maternal interface. Nat. Med. 2006;12(9):1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 49.Fanning TG, Hu WS, Cardiff RD. Analysis of tissue-specific methylation patterns of mouse mammary tumor virus DNA by two-dimensional Southern blotting. J. Virol. 1985;54(3):726–730. doi: 10.1128/jvi.54.3.726-730.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu WS, Fanning TG, Cardiff RD. Mouse mammary tumor virus: specific methylation patterns of proviral DNA in normal mouse tissues. J. Virol. 1984;49(1):66–71. doi: 10.1128/jvi.49.1.66-71.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013;45(3):325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ■.Showed that low methylation in mouse placenta may allow endogenous retroviruses to act as enhancers in that tissue.
- 52.Carter AM, Enders AC. Comparative aspects of trophoblast development and placentation. Reprod. Biol. Endocrinol. 2004;2:46. doi: 10.1186/1477-7827-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carter AM, Pijnenborg R. Evolution of invasive placentation with special reference to non-human primates. BestPract. Res. Clin. Obstet. Gynaecol. 2011;25(3):249–257. doi: 10.1016/j.bpobgyn.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 54.Carter AM. Animal models of human placentation – a review. Placenta. 2007;28(Suppl. A):S41–S47. doi: 10.1016/j.placenta.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 55.van Dijk M, Visser A, Posthuma J, Poutsma A, Oudejans CB. Naturally occurring variation in trophoblast invasion as a source of novel (epigenetic) biomarkers. Front. Genet. 2012;3:22. doi: 10.3389/fgene.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 2013;5(186):186ra166. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kulis M, Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 58.Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat. Rev. Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barker DJ. The developmental origins of adult disease. Eur. J. Epidemiol. 2003;18(8):733–736. doi: 10.1023/a:1025388901248. [DOI] [PubMed] [Google Scholar]
- 60.Barker DJ, Eriksson JG, Forsen T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int. J. Epidemiol. 2002;31(6):1235–1239. doi: 10.1093/ije/31.6.1235. [DOI] [PubMed] [Google Scholar]
- 61.Lau AW, Brown CJ, Penaherrera M, S, Kalousek DK, Robinson WP. Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am. J. Hum. Genet. 1997;61(6):1353–1361. doi: 10.1086/301651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lun FM, Chiu RW, Sun K, et al. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin. Chem. 2013 doi: 10.1373/clinchem.2013.212274. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 63.Cullum R, Alder O, Hoodless PA. The next generation: using new sequencing technologies to analyse gene regulation. Respirology. 2011;16(2):210–222. doi: 10.1111/j.1440-1843.2010.01899.x. [DOI] [PubMed] [Google Scholar]
- 64.Gargiulo G, Minucci S. Epigenomic profiling of cancer cells. Int. J. Biochem. Cell Biol. 2009;41(1):127–135. doi: 10.1016/j.biocel.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 65.Nagarajan RP, Fouse SD, Bell RJ, Costello JF. Methods for cancer epigenome analysis. Adv. Exp. Med. Biol. 2013;754:313–338. doi: 10.1007/978-1-4419-9967-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J. Med. Genet. 2011;48(11):721–730. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 67.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 68.Harris RA, Wang T, Coarfa C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28(10):1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 70.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanay A, O’Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc. Natl Acad. Sci. USA. 2007;104(13):5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP. Redefining CpG islands using hidden Markov models. Biostatistics. 2010;11(3):499–514. doi: 10.1093/biostatistics/kxq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 75.Ginno PA, Lim YW, Lott PL, Korf IF, Chedin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23(10):1590–600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]