Abstract
We describe here the development of site-specific antibody-polymer conjugates (APCs) for the selective delivery of small interference RNAs (siRNAs) to target cells. APCs were synthesized in good yields by conjugating an aminooxy-derivatized cationic block copolymer to an anti-HER2 Fab or full length IgG by means of genetically encoded para-acetyl phenylalanine (pAcF). The APCs all showed comparable binding affinity to HER2 as their native counterparts and no significant cellular cytotoxicity. Mutant S202-pAcF Fab and Q389-pAcF IgG polymer conjugates specifically delivered siRNAs to HER2+ cells and mediated potent gene silencing at both the mRNA and protein levels. However, a mutant A121-pAcF IgG polymer conjugate, despite its high binding affinity to HER2 antigen, did not induce a significant RNA interference response in HER2+ cells, presumably due to steric interference with antigen binding and internalization. These results highlight the importance of conjugation site on the activity of antibody-polymer based therapeutics and suggest that such chemically-defined APCs may afford a useful targeted delivery platform for siRNAs or other nucleic acid based therapies.
Keywords: antibody-polymer conjugate, unnatural amino acid, siRNA delivery, site-dependence
1. INTRODUCTION
The selective silencing of gene expression by small interference RNA (siRNA) is a promising approach for the treatment of a variety of human diseases, including cancer, metabolic, neurodegenerative and infectious disease.1 However, a major obstacle to the clinical application of RNA interference (RNAi) therapy is the lack of efficient methods for the delivery of siRNAs to the target cells.1g To improve selective cellular uptake, decrease the overall dosage of siRNAs required for effective RNAi and minimize off-target silencing in other tissues, a tissue specific delivery system is highly desired.2 To this end, a number of ligands that selectively bind tissue associated antigens have been explored for targeted siRNA delivery including a ScFv-9R conjugate,3 antibody-protamine fusion proteins,4 aptamer-siRNA chimeras,5 cholesterol6 and folate7-siRNA conjugates and a CpG oligonucleotide-siRNA conjugate.8 However, these systems in general suffer from problems such as siRNA degradation, toll-like receptor 7 (TLR 7) mediated immune responses, off-target gene silencing, and cytotoxicity. Various nanostructures coated with targeting ligands9 including, for example, a leukocyte-targeted lipid nanoparticle coated with anti-β7 integrin antibody8j, have also been exploited for the selective delivery of siRNAs. However, conjugation of ligands such as antibodies to the surfaces of these nanostructures using conventional nonselective electrophilic conjugation chemistry generally leads to low ligand density, decreased binding affinity and increased heterogeneity in the final materials.
In contrast to siRNAs, small molecule toxins can be efficiently delivered to tumor cells as antibody conjugates and have demonstrated impressive efficacy in the clinic (e.g., Adcetris and Kadcyla). Moreover, it has recently been shown that the site-specific conjugation of drugs to antibodies can result in improved efficacy, pharmacokinetics and therapeutic index relative to conventional nonspecific antibody-conjugates.10 One method for generating site-specific antibody-conjugates involves the genetic incorporation of unnatural amino acids (UAAs)11 into antibodies whose chemical reactivity is orthogonal to the twenty canonical amino acids. Specifically, a ketone-containing UAA, para-acetyl l-phenylalanine12 (pAcF, Fig. 1A), is genetically incorporated into the antibody at selected sites in response to the amber TAG codon using an evolved orthogonal tRNA/aminoacyl-tRNA synthetase pair specific for pAcF. The ketone group of pAcF can be selectively coupled to aminooxy derivatized ligands by formation of a stable oxime bond under mild reaction conditions. Indeed, several site-specific antibody-conjugates armed with various effector functions have been efficiently synthesized13 by this method and have shown potent and selective in vitro and in vivo activity against tumor cells.
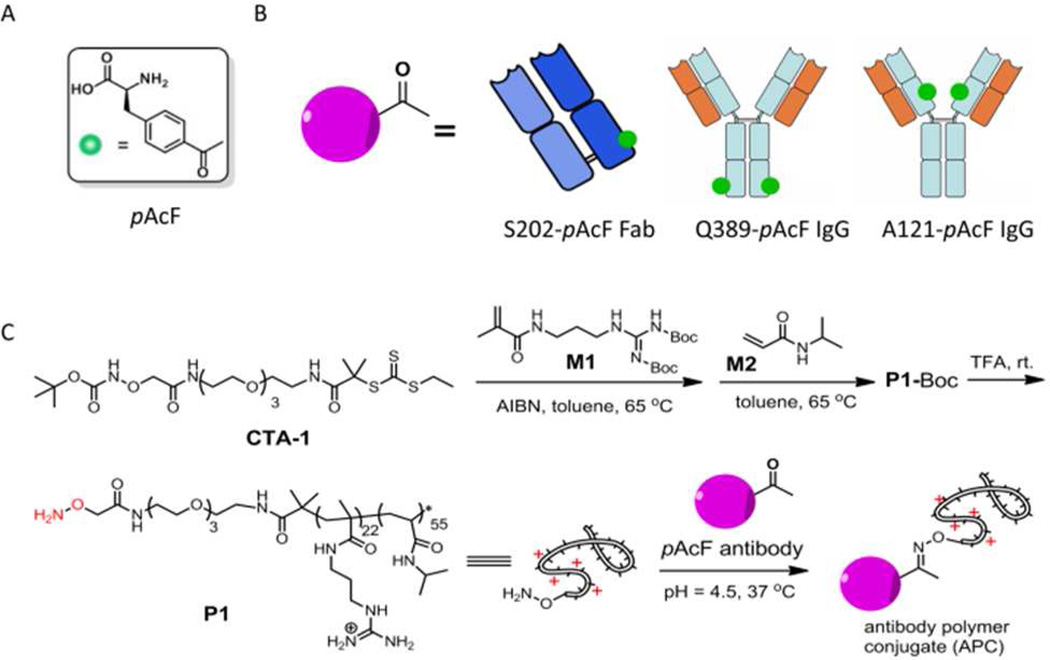
Figure 1.
(A) Structure of pAcF. (B) Schematic illustration of pAcF mutated anti-HER2 antibodies, S202-pAcF Fab, Q389-pAcF IgG and A121-pAcF IgG. (C) Synthetic scheme for the antibody-polymer conjugates.
On the basis of the above studies, we reasoned that the site-specific conjugation of cationic polymers to antibodies which bind selectively to tumor associated antigens and are efficiently internalized, might potentially provide a targeted siRNA delivery system. In this case, the cationic polymer acts to both reversibly bind the siRNA as well as to facilitate the release of the payload from the endosomal/lysosomal compartments in the cell. When combined with modern controlled polymerization technologies, these site-specific antibody-polymer conjugates (APCs) may offer advantages over more conventional targeted polymeric siRNA delivery systems in that they provide more homogenous, chemically defined structures that can be optimized with respect to affinity, physical properties, and pharmaceutics, efficacy, and ease of formulation.
Here we describe the synthesis of three site-specific APCs (S202-Fab-P1, Q389-IgG-P1 and A121-IgG-P1) through the keto group of the coupling reaction between an aminooxy-tethered cationic polymer P1 and the pAcF mutant anti-HER2 antibodies, S202-pAcF Fab, Q389-pAcF IgG and A121-pAcF IgG (Fig. 1B). Furthermore, we show that siRNA complexes of S202-Fab-P1 and Q389-IgG-P1 can be efficiently and selectively delivered to HER2+ cancer cells without significant cellular cytotoxicity. Surprisingly, we observed that the site of conjugation (e.g., Q389-IgG-P1 vs A121-IgG-P1) can have a very significant impact on the in vitro activity of the APC/siRNA complexes. This is the first example of a site-specific APC for targeted siRNA delivery and underscores the importance of conjugation site on the biological activity of the APC/siRNA complexes.
2. RESULTS AND DISCUSSION
2.1 Synthesis and Characterizations of HER2-Fab-P1
Trastuzumab (Herceptin), a humanized antibody used for the treatment of HER2+ breast cancers, binds to the extracellular juxta-membrane domain of HER2, a member of the epidermal growth factor receptor family that is highly overexpressed in approximately one third of breast cancers, and is rapidly internalized into tumor cells.14 On the basis of the Herceptin crystal structure, three solvent exposed residues (S202 on the light chain; A121 and Q389 on the heavy chain) of which showed minimal perturbation on antibody expression and binding affinity in previous studies, were selected for substitution by pAcF.12a,15 To site-specifically incorporate pAcF into the anti-HER2 Fab fragment at position S202 on the light chain (S202-pAcF Fab), an orthogonal amber suppressor tRNA/aaRS pair, derived from the tyrosyl Methanococcus jannaschii pair and evolved to selectively incorporate pAcF,15 was coexpressed with the anti-HER2 Fab gene containing a TAG codon at S202 on the light chain in the presence of pAcF. The yield of the mutant S202-pAcF Fab was ~2 mg/L when grown in shake flasks and > 400 mg/L in high-density fermentors after periplasmic lysis and protein G purification. To generate an anti-HER2 IgG with pAcF substituted for A121 (A121-pAcF IgG) or Q389 (Q389-pAcF IgG) on the heavy chain, an amber codon was substituted in the full-length anti-Her2 IgG1 gene at either heavy-chain residue A121 (HC-A121X) or Q389 (HC-Q389X) (a His-tag was added at the C-terminus of the heavy chain separately for the Q389-pAcF IgG). The mutant A121-pAcF and Q389-pAcF IgGs were produced in suspension Chinese hamster ovary (CHO-K1) cells by transient trans-fection in yields of ~3 mg/L.
We next synthesized the cationic polymer P1 with an ami-nooxy group at one terminus. P1 contains a cationic domain, consisting of side-chain guanidinium groups, which has been shown to endow membrane penetrating activity to polymers regardless of their backbones,16 and a poly(N-isopropylacrylamide) (PNIPAAm) domain to modulate the overall hydrophobicity of P1. Briefly, we first synthesized CTA-1 (Fig. 1C) as a chain transfer agent (CTA) for reversible addition-fragmentation chain-transfer polymerization (RAFT) (Scheme. S1). CTA-1 contains a Boc-protected ami-nooxy group, which provides a reactive handle for conjugation with antibodies after deprotection. Successive polymerization of monomers M1 and M2 (Fig. 1C) in the presence of CTA-1 and AIBN yielded block copolymer P1-Boc. Gel permeation chromatography (GPC) showed the molecular weight (MW) and polydispersity (PDI) of P1-Boc to be 11.2 kDa and 1.29, respectively (Fig. S1). Deprotection of P1-Boc by TFA and subsequent extensive dialysis against ultra-pure water yielded the highly water-soluble, cationic polymer P1 bearing an ami-nooxy group. The MW of P1 is ~10.1 kDa and the average degree of polymerization (DP) of M1 and M2 are ~ 22 and 55, respectively, as determined by 1H NMR spectroscopy (Fig. S2). Overall, the MW of P1 (determined by 1H NMR) agreed well with the MW of its precursor P1-Boc (determined by GPC).
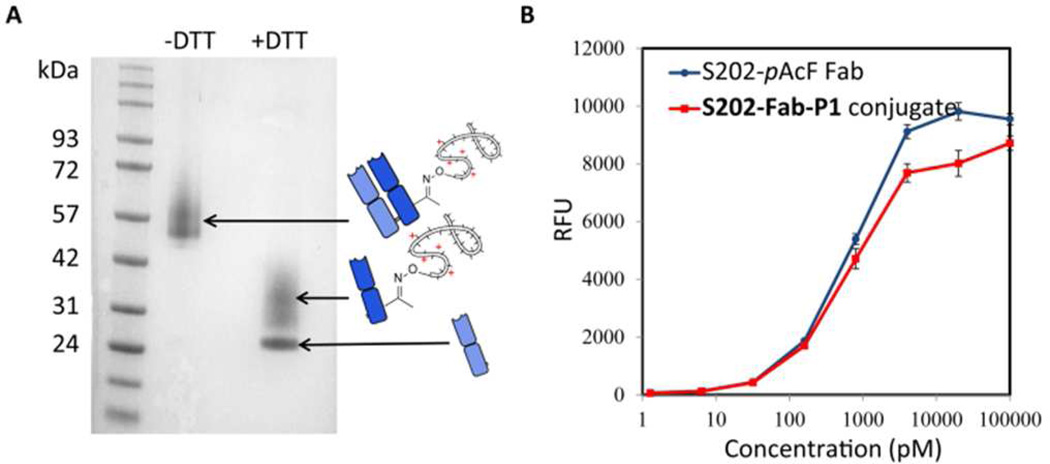
We next generated an APC (S202-Fab-P1) by incubating the S202-pAcF Fab (~ 5 mg/mL) and P1 (~100 mg/mL) in 100 mM acetate buffer (pH 4.5) at 37 °C for 3–5 days. Purification by protein G and subsequent size exclusion chromatography removed unreacted starting materials and afforded the pure conjugate in about 60% yield. SDS-PAGE gel (Fig.2A) analysis of S202-Fab-P1 showed a diffuse band from ~50–65 kDa under non-reducing conditions, indicating formation of the APC. Under reducing conditions (10 mM DTT), a discrete band at ~24 kDa and a diffuse band at ~26–42 kDa were observed for the APC, which correspond to the native heavy chain and the light chain-P1 conjugate, respectively. An enzyme-linked immunosorbent assay (ELISA) showed S202-Fab-P1 bound to HER2 antigen with similar affinity to S202-pAcF Fab (EC50 values of 0.6 ± 0.2 nM and 1.2 ± 0.3 nM, respectively) (Fig. 2B), indicating that the binding of S202-pAcF Fab to antigen was not impaired by P1 conjugation. The cationic P1 of S202-Fab-P1 should form a complex with the negatively charged siRNAs. As expected, a gel retardation assay showed that S202-Fab-P1 binds siRNA completely at molar ratio of 1.5/1 or higher after 1 min incubation at room temperature in PBS (pH = 7.4) (Fig. S3). Dynamic light scattering (DLS) further showed that a complex with average size about 50–60 nm formed when S202-Fab-P1 and siRNA were mixed at a 2/1 molar ratio under the same conditions (Fig. S4).
Figure 2.
SDS-PAGE gel and ELISA assay of S202-Fab-P1. (A) S202-Fab-P1 was analyzed by SDS-PAGE under both non-reducing (−DTT) and reducing (+DTT, 10 mM) conditions and stained with Coomassive brilliant blue. (B) ELISA assay of anti-HER2 S202-pAcF Fab and S202-Fab-P1 binding to HER2 antigen. An anti-human Ig kappa chain-HRP conjugate was used as the secondary antibody and binding affinity was assayed using fluoroscence by QuantaBlu Fluorogenic Peroxidase Substrate.
2.2 S202-Fab-P1 Selectively Delivers FITC-siRNA to HER2+ Cells
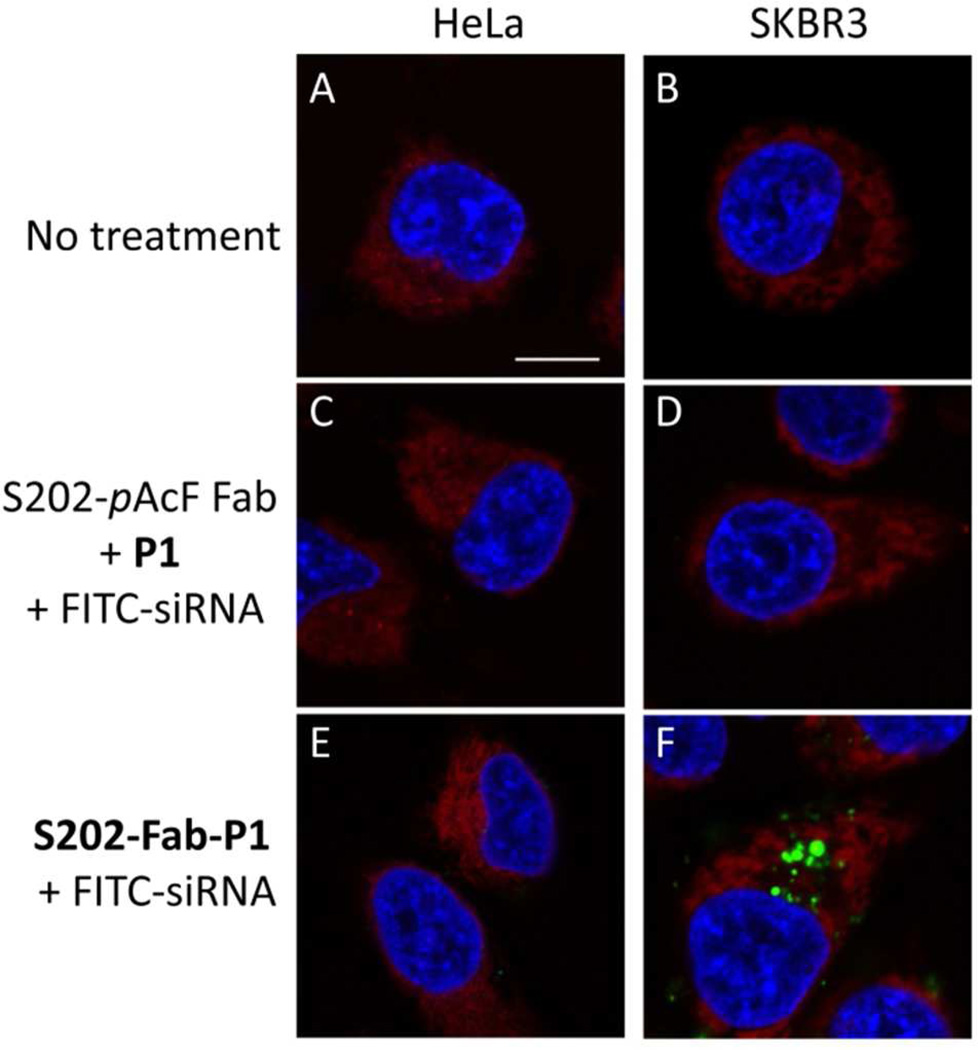
To test the ability of S202-Fab-P1 to selectively deliver siRNAs to the target cells, either the APC (200 nM) or a control mixture of anti-HER2 S202-pAcF Fab and P1 (200 nM of each) were mixed with FITC-siRNA (from Dharmacom|Thermo Scientific Inc., 50 nM) at room temperature and then added to SKBR3 (HER2+ human breast cancer) or HeLa (HER2− human cervical cancer) cell cultures. Four hours after treatment, cells were washed multiple times with cold PBS, labeled with Hoechst 33342 (blue) and ER-Tracker (red), and observed under a LSM 710 confocal microscope. Unlike the mixture of S202-pAcF Fab and P1, which showed non-detectable FITC fluorescence in both SKBR-3 and HeLa cells, S202-Fab-P1 affords strong FITC fluorescence in HER2+ positive SKBR-3 cells, but not in HER2− HeLa cells (Fig. 2). Cell viability assays showed that treatment with complexes of S202-Fab-P1/scrambled-siRNA resulted in no obvious cytotoxicity at concentrations up to 300 nM/100 nM (Fig. S5). These results indicate that S202-Fab-P1 can effectively deliver siRNAs to the HER2+ cells at concentrations where it is not toxic.
2.3 S202-Fab-P/siRNA Complexes Selectively Knockdown Genes to HER2+ Cells in a HER2-dependent Manner
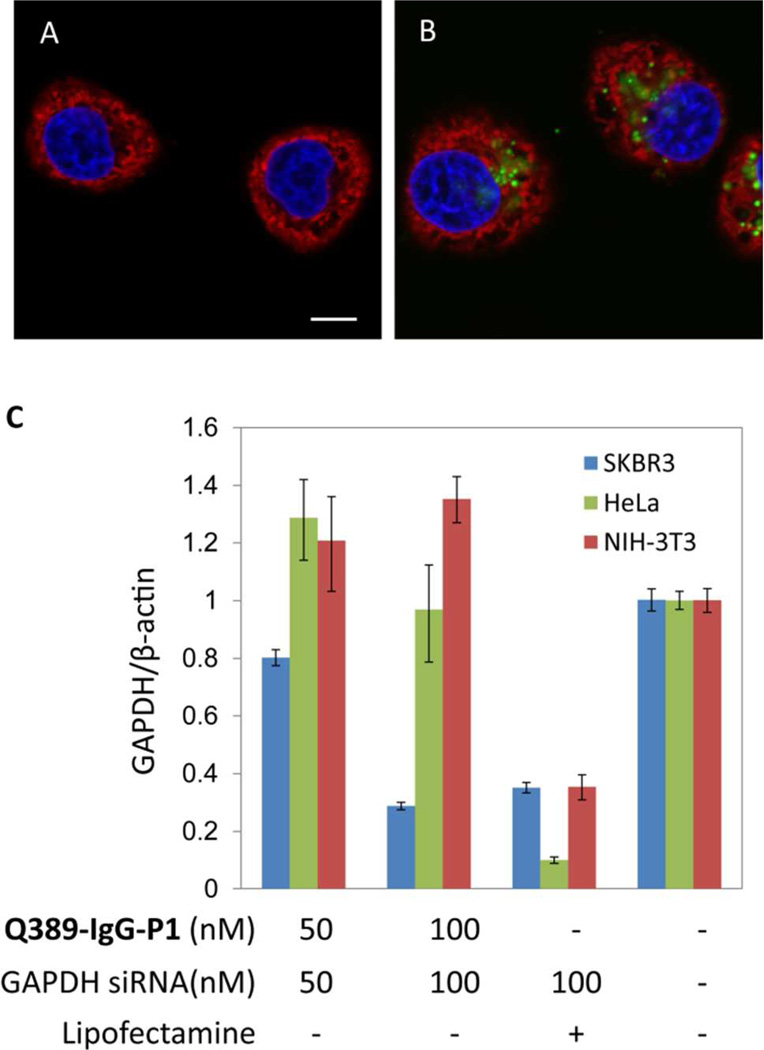
Next, we determined whether the siRNA delivered to the HER2+ cells by S202-Fab-P1 can selectively induce gene silencing. Various concentrations of S202-Fab-P1 were mixed with 50 nM GAPDH specific siRNA (GAPDH-siRNA) in serum-free Opti-MEM and then incubated with SKBR-3 cells at 37 °C for 4 h. Relative GAPDH mRNA levels (normalized to β-actin) were measured by quantitative real-time polymer-ase chain reaction (qRT-PCR) 24 h after treatment. As shown in Fig. 4A, S202-Fab-P1/siRNA ratios of 100 nM/50 nM, 150 nM/50 nM, and 200 nM/50 nM induce a concentration dependent silencing of GAPDH (blue bars, lane 1–3); and 300 nM/100 nM ratio of S202-Fab-P1/GAPDH-siRNA complex resulted in 88% knockdown of GAPDH mRNA (Fig. 4A, blue bars, lane 4). In contrast, neither the complex of a scrambled-siRNA with S202-Fab-P1, nor the GAPDH-siRNA delivered by the S202-pAcF Fab and P1 mixture, showed any significant silencing of mRNA expression (Fig. 4A, blue bars, lane 5 and 6). Moreover, treatment with the S202-Fab-P1/GAPDH-siRNA complex failed to induce gene silencing of GAPDH in HeLa (Fig. 4A, greens bars) and NIH-3T3 (Fig. 4A, red bars) cells, two HER2− cell lines. Additionally, this selectivity was well maintained at higher siRNA dosages by using relative lower APC/siRNA ratios. To confirm gene silencing by the S202-Fab-P1/siRNA complex at the protein level, western blot analysis was performed. Consistent with the qRT-PCR results, treatment with increased concentrations of the S202-Fab-P1/GAPDH-siRNA complex resulted in a dose dependent decrease of GAPDH protein levels in SKBR-3 cells, and was more efficient in gene silencing than commercial lipofectamine transfection at the same siRNA concentration (Fig. 4B, top two lanes). Again, no significant reduction in GAPDH protein levels was observed for the S202-Fab-P1/GAPDH-siRNA complex at the same concentrations in HER2− HeLa cells (Fig. 4B bottom two lanes) and NIH-3T3 cells (Fig. S6). We further confirmed that the gene silencing induced by the S202-Fab-P1/GFP-siRNA complex in SKBR-3 cells was HER2 dependent in a competition assay. When SKBR-3 cells were pretreated with 10 µM anti-HER2 IgG, GAPDH mRNA levels decreased only 30% relative to an 85% reduction by the S202-Fab-P1/GFP-siRNA complex at 300 nM/100 nM in the absence of competing antibody (Fig. 4C). Overall, these results indicate that the S202-Fab-P1 conjugate can selectively deliver siRNA to HER2+ cells and efficiently suppress expression of the targeted gene at both the mRNA and protein level in a HER2 dependent manner.
Figure 4.
Cell selective RNA silencing induced by the S202-Fab-P1/siRNA complexes. (A) CYBR-Green quantitative real-time PCR (qRT-PCR) analysis of the gene silencing at the mRNA level in HER2+ and HER2 cells; the S202-Fab-P1/GAPDH-siRNA complexes reduce relative GAPDH mRNA levels in SKBR-3 (blue), but not NIH-3T3 (red) or HeLa (green) cells. 24 h after treatment, cells were lysed for qRT-PCR analysis; the relative expression levels of GAPDH mRNA were normalized to β-actin and compared with the untreated group. (B) Western blot analysis of gene silencing at the protein level in HER2+ and HER2 cells; the S202-Fab-P1/GAPDH-siRNA complexes reduce protein levels of GAPDH in SKBR-3 but not HeLa cells. 72 h post treatment, cells were lysed and GAPDH levels were measured; γ-tubulin was served as the internal control. (C) qRT-PCR analysis of gene silencing at the mRNA level in SKBR-3 cells for the HER2 competition study; the results showed that the RNA silencing by the S202-Fab-P1/siRNA complex in SKBR-3 is HER2 dependent; SKBR-3 cells were treated with 10 µM anti-HER2 IgG and incubated in Opti-MEM for 1 h before they were treated with the S202-Fab-P1/siRNA complex at 300 nM/100 nM; 4 h later, the medium was replaced with complete medium with 10% FBS for all the treatment groups; cells were harvested for qRT PCR 24 h post treatment.
To show that S202-Fab-P1 is a general delivery vehicle for siRNAs to HER2+ cells, we tested a number of other siRNAs against different targets. As shown in Fig. S7, a dose dependent (up to 72%) knockdown of MDM2 at both the mRNA and protein levels in SKBR-3 cells was observed when S202-Fab-P1 (300 nM) was used to deliver a siRNA against MDM2 (MDM2-siRNA, 100 nM), a well-known negative transcriptional regulator of the tumor suppressor protein p53. Up to 77 % knockdown of DNAJB11 mRNA in SKBR-3 cells was also observed with the complex formed by S202-Fab-P1 (300 nM) and a siRNA targeting DNAJB11 (DNAJB11-siRNA, 100 nM), a co-chaperon protein belonging to the DNAJ/HSP40 family (Fig. S8). Additionally, no significant GFP silencing was observed in a stable 293T-GFP cell line (HER2−) treated with the complex of S202-Fab-P1 (300 nM) and a siRNA against GFP (GFP-siRNA, 100 nM) (Fig. S9). Together, these results clearly demonstrate that S202-Fab-P1 is able to selectively knockdown a variety genes in HER2+ cells.
2.4 APCs Induced Gene Silencing is Site-dependent
We next synthesized site-specific P1 conjugates of the full length anti-HER2 Q389-pAcF and A121-pAcF mutant IgGs to determine if higher binding affinity and bivalency leads to more potent mRNA silencing. Two conjugates, Q389-IgG-P1 and A121-IgG-P1, were synthesized and purified as described above (Fig. 1C and Fig. S12). ELISA analysis showed that the two conjugates have similar binding affinity to HER2 antigen with IC50 values of 0.35 nM (A121-IgG-P1) and 0.87 nM (Q389-IgG-P1) affinities which are comparable to their parental antibody (Fig. S13). Gel retardation assays showed complete siRNA binding by the two IgG-P1 conjugates at an APC/siRNA molar ratio of ~0.4/1 (Fig. S14); DLS analysis confirmed the formation of the complexes (Fig. S15). Surprisingly, confocal microscopy showed that A121-IgG-P1 (50 nM) failed to deliver FITC-siRNA (50 nM) to SKBR-3 cells (Fig. 5A). Consistent with this result, the A121-IgG-P1/GAPDH-siRNA complex did not induce RNA silencing in SKBR-3 at concentrations as high as 300 nM/100 nM, as determined by qRT-PCR (Fig. S16). In contrast, Q389-IgG-P1 delivered the FITC-siRNA to SKBR-3 cells under the same conditions used for A121-IgG-P1 (Fig. 5B). Moreover, Q389-IgG-P1 delivered the GAPDH-siRNA efficiently to SKBR-3 cells but not HeLa cells and NIH-3T3 cells, as determined by the selective mRNA silencing (Fig. 5C) and protein knockdown (Fig. S17). The RNA silencing effect of the Q389-IgG-P1/GAPDH-siRNA complex at 100 nM/100 nM was comparable to that of the S202-Fab-P1/GAPDH-siRNA at 300 nM/100 nM, indicating Q389-IgG-P1 is more potent than S202-Fab-P1. Considering the conjugation site of A121-IgG-P1 (CH1 region) is much closer to the antigen binding site than for Q389-IgG-P1 (CH3 region), the different efficacies of siRNA delivery by these two APCs is likely due to steric interference of the antigen binding site of A121-IgG-P1 but not Q389-IgG-P1 when the APC/siRNA complexes are formed.
Figure 5.
Site-dependent cell internalization and RNA silencing for A121-IgG-P1 and Q389-IgG-P1. (A–B), Confocal microscopy of FITC-siRNA delivered to SKBR-3 cells by A121-IgG-P1 (A) and Q389-IgG-P1 (B). (C) CYBR-Green qRT-PCR analysis of gene silencing at the mRNA level in HER2+ and HER2 cells ; the results showed that the Q389-IgG-P1/GAPDH-siRNA complex reduces relative GAPDH mRNA levels in SKBR-3 (blue), but not NIH-3T3 (red) and HeLa (green) cells.
3. CONCLUSIONS
In conclusion, we have demonstrated that site-specific APCs can be readily synthesized under mild conditions in good yields by the genetic incorporation of an UAA with bioorthogonal reactivity. These APCs have high binding affinity to HER2 antigen that is comparable to their parental antibodies. S202-Fab-P1 and Q389-IgG-P1 effectively and selectively deliver functional siRNAs to HER2+ cancer cells with no obvious toxicity. Significant gene silencing at both the mRNA and protein levels was observed using a number of different siRNAs. We also demonstrate that the conjugation site of the APCs plays a critical role in determining the efficient internalization and silencing of the APC/siRNA complexes. We are currently further optimizing the structure and MW of the polymer as well as testing siRNA delivery of these novel APCs in vivo. This site specific APC platform may provide a novel approach for the selective delivery of siRNAs or other nucleic acid therapeutic agents.
4. EXPERIMENTAL SECTIONS
Synthesis of P1
CTA-1 (12 mg, 0.02 mmol), M1 (150 mg, 0.39 mmol) and AIBN (2 mg, 0.012 mmol) were added to a 25 mL two-neck round bottom flask. Under argon, the mixture was dissolved in toluene (3 mL) and heated to 65 °C for 3 days; 1 H NMR indicated more than 95% M1 was consumed. M2 (NIPAAm, 90 mg, 0.8 mmol) was then added to the solution via syringe. The reaction was stirred under argon at 65 °C for another 2 days; 1 H NMR confirmed the consumption of both M1 and M2. An aliquot (10 µL) of the P1-Boc solution was injected directly into the GPC for MW analysis (Fig. S1) and the rest of the solution was dried under vacuum. The residual oil was dissolved in trifluoroacetic acid (TFA, 3 mL) and stirred at rt overnight to remove the Boc group. TFA was removed under vaccum and the residue was dissolved in 5-mL ultrapure water and extensively dialyzed against ultrapure water to remove any small molecular impurities. Water was changed every 4–6 hours for a total of 4 times. The solution was then lyophilized to afford a white fluffy powder (126 mg, yield 73%). The 1H NMR spectrum of P1 is shown in Fig. S2.
Mutation and expression of the anti-HER2 Q389-pAcF IgG
The Herceptin gene containing a TAG codon at site Q389 (HC-Q389X) and a His-tag at the C-terminus of the heavy chain was amplified by PCR and inserted into a plasmid containing an orthogonal Escherichia coli tyrosyl-derived tRNA/aaRS pair that incorporates pAcF. The mutant Q389-pAcF IgG was expressed in suspension CHO cells using Free Style Max (Life Technologies, Carlsbad, CA) as a transfection reagent. For a 30 mL scale transfection, CHO cells were seed-ed at 0.5×106/mL the day before transfection. On the day of transfection, cells were counted before transfection to ensure a cell density of 1.0×106/mL. pAcF in PBS was added into cell culture flask 15 min before transfection (final concentration 1.3 mM). To two separate tubes each containing 0.6 mL Opti-PRO media was added 45 µg of the mutant pLou plasmid and 37.5 µL of MAX reagent separately. The MAX/OptiPRO solution was then added to the DNA/OptiPRO solution and mixed by swirling the tube gently. The complex was incubated for 15 min and then poured into 30 mL of the CHO cell culture. The flask was transferred to 32 °C incubator shaking at 125 rpm. 3 mL 10× cell boost 5 (70 g/L stock solution; Hy-Clone) was added to the cell culture 24 h after transfection; the medium was harvested 7 days after transfection by centrifugation. The supernatant was filtered with a 0.22 µM filter and the mutant Q389-pAcF IgG was purified by Ni-NTA column (Qiagen, Valencia, CA)in a yield of ~2.5–3 mg/L. The structure and molecular weight of the antibody were confirmed by SDS-PAGE gel (Fig. S10) and ESI-MS (Fig. S11)
Synthesis of the S202-Fab-P1 conjugate
The mutant S202-pAcF Fab was buffer exchanged to 100 mM acetate buffer (pH, 4.5) and concentrated to ~ 5 mg/mL with a 10 kDa Amicon concentrator before reaction. P1 (18 mg, ~ 1.8 µmol – ONH2) was dissolved in 100 mM acetate buffer (pH 4.5, 50 µL) in a 0.5-mL eppendorf tube. The S202-pAcF Fab (5.2 mg/mL, 200 µL) was slowly added to the P1 solution. The mixture was incubated at 37 °C for ~3–5 days and then quenched by slow addition to protein G column binding buffer (50 mM NaOAc, pH=5.2, 1.5 mL). The mixture was passed through a bench-top protein G column (to increase yield, repeat this for another three times). The column was washed extensively with binding buffer (~ 100 mL) and eluted with protein G elution buffer (100 mM glycine, pH =3.0, 10 mL). The eluent was immediately neutralized with Tris·HCl buffer (1.0 M, pH 7.4, 1.0 mL) and concentrated with a 10 kDa Amicon concentrator to ~ 0.5 mL. The conjugate was further purified by size exclusion chromatography (Superdex 200, 10/300 GL, GE healthcare) on a FPLC system using 1× dPBS as mobile phase. The desired fractions were combined and concentrated with a 10 kDa Amicon concentrator to give S202-Fab-P1 (0.5 mg/mL of antibody × 1.2 mL, determined by both nanodrop absorption at 280 nm and BCA protein detection kit) in a yield of ~60%. Q389-IgG-P1 and A121-IgG-P1 were synthesized following the same protocol; the yield of Q389-IgG-P1 and A121-IgG-P1 after purification averaged 40%. The conjugates were confirmed by SDS-PAGE gel (Fig. S12).
Confocal microscopy
SKBR-3 and HeLa cells were seeded separately in two 35 mm 4-chamber glass bottom dishes at 100,000 and 80,000 cells/well, respectively. The dishes were incubated at 37 °C for 24 h to reach 50–70 % confluence. Before treatment, the complete medium was removed and cells were washed with PBS once and incubated with 400 µL Opti-MEM (Life Technologies, Carlsbad, CA). To make the S202-Fab-P1/FITC-siRNA complexes, S202-Fab-P1 and FITC-siRNA were diluted to desired concentrations in 50 µL Opti-MEM separately. Equal volumes of S202-Fab-P1 and FITC-siRNA in Opti-MEM were mixed together and incubated at RT for 10–20 min. The control mixture of anti-HER2 S202-pAcF Fab, P1 and FITC-siRNA was prepared in the same way. The S202-Fab-P1/FITC-siRNA complex and the control mixture in 100 µL Opti-MEM were added to SKBR-3 and HeLa cells and incubated at 37 °C for 4 h. Opti-MEM was removed and cells were stained with ER tracker red (1/2000 dilution) and Hoechst (1/5000 dilution) in PBS (500 µL). After incubation at RT for 10 min, the medium was removed and cells were washed with cold PBS multiple times and imaged.
siRNA delivery
Cells were incubated in 24-well plates in DMEM supplied with 10 % FBS until they reached 50–70 % confluence. Complete medium was removed before treatment and cells were washed with PBS and replaced with 400 µL Opti-MEM (Life Technologies, Carlsbad, CA). To make the S202-Fab-P1/siRNA complexes, S202-Fab-P1 and the appropriate siRNAs were diluted to the desired concentrations in 50 µL Opti-MEM separately. Equal volumes of S202-Fab-P1 and siRNA in Opti-MEM were mixed together and incubated at RT for 10–20 min. The S202-Fab-P1/siRNA complexes in 100 µL Opti-MEM were then added to cells and incubated at 37 °C for 4 hours. Positive controls were performed using Lipofectamine® RNAi MAX (Life Technologies, Carlsbad, CA) following manufacturer’s protocol. The Opti-MEM medium was replaced by DMEM supplied with 10% FBS without antibiotics 4 h after treatment. The cells were analyzed by either qRT-PCR (24–48 hours post treatment) or western blot (48–72 hours post treatment) at appropriate time points.
Supplementary Material
Figure 3.
Confocal microscopy of internalization of siRNA mediated by S202-Fab-P1. HeLa (A, C and E) and SKBR-3 (B, D and F) cells were treated with buffer (A and B), 200 nM S202-pAcF Fab + 200 nM P1 + 50 nM siRNA-FITC (C and D) or 200 nM S202-Fab-P1 + 50 nM siRNA-FITC (E and F) for 4 h. Cells were then stained with Hoechst 33342 (blue) and ER-Tracker (red) and imaged with a Leica 710 confocal microscope. Bar = 10 micron.
ACKNOWLEDGMENT
This work is supported by NIH grant R01 GM062159. H.L. is The Jake Wetchler Foundation Fellow for Pediatric Innovation of the Damon Runyon Cancer Research Foundation (DRG-2099-11). We thank Dr. M.G. Finn and his group for allowing us using their GPC and DLS facilities.
Footnotes
ASSOCIATED CONTENT
Additional experimental details and supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.(a) Yu DB, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, Crooke ST, Prakash TP, Corey DR. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Watts JK, Corey DR. J. Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lima WF, Prakash TP, Murray HM, Kinberger GA, Li WY, Chappell AE, Li CS, Murray SF, Gaus H, Seth PP, Swayze EE, Crooke ST. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]; (d) Stanton MG, Colletti SL. J. Med. Chem. 2010;53:7887–7901. doi: 10.1021/jm1003914. [DOI] [PubMed] [Google Scholar]; (e) Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Bennett CF, Swayze EE. Annu. Rev. Pharmacol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]; (g) Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Jeong JH, Mok H, Oh YK, Park TG. Bioconjugate Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]; (i) Zimmermann TS, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 2.(a) Zhou JH, Rossi JJ. Oligonucleotides. 2011;21:1–10. doi: 10.1089/oli.2010.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Peer D, Lieberman J. Gene Ther. 2011;18:1127–1133. doi: 10.1038/gt.2011.56. [DOI] [PubMed] [Google Scholar]
- 3.Kumar P, Ban HS, Kim SS, Wu HQ, Pearson T, Greiner DL, Laouar A, Yao JH, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Yao YD, Sun TM, Huang SY, Dou S, Lin L, Chen JN, Ruan JB, Mao CQ, Yu FY, Zeng MS, Zang JY, Liu Q, Su FX, Zhang P, Lieberman J, Wang J, Song EW. Sci. Transl. Med. 2012:4. doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]; (b) Song EW, Zhu PC, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]; (c) Peer D, Zhu PC, Carman CV, Lieberman J, Shimaoka M. P. Natl. Acad. Sci. USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, Giangrande PH. Nucleic Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, Giangrande PH. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) McNamara JO, Andrechek ER, Wang Y, D Viles K, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 6.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 7.(a) Dohmen C, Fröhlich T, Lächelt U, Röhl I, Vornlocher H-P, Hadwiger P, Wagner E. Mol. Ther. Nucleic Acids. 2012;1:e7. doi: 10.1038/mtna.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Thomas M, Kularatne SA, Qi LW, Kleindl P, Leamon CP, Hansen MJ, Low PS. Ann. Ny. Acad. Sci. 2009;1175:32–39. doi: 10.1111/j.1749-6632.2009.04977.x. [DOI] [PubMed] [Google Scholar]; (c) Zhang KX, Wang QQ, Xie YH, Mor G, Sega E, Low PS, Huang YQ. RNA. 2008;14:577–583. doi: 10.1261/rna.739308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, Lee H, Scuto A, Liu Y, Yang CM, Deng JH, Soifer HS, Raubitschek A, Forman S, Rossi JJ, Pardoll DM, Jove R, Yu H. Nat. Biotechnol. 2009;27:925–932. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Bioconjugate Chem. 2008;19:2156–2162. doi: 10.1021/bc800249n. [DOI] [PubMed] [Google Scholar]; (b) Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, Hagstrom JE, Wolff JA. P. Natl. Acad. Sci. USA. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Bioconjugate Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]; (d) Choi CHJ, Alabi CA, Webster P, Davis ME. P. Natl. Acad. Sci. USA. 2010;107:1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zhang K, Hao L, Hurst SJ, Mirkin CA. J. Am. Chem. Soc. 2012;134:16488–16491. doi: 10.1021/ja306854d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lee J, Yun KS, Choi CS, Shin SH, Ban HS, Rhim T, Lee SK, Lee KY. Bioconjugate Chem. 2012;23:1174–1180. doi: 10.1021/bc2006219. [DOI] [PubMed] [Google Scholar]; (g) Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Chen YC, Zhu XD, Zhang XJ, Liu B, Huang L. Mol. Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Sato Y, Murase K, Kato J, Kobune M, Sato T, Kawano Y, Takimoto R, Takada K, Miyanishi K, Matsunaga T, Takayama T, Niitsu Y. Nat. Biotechnol. 2008;26:431–442. doi: 10.1038/nbt1396. [DOI] [PubMed] [Google Scholar]; (j) Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Shen BQ, Xu KY, Liu LN, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu SF, Mai E, Li DW, Tibbitts J, Baudys J, Saadi OM, Scales SJ, McDonald PJ, Hass PE, Eigenbrot C, Nguyen T, Solis WA, Fuji RN, Flagella KM, Patel D, Spencer SD, Khawlil LA, Ebens A, Wong WL, Vandlen R, Kaur S, Sliwkowski MX, Scheller RH, Polakis P, Junutula JR. Nat. Biotechnol. 2012;30:184–189. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]; (b) Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, Lu YM, Meng YG, Ng C, Yang JH, Lee CC, Duenas E, Gorrell J, Katta V, Kim A, McDorman K, Flagella K, Venook R, Ross S, Spencer SD, Wong WL, Lowman HB, Vandlen R, Sliwkowski MX, Scheller RH, Polakis P, Mallet W. Nat. Biotechnol. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]; (c) Wu AM, Senter PD. Nat. Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 11.(a) Wu X, Schultz PG. J. Am. Chem. Soc. 2009;131:12497–12515. doi: 10.1021/ja9026067. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 12.(a) Wang L, Zhang ZW, Brock A, Schultz PG. P. Natl. Acad. Sci. USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu WS, Brock A, Chen S, Chen SB, Schultz PG. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 13.(a) Kazane SA, Axup JY, Kim CH, Ciobanu M, Wold ED, Barluenga S, Hutchins BA, Schultz PG, Winssinger N, Smider VV. J. Am. Chem. Soc. 2013;135:340–346. doi: 10.1021/ja309505c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kazane SA, Sok D, Cho EH, Uson ML, Kuhn P, Schultz PG, Smider VV. P. Natl. Acad. Sci. USA. 2012;109:3731–3736. doi: 10.1073/pnas.1120682109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, Lu YC, Tran H, Seller AJ, Biroce SL, Szydlik A, Pinkstaff JK, Tian F, Sinha SC, Felding-Habermann B, Smider VV, Schultz PG. P. Natl. Acad. Sci. USA. 2012;109:16101–16106. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kim CH, Axup JY, Dubrovska A, Kazane SA, Hutchins BA, Wold ED, Smider VV, Schultz PG. J. Am. Chem. Soc. 2012;134:9918–9921. doi: 10.1021/ja303904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. Oncologist. 2009;14:320–368. doi: 10.1634/theoncologist.2008-0230. [DOI] [PubMed] [Google Scholar]
- 15.Hutchins BM, Kazane SA, Staflin K, Forsyth JS, Felding-Habermann B, Schultz PG, Smider VV. J. Mol. Biol. 2011;406:595–603. doi: 10.1016/j.jmb.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Zhou P, Wang MM, Du L, Fisher GW, Waggoner A, Ly DH. J. Am. Chem. Soc. 2003;125:6878–6879. doi: 10.1021/ja029665m. [DOI] [PubMed] [Google Scholar]; (b) Luedtke NW, Carmichael P, Tor Y. J. Am. Chem. Soc. 2003;125:12374–12375. doi: 10.1021/ja0360135. [DOI] [PubMed] [Google Scholar]; (c) Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. P. Natl. Acad. Sci. USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.