Abstract
Although the role of E proteins in the thymocyte development is well documented, much less is known about their function in peripheral T cells. Here we demonstrated that CD4 promoter-driven transgenic expression of Id1, a naturally occurring dominant-negative inhibitor of E proteins, can substitute for the co-stimulatory signal delivered by CD28 to facilitate the proliferation and survival of naïve CD4+ cells upon anti-CD3 stimulation. We next discovered that IL-2 production and NF-κB activity after anti-CD3 stimulation were significantly elevated in Id1-expressing cells, which may be, at least in part, responsible for the augmentation of their proliferation and survival. Taken together, results from this study suggest an important role of E and Id proteins in peripheral T cell activation. The ability of Id proteins to by-pass co-stimulatory signals to enable T cell activation has significant implications in regulating T cell immunity.
Keywords: Inhibitor of differentiation, E protein, CD4+ T cell, co-stimulation, T cell activation
1. Introduction
Activation of naïve T cells is crucial for T cell expansion and effecter cell differentiation. This requires two distinct signals: an antigen-specific signal arising from T cell receptor (TCR) engagement with MHC bound peptides displayed on antigen-presenting cells (APC) and a co-stimulatory signal originating from co-stimulatory receptors such as CD28 and their ligands including B7 molecules expressed on APC [1]. These signaling cascades trigger cell division and promote cell survival. The downstream events involve the activation of RAS/MAP kinase and PI3 kinase pathways as well as the mobilization of intracellular calcium [2;3], which ultimately lead to the activation of several transcription factors such as NF-κB, AP-1 and NFAT [4;5;6]. As a result, expression of IL-2 and activation-associated cell surface molecules are induced [7].
Cross-talks between the signaling events from TCR/co-stimulatory receptors and various transcription factors occur at multiple levels. One such family of transcription factors are the basic helix-loop-helix transcription factors, including E2A, E2-2 and HEB, which are collectively called E proteins [8;9]. E proteins share extensive homology, bind to specific DNA sequences called E box and activate transcription of similar target genes. In T cells, both E2A and HEB genes are abundantly expressed [10]. However, the function of E proteins can be eliminated by their naturally occurring dominant-negative inhibitors, inhibitor of differentiation (Id) 1 to Id4 [8;9]. Id proteins are helix-loop-helix proteins that heterodimerize with all E proteins and inhibit their DNA binding function. Id proteins have been used as excellent tools to ablation the function of multiple E proteins [11;12].
The role of E proteins in the commitment, growth and differentiation of T cells in the thymus has been extensively investigated [10;13;14]. Previous studies revealed that E proteins play important roles in T-cell development at multiple checkpoints [11;14–18]. For example, E proteins are essential for the differentiation of earliest T cell progenitors since deletion of both E2A and HEB genes or ectopic expression of Id1 results in developmental arrest at the transition from the CD4 and CD8 double negative stage 1 (CD44+CD25−) to stage 2 (CD44+CD25−). E proteins are thought to control the threshold of TCR β selection as down-regulation of E proteins enable DN T cells to progress to the double positive stage without pre-TCR signaling. E proteins and Id proteins have also been shown to regulate the differentiation of CD4 or CD8 single positive cells in the thymus [16;19]. Furthermore, loss of Id3 has been found to impact the differentiation of T regulatory cells [20]. However, little is known about the role of E proteins in the expansion and survival of peripheral naïve CD4+ T cells.
We have generated a new line of transgenic mice, CD4-Id1, where Id1 expression is driven by the promoter and enhance of the Cd4 gene. In these transgenic mice, the T cell developmental profiles in the thymus and periphery appear largely unchanged in comparison to wild type control mice. Therefore, this strain of mouse provides an excellent model for the investigation of the implication of E proteins in the functionality of peripheral CD4+ T cells. Indeed, we found that ectopic expression of Id1 facilitated T cell proliferation and survival upon TCR engagement in the absence of co-stimulatory signals, possibly due to enhanced IL-2 production and NF-κB activation. These results suggest an important role of E and Id proteins in the regulation of peripheral T cell activation.
2. Materials and Methods
2.1 Mice
The CD4-Id1 transgenic strain was created by injecting an Id1-expressing construct into the oocytes of FVB/N mice. One of the several transgenic lines was then backcrossed onto the C57BL/6 background for 6–8 generations and littermates were used as wild type controls. The construct was generated by inserting the Id1 cDNA, which contains an HA tag fused at the 3’ end of the coding sequence, into the CD4 transgenic vector [21].
2.2 Culture medium, antibodies and reagents
RPMI1640 medium containing 10% FCS were used for CD4 naïve T cell culture. The antibodies and reagents used for cell culture were: anti-mouse CD3 (145-2C11), anti-mouse CD28 (37.51), anti-mouse IL2 (BD BioSciences, San Jose, CA). Recombinant mouse IL-2 was purchased from R&D Systems (Minneapolis, MN). The following antibodies were used for flow cytometry and cell sorting: anti-CD4-PerCP, anti-CD4-PECy7, anti-CD8-APC, anti-CD25-APC, anti-CD44-FITC, anti-CD62L-PE, and anti-BrdU-FITC (BD Biosciences).
2.3 Naïve CD4 cell sorting and stimulation
Lymphocytes from lymph nodes were stained with fluorochrome-conjugated antibodies for 30 minutes at 4°C. Cells were then washed twice and resuspended at a density of 1×108 cells/ml. CD4+CD62LhiCD44loCD25− cells were sorted using BD FACS Aria II. The anti-CD3 antibody was coated onto 48-well flat-bottom plates at 1 µg/ml in PBS overnight and then washed once with PBS. Naïve CD4 cells were then placed into the wells at a density of 1×106 cells /ml with or without anti-CD28 (2 µg/ml) in the culture. The cells were incubated at 37°C containing 5% carbon dioxide for desired length of time.
2.4 BrdU incorporation
Cells were incubated in media containing 0.1 mM 5-bromo-2-deoxyuridine (BrdU; Sigma-Aldrich, Saint Louis, MO) for 1 hour, fixed with 4% paraformaldehyde in PBS, pH 7.4, for 10 minutes at 25°C, washed with PBS, treated with 4 N hydrochloric acid for 15 minutes, and then neutralized in 0.1 M sodium borate, pH 8.5 for 20 minutes. Cells were incubated with rat anti-BrdU antibody (0.5 µg/ml; BD Biosciences) in PBS containing 0.2% Triton X-100 for 30 minutes at 25°C. Cells were then washed and resuspended into PBS. BrdU staining was quantified using flow cytometry with BD FACSCalibur.
2.5 3H-TdR (thymidine) incorporation
To measure proliferation, naïve CD4+ cells were cultured in 96-well plates in the presence of anti-CD3 or anti-CD3/CD28 for 48 hours. 3H-TdR (thymidine) was added to the culture at 1 µCi/well 12 hours before harvest. Ice-cold 10% trichloroacetic acid was then added to the dishes and incubated on ice for 15 minutes. After washing with trichloroacetic acid and methanol, the cells were solubilized in 0.2 N NaOH and radioactivity was measured in a scintillation counter.
2.6 Assessment of Survival
After stimulation of naïve CD4 cells for desired length of time, cells were labeled with 5 µl of anti-Annexin V-FITC in 20 µl of binding buffer according to manufacturers’ instruction (eBioscience, San Diego, CA). Samples were mixed gently and incubated at room temperature for 15 minutes. Immediately before analysis using flow cytometry, 2 µl of propidium iodide (PI, 1 mg/ml) were added to each sample. A minimum of 10,000 cells within the gated region were analyzed.
2.7 Statistical Analysis
Statistical analysis of the data was carried out using Student’s t test.
3. Results
3.1 Id1 expression promotes anti-CD3-induced activation of naïve CD4+ T cells
To express Id1 in T cells post β selection, we inserted the Id1 cDNA downstream of a transgenic vector containing the enhancer and promoter of the CD4 gene (suppl. Fig. 1A). Flow cytometry analyses of thymocytes and splenocytes revealed no significant developmental blocks in these mice except a slight increase in immature CD8+ thymocytes (suppl. Fig. 1B and C). We observed a significant increase in CD44hiCD62Lhi and CD44hiCD62lo CD4+ T cells in lymph nodes (suppl. Fig. 1D) as well as spleen (data not shown) in aged (over 4 month-old) but not in young transgenic mice. The increased accumulation of these two subpopulations, which represent central and effector memory cells, respectively [22], suggests that E and Id proteins may be implicated in the regulation of peripheral T cell activation and differentiation.
To determine the role of Id in the activation of peripheral CD4 T cells, we sorted CD4+CD62LhiCD44loCD25− naïve T cells from CD4-Id1 transgenic mice and wild type littermates. The sorted cells were then cultured in the presence of plate-bound anti-CD3 antibodies with or without anti-CD28 antibodies. We examined T cell activation by measuring cell proliferation and expression of surface markers.
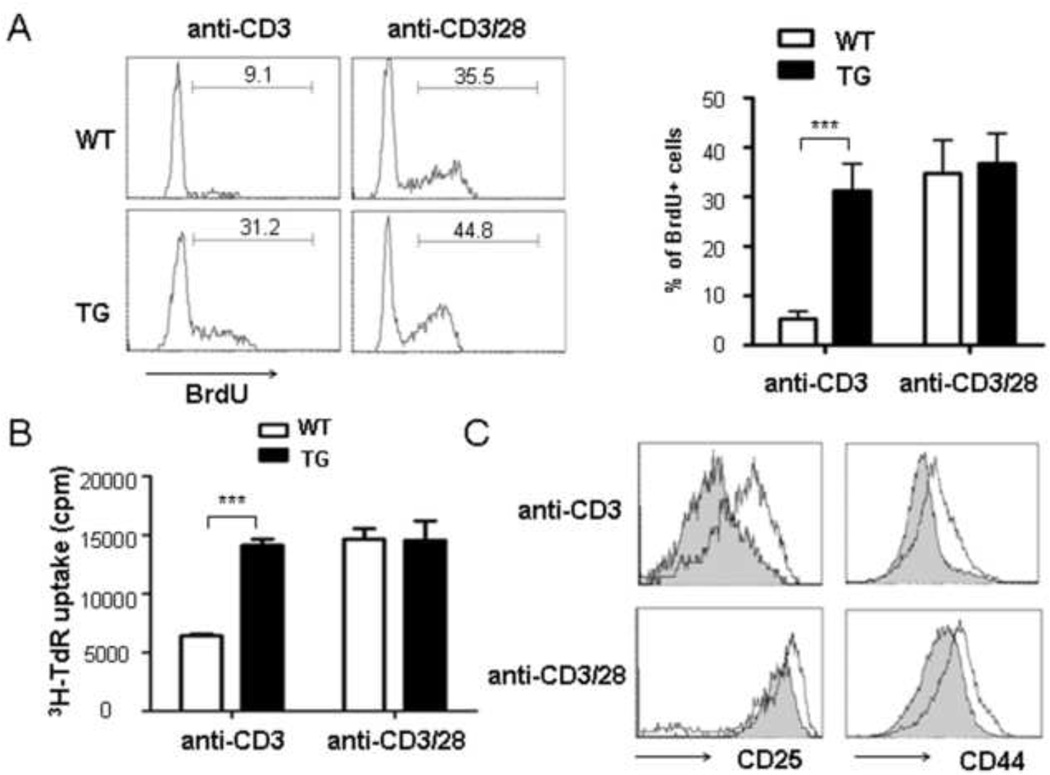
The proliferation of wild type and transgenic cells was first measured BrdU uptake by cultured cells using intracellular staining and found that BrdU incorporation by Id1-expressing cells was at least 5-fold higher than WT cells when they were treated with anti-CD3 alone (Fig. 1A). In contrast, when cultured in the presence of both anti-CD3 and anti-CD28, both wild type and transgenic cells incorporated BrdU robustly. Alternatively, we performed the 3H-thymidine incorporation assay and obtained similar results (Fig. 1B).
Fig 1. Id1 expression promotes anti-CD3-induced activation of naïve CD4-Id1tg CD4+ T cells.
Naïve CD4+ T cells from wild type (WT) or CD4-Id1tg (TG) mice were stimulated with anti-CD3 (1.0 µg/ml) with or without anti-CD28 (2.0 µg/ml). (A) BrdU incorporation. Cells were cultured for 3 days and BrdU was added to the culture in the last hour. Representative histograms are shown (left) and the results from 4 independent experiments are presented as mean±SD (right). (B) 3H-TdR uptake. Cells were cultured for 48 hours and 3H-TdR was added 12 hours before harvest. Results from three independent experiments are presented as mean±SEM. (C) CD44 and CD25 expression was compared between anti-CD3 or anti-CD3/CD28 stimulated WT (shaded) and TG (line) CD4+ T cells. Results from one representative experiment out of the three are shown. ***p<0.001.
T cell activation is known to be accompanied by the up-regulation of CD44 and CD25 [23;24]. As shown in Fig. 1C, the levels of CD44 and CD25 on Id1-expressing cells treated with anti-CD3 alone were significantly higher than those on wild type control cells and approached those on cells stimulated with both anti-CD3 and anti-CD28.
3.2 Id1 expression enhances the survival of anti-CD3-activated T cells in the absence of co-stimulation
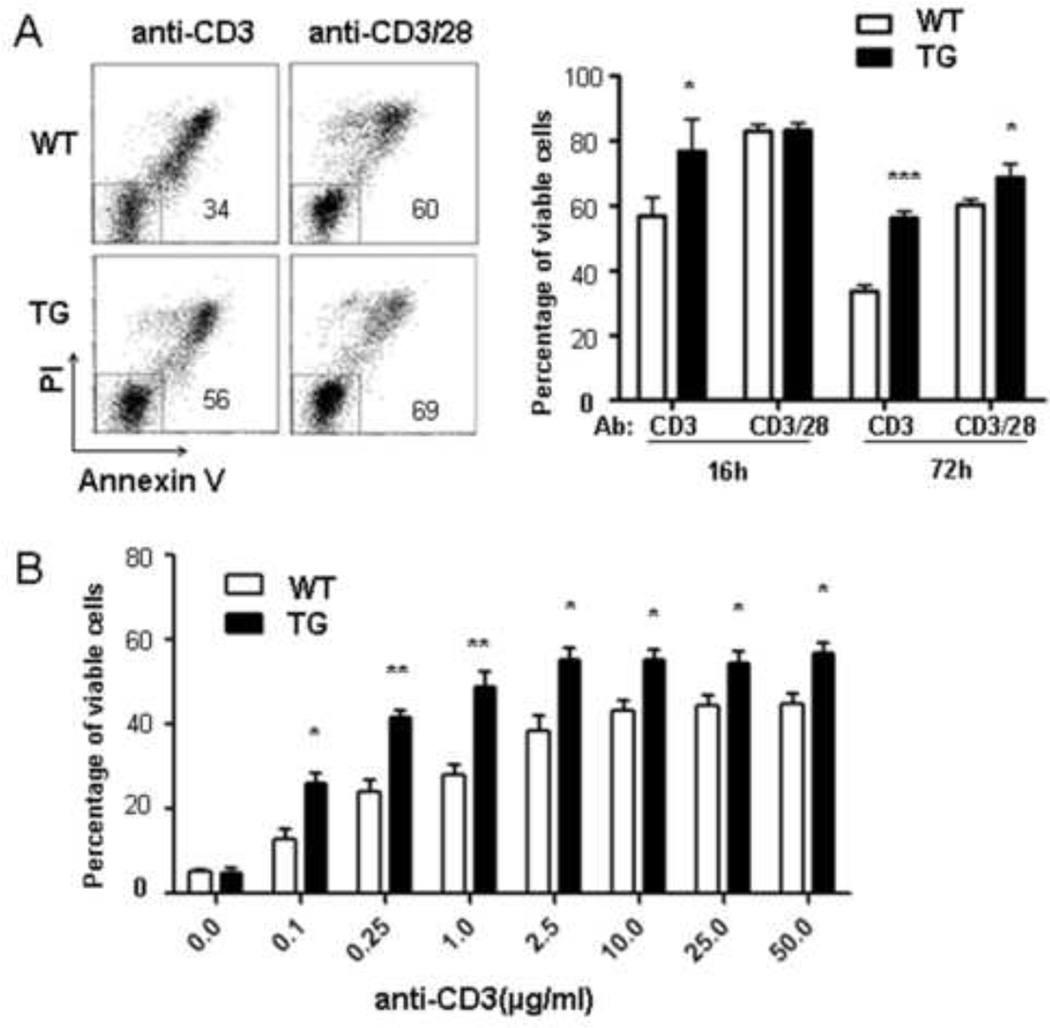
Since co-stimulatory signal is important for the survival of T cell during TCR activation [25], we examined the impact of Id expression on cell survival upon anti-CD3 stimulation. Sorted naïve CD4+ cells were cultured in the presence of anti-CD3 along with or without anti-CD28 and then stained with PI and Annexin-V. Cell survival was determined by quantifying the percentage of the PI–Annexin-V− population at 16 hours and 72 hours after the initiation of the cultures (Fig. 2A). We found that a significantly larger fraction of Id1-expressing cells than wild type cells survived when treated with 1 µg/ml of anti-CD3 alone. However, both wild type and transgenic cells showed high levels of survival when stimulated with anti-CD3 and anti-CD28.
Fig 2. Id1 expression enhances survival of anti-CD3-activated T cells.
(A) Naïve CD4+ T cells from WT or TG mice were stimulated as in Fig. 1A and analyzed by PI and Annexin V staining. Representative dot plots after culturing for 72 hours are shown. The number in the plot indicates the percentage of PI−Annexin V− live cells. Results from four independent experiments obtained after culturing for 16 and 72 hours are summarized as mean±SD. (B) T cells survival was analyzed 72 hours after stimulation with different concentrations of anti-CD3. Data shown are from four independent experiments statistical analyses were done by comparing WT to TG cells. *p<0.05; **p<0.01; ***p<0.001.
To further examine the survival response to anti-CD3 stimulation, we tested the effect of different concentrations of anti-CD3. Although increasing the amount of anti-CD3 enhanced the viability of cells of both genotypes, Id1-expressing cells consistently showed better survival at any given concentration of anti-CD3 (Fig. 3B), which suggest that Id1 provided the cells with survival signals normally delivered by anti-CD28.
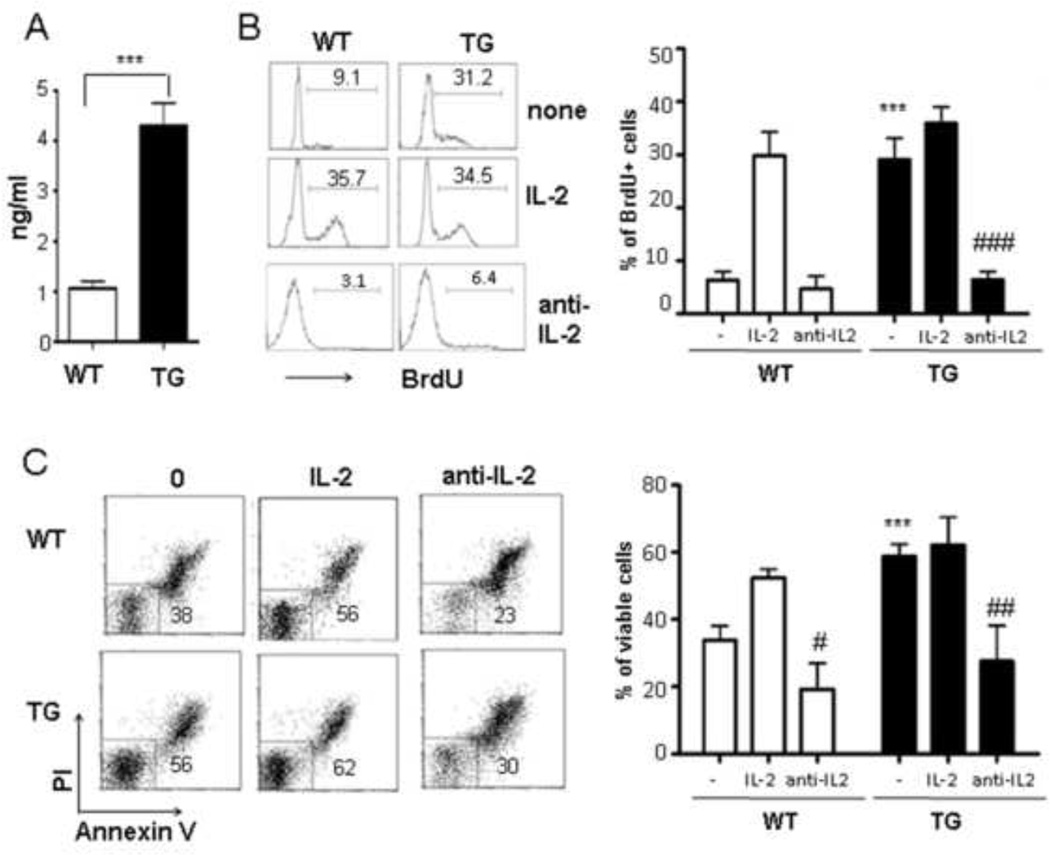
Fig 3. IL-2 contributes to the enhanced proliferation and survival of anti-CD3-activated Id1tg CD4+ T cells.
(A) IL-2 levels in culture supernatants 2 days after anti-CD3 stimulation was measured by ELISA in triplicates. A representative of three experiments is shown as mean±SEM. (B) Cell proliferation assay of WT and TG naïve CD4+ T cells stimulated with anti-CD3 (1 µg/ml) for 3 days with or without IL-2 (2 ng/ml) or anti-IL-2 neutralizing antibodies (10µg/ml). (C) Cell survival was analyzed by PI/Annexin V staining. Cells were stimulated with anti-CD3 (1 µg/ml) for 3 days with or without IL-2 (5 ng/ml) or anti-IL-2 antibodies. Data shown are representatives of 4 experiments. *** p<0.001 indicates comparison between WT and TG cells. # p<0.05, ## p<0.01 and ### p<0.001 show comparison between with or with anti-IL-2 treatment.
3.3 IL-2 contributes to the enhanced proliferation and survival of Id1-expressing CD4+ T cells upon anti-CD3 stimulation
Since induction of cytokine secretion such as IL-2 by anti-CD28 co-stimulation plays an important role in CD4 T cell activation and survival [26;27], we determined the effect of Id1 on IL-2 production by culturing naïve T cells in the presence of anti-CD3 for 2 days and measuring secreted IL-2 using ELISA (Fig. 4A). Id1 transgenic cells produced over 4 fold higher levels of IL-2 than wild type cells. However, it should be pointed out that this level of IL-2 secretion in Id1 transgenic cells upon anti-CD3 stimulation was only about 20% of the IL-2 levels secreted by wild type and Id1 transgenic cells upon stimulation with anti-CD3 plus anti-CD28 (data not shown).
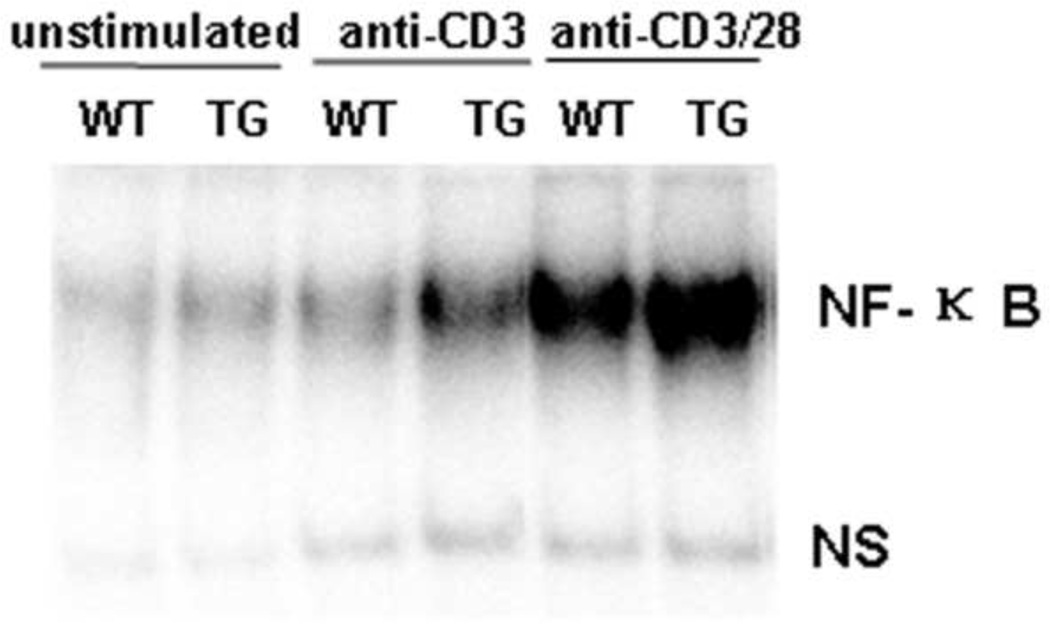
Fig 4. Enhanced NF-κB activation in Id1tg CD4+ T cells upon anti-CD3 stimulation.
Sorted WT and TG naïve CD4+ naïve T cells were treated with anti-CD3 (2 µg/ml) with or without anti-CD28 (2 µg/ml) for 18 hours and then harvest for nuclear extract. An aliquot was reserved for no treatment control. EMSA was done with 32P-labeled specific NF-κB probe as previously described [18]. NF-κB binding complexes and a non-specific band (ns), which serves as an internal control, are as marked.
To test if the elevated IL-2 secretion is responsible for the enhanced proliferation and survival, we evaluated the effects of IL-2 addition and subtraction in anti-CD3 stimulated cultures. We found that Id1-expressing cells proliferated robustly without exogenous IL-2 compared to wild type cells as indicated by the percentage of BrdU+ cells. However, addition of 2 ng/ml of IL-2 was sufficient to override the difference and wild type cells incorporated BrdU at levels comparable to those by Id1 transgenic cells cultured with or without 2 ng/ml of IL-2 (Fig. 4B).
Interestingly, the concentration of IL-2 present in the culture supernatants of Id1-expressing cells was found to be about 4 ng/ml (Fig. 4A), which is above the threshold of IL-2 necessary for efficient BrdU uptake. To test if this endogenous level of IL-2 produced by Id1 transgenic cells is responsible for their ability to proliferate, we added neutralizing antibodies against IL-2 to cultures set up with either wild type or transgenic naïve T cells. The inhibitory effect of the neutralizing antibodies on wild type cells was insignificant but profound on Id1 transgenic cells (Fig. 4B).
In addition, we also examined the contribution of IL-2 to cell survival (Fig. 4C). In the absence of exogenous IL-2, Id1 expression provided a huge survival advantage and this gap was eliminated by addition of 5 ng/ml of IL-2. IL-2 neutralizing antibodies significantly diminished the ability of Id1 expression to facilitate the survival of transgenic cells, resulting in a similar percentage of viable Id1 transgenic cells as their wild type counterparts (Fig. 4C). These data thus suggest that IL-2 serves as a mediator for co-stimulatory effects of Id1 on both proliferation and survival of naïve CD4+ T cells.
3.4 Enhanced NF-κB activation in Id1-expressing CD4+ T cells upon anti-CD3 stimulation
CD28 signaling not only promotes cytokine secretion but also activates transcription factors such as NF-κB, which plays critical roles in the proliferation and survival of peripheral CD4 T cells [28 31]. To test if Id1 expression could substitute for anti-CD28 co-stimulatory signal by augmenting NF-κB activity, we measured the DNA binding activity of these factors to a canonical NF-κB recognition sequence. Naïve CD4 cells were sorted and stimulated with anti-CD3 alone or anti-CD3 together with anti-CD28 for 18 hours. Nuclear extracts were then prepared and used in an electrophoretic mobility shift assay (EMSA) with an NF-κB specific probe. Although stimulation with anti-CD3 and anti-CD28 dramatically increased the binding activity in both wild type and transgenic cells, anti-CD3 alone induced NF-κB activation in Id1 transgenic but not wild type cells (Fig. 4). Therefore, Id1 expression seems to potentiate NF-κB activation when stimulated with anti-CD3 alone.
4. Discussion
The principal finding of this study is the ability of ectopic Id1 expression to promote peripheral naïve T cell proliferation and survival in the absence of CD28-mediated co-stimulation. Consistent with the data obtained in culture, Id1 transgenic mice were found to exhibit increased expression of activation markers such as CD44 and CD25 and harbor high percentages of T cells with effector/memory phenotypes. These findings significantly extend previous data that co-stimulation-independent proliferation occurs in CD4+ thymocytes from lck-Id1 transgenic mice, which exhibits several blocks in T cell development [32]. Since signaling from TCR and CD28 act synergistically to activate downstream signaling pathways and transcription factors, Id1 expression may augment these activities, thus allowing the cells to reach the threshold required for T cell activation without triggers from CD28.
From the physiological point of view, Id3 transcription is known to be activated by pre-TCR/TCR signaling in a Ras/MAP kinase-dependent manner [33;34]. Indeed, we have found Id3 up-regulation heavily depends on anti-CD3 and anti-CD28 co-stimulation (data not shown). Since Id1 and Id3 have redundant functions, ectopic expression of Id1 in our animal model could mimic the situation where Id3 is up-regulated. The ability of Id1 to replace co-stimulation in T cell activation suggests that up-regulation of Id3 may be one of the key downstream events necessary for T cell activation.
With regard to the mechanisms whereby Id1 exerts its co-stimulatory effects, we have found that upon anti-CD3 stimulation, Id1-expressing cells produced a 4-fold higher level of IL-2 than controls (Fig. 4A), resulting in a concentration in the culture supernatant, more than sufficient to support a robust proliferation of naïve T cells, as well as their survival. Consistently, incubation with neutralizing antibodies against IL-2 diminished the proliferative and survival advantage of Id1-expressing cells. Considering that one of the major downstream events of CD28 signaling is the up-regulation of IL-2 expression [26;27;35], the effect of Id1 on IL-2 production could explain why Id1 expression by-passed the need for co-stimulation with anti-CD28. IL-2 is well known to play an important role in T cell homeostasis and can certainly fulfill its role as an effector of co-stimulation [7;35]. Interestingly, Id1-stimulated IL-2 production appears to be specific for peripheral naïve CD4+ T cells since this phenomenon was not observed in Id1-expressing thymocytes [32].
The second mechanism involved in the co-stimulatory effect of Id1 expression is the enhancement of NF-κB activation. Anti-CD3 alone was able to activate NF-κB in Id1-expressing but not wild type naïve T cells. This result is similar to that found in Id1-expressing CD4+ thymocytes. Qi and Sun have previously shown that inhibition of NF-κB activation dramatically reduced the proliferative potential of these CD4+ thymocytes [32]. Furthermore, Id1 has be shown to augment NF-κB activity in primary DP or a DP cell line by promoting the activation of IκB kinases [36]. Like IL-2 production, activation of NF-κB is also a major downstream event of CD28 signaling [8]. NF-κB is crucial for facilitating cell survival, which can explain the increased viability of Id1-expressing naïve T cells during TCR activation and activation induced cell death (data not shown). Therefore, the ability of Id1 to augment NF-κB activity independent of exogenous co-stimulatory signals is likely involved in its effects on T cell activation in the absence of anti-CD28 antibodies.
Since Id1 is an inhibitor of E protein-mediated transcription, its co-stimulatory effects would likely involve repression of E protein-target genes, whose products may function to oppose the effects of CD28-mediated signaling. However, the identity of these genes has been elusive. Given that Id1 expression has been linked to increased IL-2 production and NF-κB activation as well as augmentation of MAP kinase activities in anti-CD3 stimulated DP thymocytes or naïve CD4+ T cells [37], it is possible that Id1 affects events higher up in the hierarchy of the co-stimulatory pathway. CD4-Id1 transgenic mice, which have intact thymic and peripheral T cell systems, will aid the investigation into the mechanisms whereby Id1 exerts its co-stimulatory effects, which will shed light on the importance of E proteins in controlling normal immunity and autoimmunity.
Supplementary Material
(A) Id1 transgenic construct. Id1 coding sequence was inserted downstream of exon 2 of the CD4 gene, followed by the SV40 polyadenylation site (SV40 Poly A). CD4 promoter (Prt) and enhancer (Enh) are designated by ovals as indicated. Black rectangles represent exons and the thin line in between shows the intron. The graph is not drawn to scale. (B) FACS analysis of thymocytes. Thymocytes from wild type (WT) and CD4-Id1tg (TG) mice were stained for CD4, CD8 and TCRβ. Dot plots show the representative CD4/CD8 staining profiles. The number in each quadrant indicates the percentage of cells in the gate and the absolute number of each subset is presented as mean ± SD on the right (n=4). The histograms show TCRβ staining of CD4 and CD8 SP thymocytes from WT (shaded) and TG (line) mice. (C) CD4/CD8 staining of cells from the spleen and lymph nodes. The percentage of gated populations is shown in the FACS plots. The numbers of CD4SP and CD8SP cells are presented as mean ± SD on the right (n=4). (D) CD44/ CD62L staining of CD4+ T cells from the lymph node of 4- month-old mice. The percentage of CD44hiCD62Llo and CD44hi CD62Lhi subsets is presented as mean ± SD on the right (n=4). ** p<0.01; * p<0.05; ns, not significant.
Acknowledgements
This work was supported by grants from National Basic Research Program of China (2011CB946100), National Natural Sciences Foundation of China (30830091, 31000393) and the 111 Project of China (B07001), and from the National Institute of Health (AI56129), and from Beijing Natural Science Foundation ( 7132114). XHS holds the Lew and Myra Chair in Biomedical Research.
We thank for Dr. Darryll D. Dudley for the generation of CD4-Id1 transgenic mice.
Abbreviations
- BrdU
5-bromo-2-deoxyuridine
- DN
double negative
- DP
double positive
- ELISA
Enzyme-linked immunosorbent assay
- EMSA
electrophoretic mobility shift assay
- Id
Inhibitor of differention
- IKK
IκB Kinase
- IL
interleukin
- PI
propidium iodide
- TCR
T cell receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shaw AS. T-cell activation and immunologic synapse. Immunol.Res. 2005;32:247–252. doi: 10.1385/IR:32:1-3:247. [DOI] [PubMed] [Google Scholar]
- 2.Abraham RT, Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat.Rev.Immunol. 2004;4:301–308. doi: 10.1038/nri1330. [DOI] [PubMed] [Google Scholar]
- 3.Crabtree GR, Clipstone NA. Signal transmission between the plasma membrane and nucleus of T lymphocytes. Annu.Rev.Biochem. 1994;63:1045–1083. doi: 10.1146/annurev.bi.63.070194.005145. [DOI] [PubMed] [Google Scholar]
- 4.Cheng J, Montecalvo A, Kane LP. Regulation of NF-kappaB induction by TCR/CD28. Immunol.Res. 2011;50:113–117. doi: 10.1007/s12026-011-8216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu.Rev.Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro VS, Truitt KE, Imboden JB, Weiss A. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol.Cell Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimmi CD, Freeman GJ, Gribben JG, Sugita K, Freedman AS, Morimoto C, Nadler LM. B-cell surface antigen B7 provides a costimulatory signal that induces T cells to proliferate and secrete interleukin 2. Proc.Natl.Acad.Sci.U.S.A. 1991;88:6575–6579. doi: 10.1073/pnas.88.15.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat.Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 9.Sun XH. Multitasking of helix-loop-helix proteins in lymphopoiesis. Adv.Immunol. 2004;84:43–77. doi: 10.1016/S0065-2776(04)84002-1. [DOI] [PubMed] [Google Scholar]
- 10.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol.Cell.Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim D, Peng XC, Sun XH. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol.Cell.Biol. 1999;19:8240–8253. doi: 10.1128/mcb.19.12.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun X-H. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 13.Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J.Exp.Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones ME, Zhuang Y. Stage-specific functions of E-proteins at the beta-selection and T-cell receptor checkpoints during thymocyte development. Immunol.Res. 2011;49:202–215. doi: 10.1007/s12026-010-8182-x. [DOI] [PubMed] [Google Scholar]
- 16.Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, Zhuang Y. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity. 2012;36:348–361. doi: 10.1016/j.immuni.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, Xu M, Nie L, Peng XC, Jimi E, Voll RE, Nguyen T, Ghosh S, Sun XH. Helix-loop-helix proteins regulate pre-TCR and TCR signaling through modulation of Rel/NF-kappaB activities. Immunity. 2002;16:9–21. doi: 10.1016/s1074-7613(02)00264-9. [DOI] [PubMed] [Google Scholar]
- 18.Wang HC, Perry SS, Sun XH. Id1 attenuates Notch signaling and impairs T-cell commitment by elevating Deltex1 expression. Mol.Cell Biol. 2009;29:4640–4652. doi: 10.1128/MCB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruyama T, Li J, Vaque JP, Konkel JE, Wang W, Zhang B, Zhang P, Zamarron BF, Yu D, Wu Y, Zhuang Y, Gutkind JS, Chen W. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat.Immunol. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 22.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 23.Haynes BF, Telen MJ, Hale LP, Denning SM. CD44--a molecule involved in leukocyte adherence and T-cell activation. Immunol.Today. 1989;10:423–428. doi: 10.1016/0167-5699(89)90040-6. [DOI] [PubMed] [Google Scholar]
- 24.Minami Y, Kono T, Miyazaki T, Taniguchi T. The IL-2 receptor complex: its structure, function, and target genes. Annu.Rev.Immunol. 1993;11:245–268. doi: 10.1146/annurev.iy.11.040193.001333. [DOI] [PubMed] [Google Scholar]
- 25.Boise LH, Minn AJ, Noel PJ, June CH, Accavitti MA, Lindsten T, Thompson CB. CD28 costimulation can promote T cell survival by enhancing the expression of Bcl-XL. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 26.Martin PJ, Ledbetter JA, Morishita Y, June CH, Beatty PG, Hansen JA. A 44 kilodalton cell surface homodimer regulates interleukin 2 production by activated human T lymphocytes. J.Immunol. 1986;136:3282–3287. [PubMed] [Google Scholar]
- 27.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 28.Baldwin AS. Regulation of cell death and autophagy by IKK and NF-kappaB: critical mechanisms in immune function and cancer. Immunol.Rev. 2012;246:327–345. doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- 29.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu.Rev.Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 31.Weil R, Israel A. Deciphering the pathway from the TCR to NF-kappaB. Cell Death.Differ. 2006;13:826–833. doi: 10.1038/sj.cdd.4401856. [DOI] [PubMed] [Google Scholar]
- 32.Qi Z, Sun XH. Hyperresponse to T-cell receptor signaling and apoptosis of Id1 transgenic thymocytes. Mol.Cell Biol. 2004;24:7313–7323. doi: 10.1128/MCB.24.17.7313-7323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras- ERK MAPK cascade. Nat.Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 34.DeRyckere D, Mann DL, DeGregori J. Characterization of transcriptional regulation during negative selection in vivo. J.Immunol. 2003;171:802–811. doi: 10.4049/jimmunol.171.2.802. [DOI] [PubMed] [Google Scholar]
- 35.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat.Rev.Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Liou HC, Sun XH. Id1 potentiates NF-kappaB activation upon T cell receptor signaling. J.Biol.Chem. 2006;281:34989–34996. doi: 10.1074/jbc.M608078200. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Wang HC, Sun XH. Id1 induces apoptosis through inhibition of RORgammat expression. BMC.Immunol. 2008;9:20. doi: 10.1186/1471-2172-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Id1 transgenic construct. Id1 coding sequence was inserted downstream of exon 2 of the CD4 gene, followed by the SV40 polyadenylation site (SV40 Poly A). CD4 promoter (Prt) and enhancer (Enh) are designated by ovals as indicated. Black rectangles represent exons and the thin line in between shows the intron. The graph is not drawn to scale. (B) FACS analysis of thymocytes. Thymocytes from wild type (WT) and CD4-Id1tg (TG) mice were stained for CD4, CD8 and TCRβ. Dot plots show the representative CD4/CD8 staining profiles. The number in each quadrant indicates the percentage of cells in the gate and the absolute number of each subset is presented as mean ± SD on the right (n=4). The histograms show TCRβ staining of CD4 and CD8 SP thymocytes from WT (shaded) and TG (line) mice. (C) CD4/CD8 staining of cells from the spleen and lymph nodes. The percentage of gated populations is shown in the FACS plots. The numbers of CD4SP and CD8SP cells are presented as mean ± SD on the right (n=4). (D) CD44/ CD62L staining of CD4+ T cells from the lymph node of 4- month-old mice. The percentage of CD44hiCD62Llo and CD44hi CD62Lhi subsets is presented as mean ± SD on the right (n=4). ** p<0.01; * p<0.05; ns, not significant.