Abstract
Epigenetic dysregulation of gene expression is thought to be critically involved in the pathophysiology of Alzheimer’s disease (AD). Recent studies indicate that DNA methylation and DNA hydroxymethylation are 2 important epigenetic mechanisms that regulate gene expression in the aging brain. However, very little is known about the levels of markers of DNA methylation and hydroxymethylation in the brains of patients with AD, the cell-type specificity of putative AD-related alterations in these markers, as well as the link between epigenetic alterations and the gross pathology of AD. The present quantitative immunohistochemical study investigated the levels of the 2 most important markers of DNA methylation and hydroxymethylation, that is, 5-methylcytidine (5-mC) and 5-hydroxymethylcytidine (5-hmC), in the hippocampus of AD patients (n = 10) and compared these to non demented, age-matched controls (n = 10). In addition, the levels of 5-hmC in the hippocampus of a pair of monozygotic twins discordant for AD were assessed. The levels of 5-mC and 5-hmC were furthermore analyzed in a cell-type and hippocampal subregion–specific manner, and were correlated with amyloid plaque load and neurofibrillary tangle load. The results showed robust decreases in the hippocampal levels of 5-mC and 5-hmC in AD patients (19.6% and 20.2%, respectively). Similar results were obtained for the twin with AD when compared to the non-demented co-twin. Moreover, levels of 5-mC as well as the levels of 5-hmC showed a significant negative correlation with amyloid plaque load in the hippocampus (rp = −0.539, p = 0.021 for 5-mC and rp = −0.558, p = 0.016 for 5-hmC). These human postmortem results thus strengthen the notion that AD is associated with alterations in DNA methylation and hydroxymethylation, and provide a basis for further epigenetic studies identifying the exact genetic loci with aberrant epigenetic signatures.
Keywords: Alzheimer’s disease, Epigenetics, DNA methylation, DNA hydroxymethylation, Amyloid
1. Introduction
The biological mechanisms that underlie age-related dysfunction and the pathophysiology of Alzheimer’s disease (AD) are still largely unknown. Recent findings have suggested that aging and AD are associated with profound changes in the epigenetic regulation of gene expression, especially at the level of DNA methylation (Chouliaras et al., 2010a; Mastroeni et al., 2011). DNA methylation, 1 of the fundamental epigenetic mechanisms, regulates gene transcription and can result in long-term changes in cellular function (Jaenisch and Bird, 2003). Methylation of CpG di-nucleotides is catalyzed by DNA methyltransferases (Dnmts), disrupts the binding of transcription factors and recruits proteins known as methyl CpG–binding domain proteins (MBDs) that are associated with chromatin compaction and gene silencing (Cedar and Bergman, 2009). DNA hydroxymethylation is a newly described epigenetic modification derived from the oxidation of methylated cytosines by ten-eleven translocation (TET) enzymes (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Recent evidence suggested that the biological role of DNA hydroxymethylation might be different from DNA methylation, while it is highly abundant in the brain compared to other tissues (Jin et al., 2011; Munzel et al., 2010; Valinluck and Sowers, 2007; Valinluck et al., 2004).
Aging is the most important risk factor for developing AD and is associated with aberrant DNA methylation patterns. In particular, aging has been linked with global DNA hypomethylation and hypermethylation of CpG islands in various bran regions, including the frontal and temporal cortex (Christensen et al., 2009; Hernandez et al., 2011; Siegmund et al., 2007). Other recent reports have also shown alterations in the DNA hydroxymethylation marker 5-hydroxymethylcytidine (5-hmC) in the aging mouse brain (Chouliaras et al., 2012; Song et al., 2011; Szulwach et al., 2011b).
Evidence from genetic and immunohistochemical studies on AD cases further supports the notion that aberrant DNA methylation and hydroxymethylation are involved in the pathophysiology of AD. A genetic association study reported an increased risk of AD for carriers of certain genetic variants in TET1 (Morgan et al., 2008). Immunohistochemical studies showed decreased levels of the DNA methylation marker 5-methylcytidine (5-mC) and a variety of enzymes and proteins involved in the process of DNA methylation, such as Dnmt1, and methyl CpG–binding protein 2 (MeCP2), in the entorhinal cortex of AD patients, (Mastroeni et al., 2010). Similar findings of decreased 5-mC were observed in the frontal cortex of a monozygotic twin pair discordant for AD (Mastroeni et al., 2009)—the same twin pair used in the present study. Recent epigenetic studies furthermore indicate gene-specific alterations in DNA methylation in the brain, peripheral lymphocytes, and transgenic mouse models of AD neuropathology (Bakulski et al., 2012; Bihaqi et al., 2011; Bollati et al., 2011; Fuso et al., 2012; Wang et al., 2008), as reviewed by Chouliaras et al. (2010a). For example, Bakulski et al. (2012) have found disease-associated methylation differences in 948 of 27,578 CpG sites examined using a genome-wide DNA methylation array in DNA derived from the human prefrontal cortex (Bakulski et al., 2012), whereas Fuso et al. (2012) have reported a decrease in the methylation of the presenilin 1 gene (PSEN1) promoter induced by vitamin B deficiency in cell lines and transgenic mouse models of AD, carrying familial mutations of the amyloid precursor protein (APP) gene (Fuso et al., 2011).
Thus, accumulating evidence suggests a role for DNA methylation and hydroxymethylation in AD. However, it is currently unknown whether the previously reported AD-related decrements in DNA methylation and associated molecules in the entorhinal cortex are also present in the hippocampus, although alterations in DNA hydroxymethylation have not been investigated so far in AD. Furthermore, given the fact that most investigations were carried out on whole-tissue homogenates, it remains unclear whether the various hippocampal subregions and hippocampal cell types show selective vulnerability for alterations in epigenetic markers, and whether hippocampal alterations in epigenetic markers correlate with hippocampal amyloid plaque and/or neurofibrillary tangle load.
2. Methods
2.1. Human subjects and tissue processing
Postmortem brain materials were collected through the Banner Sun Health Research Institute Brain and Body Donation Program (Sun City, AZ). Blocks of fixed, paraffin-embedded hippocampal tissue (at the level of the lateral geniculate nucleus) from 10 AD cases and 10 age-matched controls were obtained from the donated brains. All tissue samples were collected under institutional review board—approved protocols and informed consents permitting use of samples for research purposes. Antemortem evaluation by board-certified neurologists and neuropsychologists and postmortem evaluation by a board-certified neuropathologist were performed for all cases as described previously (Beach et al., 2012). The diagnostic criteria followed consensus guidelines for National Institute on Aging Alzheimer’s Disease Centers. Age, sex, and postmortem interval were well matched between the groups (Table 1). The hippocampal tissue was sliced axially into 1-cm thick blocks, immersed to 4% parafolmaldehyde fixative solution for 48 hours at 4 °C and embedded in paraffin. Subsequently, the paraffin blocks were cut into 6-μm thick slices on a rotary microtome (Leica, Wetzlar, Germany), placed on microscope slides (VWR, Batavia, IL) and stored in a dark container at room temperature until further use.
Table 1.
Characteristics of Alzheimer's disease casees and non-demented controls
| Characteristic | Non-demented controls | Alzheimer’s disease cases |
|---|---|---|
| Age | Mean = 77.91, SD = 4.1 | Mean = 75.36, SD = 5.5 |
| Sex (female/male) | 3/7 | W = 3, M = 7 |
| Postmortem interval | Mean = 2.77, SD = 0.61 | Mean = 2.98, SD = 0.93 |
| Braak staging | I–III | V–VI |
| Hippocampal amyloid plaques |
Mean = 0.00, SD = 0.00 | Mean = 2.22, SD = 0.56 |
| Hippocampal neurofibrillary tangles |
Mean = 0.81, SD = 0.75 | Mean = 2.90, SD = 0.20 |
The tissue from the monozygotic twin pair was obtained from the Boston University Alzheimer’s Disease Center. Written informed consent for autopsy was obtained for both cases in compliance with institutional guidelines of Boston University. Details regarding this monozygotic twin pair discordant for AD have been described previously (Mastroeni et al., 2009). In brief, antemortem and postmortem evaluations were performed by board-certified neurologists and a neuropathologist who determined the diagnosis according to the standard NIH AD centers protocols. The AD twin died at 76 years of age, and the non-demented control (ND) twin at 79 years of age. Postmortem delay was 7.3 hours for the AD twin and 3.1 for the ND twin. They were autopsied at the same facility using the same tissue processing protocols. Because of limitations in hippocampal tissue availability, only sections containing the CA1 region were available (protocols described below). In brief, the sections were sliced axially into 1-cm thick slabs, immersion fixed for 48 hours in 4% parafolmaldehyde at 4 °C, washed in phosphate buffer (PB), and cryoprotected in ethylene glycol and glycerol. The slabs were then sectioned at 40 μm on a cryostat. Free-floating sections were stored in freezing solution (glycol/glycerol/PB) at −20 °C until further use (Mastroeni et al., 2009).
2.2. Immunohistochemistry and image analysis
For each marker, the sections were immunoreacted simultaneously using standard immunohistochemical procedures. In case of paraffin-embedded sections, deparaffination in 2 series of xylene, and rehydration in series of 100%, 96%, and 80% ethanol, and distilled water was performed before the immunohistochemical procedure. Briefly, the sections were further rinsed in phospate-buffered saline Tween-20 (PBS-T), followed by antigen retrieval using 10 mmol/L citrate buffer at 95 °C for 20 minutes, after which the sections were rinsed again with PBS-T and incubated in 1% H2O2 for 30 minutes to quench endogenous peroxidase. Subsequently, the sections were rinsed in PBS-T blocked in 3% bovine serum albumin and incubated with the primary antibodies (5-mC, diluted 1:1000, Genway Biotech, San Diego CA; 5-hmC, diluted 1:5000, Active Motif, Carlsbad, CA) overnight at room temperature. After primary antibody incubation the sections were washed again and incubated for 2 hours with the secondary antibody (goat anti-mouse alexa 488 for 5-mC and goat anti-rabbit alexa 488 for 5-hmC, 1:5000; Invitrogen, Grand Island, NY). After washing in PBS-T, the sections were counterstained with Neurotrace red fluorescent stain (Invitrogen), washed again in PBS-T, taken through Sudan Black B (Sigma Aldrich, St Louis, MO) to reduce autofluorescence, air dried overnight, and coverslipped using Vectashield mounting medium (Vector Labs, Burlingame, CA). The Neurotrace counterstain was performed so as to use neuron-specific analyses based on the characteristic morphology and size of neuronal cells when stained with this counterstain. The specificity of the 5-mC and 5-hmC primary antibodies was confirmed by using methylated or hydroxymethylated nucleotides (10 nmol/L 5-mC dCTP, 10 nmol/L 5-hmC dCTP; Zymoresearch, Irvine, CA), which brought IR levels to background signal when pre-incubated with the primary antibodies (data not shown).
An Olympus IX51 microscope (Olympus, Tokyo, Japan) was used for all imaging procedures. Images were taken with the ×40 objective using a green fluorescence filter for each staining, whereas their corresponding Neurotrace-counterstained images were taken using a red fluorescence filter. All images were taken using identical camera, microscope lens, and light settings. One hippocampal section was used per each staining, where images were taken from 5 sites in the DG, 5 sites in the CA3, and 5 sites in the CA1 per subject (total of 15 images per subject for each staining concomitant with 15 corresponding Neutrorace-counterstained images).
Fluorescence intensity analysis was performed using ImageJ software (ImageJ, U.S. National Institutes of Health, Bethesda, MD; imagej.nih.gov/ij/). The intensity measurements were corrected for background differences by dividing the measured intensities with the average intensity of a cell-free region, such as the hippocampal white matter, in each section. For each image, the fluorescence intensity of 15 individual neurons and 15 individual cells of non-neuronal origin were analyzed by delineating the nucleus of each cell and measuring the mean intensity value. Thus, the fluorescence intensity of a total of 450 cells per individual was analyzed per staining. The discrimination between cells of neuronal and non-neuronal origin was done based on their characteristic morphology and size of neurons when using the Neurotrace counterstain (Sekirnjak et al., 2003). According to these criteria, a clear differentiation between neurons and glia in the DG was not possible, and thus no cell-type specific analyses were carried on in this subregion. Of note, the measurements of fluorescence intensities were calculated in arbitrary units and do not represent absolute levels of these markers (Fig. 1).
Fig. 1.
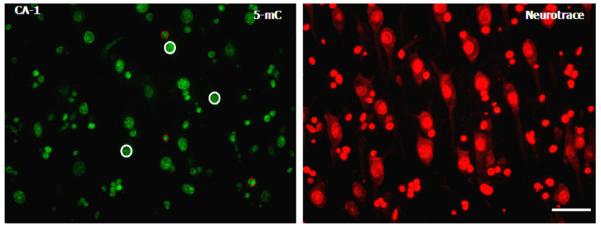
Hippocampal 5-methylcytidine (5-mC) immunoreactivity (IR) and Neurotrace counterstain. Representative photomicrograph of 5-mC IR (green) and the corresponding Neurotrace image (red) taken from the CA1 hippocampal subregion. Based on the Neurotrace morphology, a total of 15 neurons and 15 glial cells were delineated within each photomicrograph, and their fluorescence intensity was analyzed using ImageJ software (details in text). Representative neurons are highlighted with a continuous white circle, whereas glial cells are highlighted with a dotted red circle. A total of 15 images per subject for each staining concomitant with 15 corresponding Neutrorace-counterstained images were analyzed. Scale bar = 50 μm.
2.3. Statistical analysis
A multilevel linear mixed model was used for the statistical analyses. For each marker, the fluorescence intensities were used as the dependent variable and the AD diagnosis as the independent variable. Measurements of individual cells (level 1) were clustered in the different subjects (level 2). Statistical significance was set at α = 0.05. Following the multilevel analyses, stratified analyses for neuronal cells and cells of non-neuronal origin (glial cells) were performed for each hippocampal subregion (CA3, CA1-2) or just for neuronal cells (DG) separately using the same statistical model. Correlation analyses were carried out by calculating the Pearson’s correlation coefficient (rp). All statistical calculations were performed using STATA 11 (StataCorp, College Station, TX) or the Statistical Package for the Social Sciences (SPSS 17; SPSS Inc, Chicago, IL). Graphs were built in GraphPad Prism (Version 4, GraphPad Software, San Diego, CA).
3. Results
3.1. Qualitative analysis and quantitative analyses of 5-mC and 5-hmC IR in the hippocampus
Both 5-mC and 5-hmC IR were detected in all cells, and were primarily observed as punctate IR in the nucleus. Microscopic inspection suggested a striking decrease in IR of both markers in the AD hippocampus when compared to controls (Figs. 2 and 3), and a similar pattern was observed for 5-hmC IR in the hippocampus of the AD twin when compared to the ND co-twin. Quantitative image analyses were further carried out to confirm these findings. Multilevel linear model analysis revealed a decrease of 5-mC IR in the hippocampus of AD patients when compared to ND controls (−19.6%, p = 0.006). Similarly a significant decrease of 5-hmC IR (−20.2%, p = 0.012) was observed in the AD hippocampus when compared to ND controls (Fig. 4).
Fig. 2.
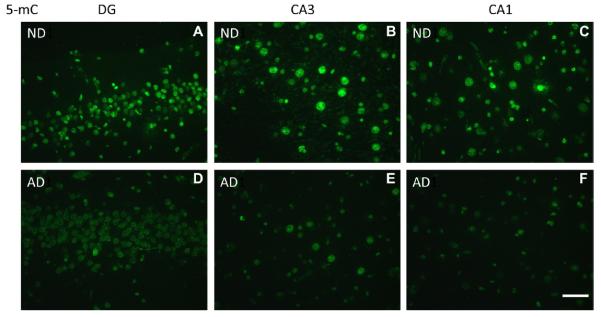
Representative photomicrographs of 5-mC immunoreactivity (IR). High magnification representative photomicrographs of the hippocampal DG, CA3, and CA1-2 regions. (A–C) Non-demented control cases (ND) and (D–F) Alzheimer’s disease cases (AD). Note: A loss of 5-mC IR is observed in AD cases when compared to ND controls in all 3 hippocampal subregions. Images were taken with a ×40 objective. Scale bar = 50 μm.
Fig. 3.
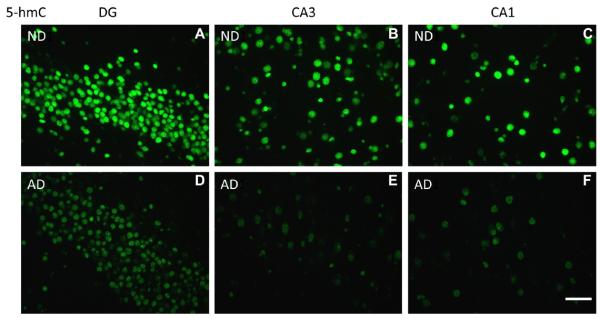
Representative photomicrographs of 5-hydroxymethylcytosine (5-hmC) immunoreactivity (IR). High magnification representative photomicrographs of the hippocampal DG, CA3, and CA1-2 regions. (A–C) Non-demented control cases (ND) and (D–F) Alzheimer’s disease cases (AD). Note: A decrease of 5-hmC IR is observed in AD cases when compared to ND controls in all 3 hippocampal subregions. Images were taken with a ×40 objective. Scale bar = 50 μm.
Fig. 4.
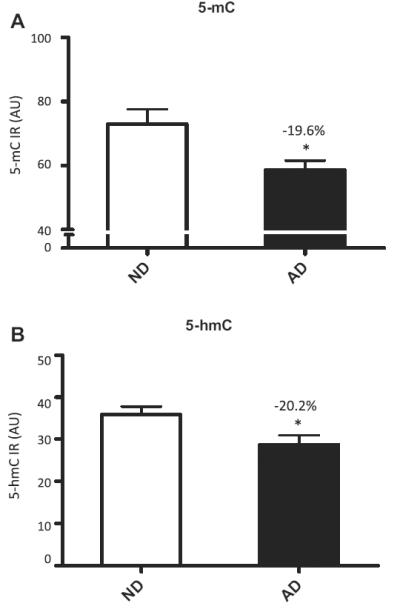
5-mC and 5-hmC hippocampal fluorescence intensities. Mean and standard error of the mean of fluorescence intensity measurements of 5-mC (A) and 5-hmC (B) immunoreactivity (IR). Pooled data from the 3 hippocampal subregions of non-demented control cases (ND; open bars) and Alzheimer’s disease cases (AD; filled bars). The percentage of decrease in each analysis and the significant effects (p < 0.05 in all cases) are indicated with an asterisk in each graph. Abbreviation: AU, arbitrary units.
3.2. 5-mC and 5-hmC in glial cells and neurons of the hippocampal subregions
Analyses of 5-mC IR in neurons and glial cells in the 3 hippocampal subregions revealed that AD patients had a significantly decreased level of 5-mC in glial cells in the CA3 (−26.9%, p = 0.016), neurons in the CA1(−21.1%, p = 0.01), and glial cells in the CA1 ( −25.7%, p = 0.003), but there were no statistically significant differences for the DG or the CA3 neurons (Fig. 5).
Fig. 5.
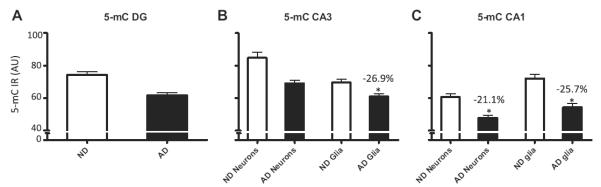
Stratified hippocampal 5-mC fluorescence intensities. Mean and standard error of the mean of fluorescence intensity measurements of 5-mC immunoreactivity (IR) (A–C). Pooled data from the non-demented control cases (ND; open bars) and Alzheimer’s disease cases (AD; filled bars) are represented separately for the DG (A), CA3 (B), and CA1-2 (C). The percentage of decrease in each analysis and the significant effects (p < 0.05 in all cases) are indicated with an asterisk in each graph. Abbreviation: AU, arbitrary units.
Analyses of 5-hmC IR in neurons and glial cells in the 3 hippocampal subregions revealed that AD patients had a significantly decreased level of 5-hmC in cells of the DG (−16.1%, p = 0.042) and glial cells in the CA3 (−34.2%, p = 0.011), whereas tendencies towards statistical significance were observed for neurons within the CA3 (−18.4%, p = 0.064) and the neurons in the CA1 (−17.8%, p = 0.083). No statistically significant differences for glia cells in the CA1 were found (Fig. 6).
Fig. 6.
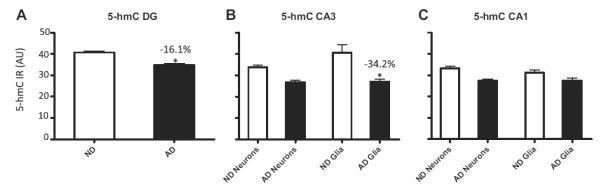
Stratified hippocampal 5-hmC fluorescence intensities. Mean and standard error of the mean of fluorescence intensity measurements of 5-hmC immunoreactivity (IR) (A–C). Pooled data from the non-demented control cases (ND; open bars) and Alzheimer’s disease cases (AD; filled bars) are represented separately for the DG (A), CA3 (B), and CA1-2 (C). Percentage of decrease in each analysis and the significant effects (p < 0.05 in all cases) are indicated with an asterisk in each graph. Abbreviation: AU, arbitrary units.
3.3. 5-hmC in the monozygotic twins discordant for AD
Analysis of 5-hmC fluorescence intensity in the twins discordant for AD revealed a 31.4% reduction of 5-hmC IR in the CA1 of the hippocampus of the AD twin. Stratified analyses showed a reduction of 39.6% in the neurons and 20.7% in the glial cells of the CA1 subregion of the AD twin compared to the ND twin (Fig. 7).
Fig. 7.
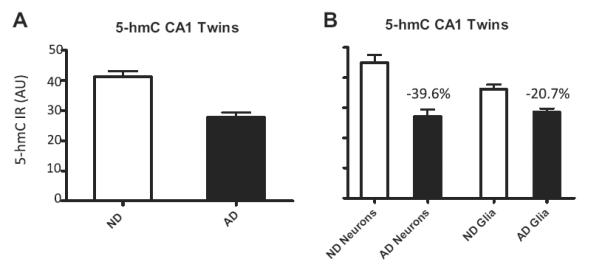
Hippocampal 5-hmC fluorescence intensities in the monozygotic twin pair discordant for Alzheimer’s disease cases (AD). Mean and standard error of the mean of fluorescence intensity measurements of 5-hmC immunoreactivity (IR) (A and B). Pooled data (A) and stratified data from neurons and glial cells (B) from the non-demented control case (ND; open bars) and Alzheimer’s disease case (twin) (AD; filled bars) are represented for the CA1 hippocampal subregion. Percentages of decrease in each analysis are indicated in each graph. Abbreviation: AU, arbitrary units.
3.4. Correlation analyses
Linear correlation analyses revealed significant negative correlations between 5-mC IR and hippocampal amyloid plaque load (rp = −0.539, p = 0.021), as well as 5-hmC IR and hippocampal amyloid plaque load (rp = −0.558, p = 0.016). Similarly, a trend toward statistical significance for a negative correlation was observed between hippocampal 5-mC IR and neurofibrillary tangle load (rp = −0.461, p = 0.054) (Fig. 8).
Fig. 8.
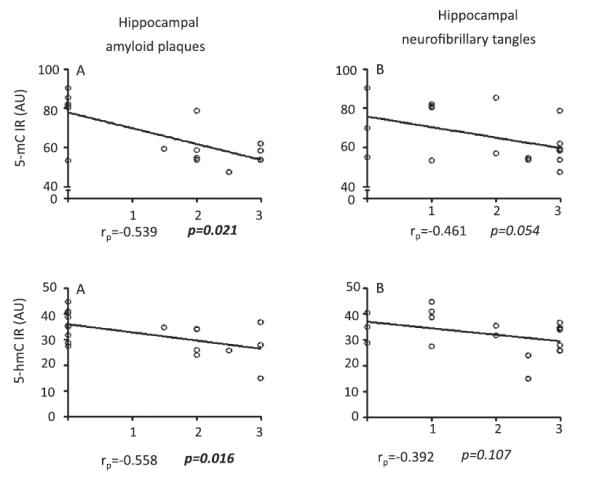
Correlation analysis between hippocampal 5-mC, 5-hmC immunoreactivity (IR) and amyloid plaque and neurofibrillary tangle loads. Spearman’s correlation coefficients and p values are noted in the bottom of each graph.
4. Discussion
Qualitative and quantitative assessment of the sections of the human hippocampus revealed an AD-related decrease of both the DNA methylation marker 5-mC and the DNA hydroxymethylation marker 5-hmC. Moreover, similar patterns of decreased 5-hmC IR were observed in the CA1 hippocampal subregion in the affected twin of a monozygotic twin pair discordant for AD.
4.1. AD is associated with decreased hippocampal 5-mC IR
In the present study, we report that 5-mC IR, as a marker for global DNA methylation, is decreased in the hippocampus of AD patients when compared to ND controls. Pooled data from all cell-types and all regions also revealed a decrease in 5-hmC IR, reflecting global DNA hydroxymethylation. Stratification of the data according to cell-type and subregions reveals that the observed differences are particularly pronounced in the CA1 neurons and glial cells and in the glial cells residing in the CA3, whereas no differences were observed in the DG.
The observed differences in 5-mC are in agreement with previous reports of a global loss of methylation in the entorhinal cortex of AD patients (Mastroeni et al., 2010) and can further be linked to aberrant methylation patterns previously associated with AD (Bakulski et al., 2012; Siegmund et al., 2007; Wang et al., 2008). Although the exact reason of the observed methylation changes is not yet clear, this might be related to AD-specific deficits in the nuclear transport of epigenetic molecules, such as Dnmt1, which is responsible for maintenance of DNA methylation (Mastroeni et al., 2013). This disturbed nucleocytoplasmic transport has been linked with the downregulation of the Ras-related nuclear protein (RAN), which plays an important role in the transport of molecules into and out of the cell nucleus (Mastroeni et al., 2013).
4.2. AD is associated with decreased hippocampal 5-hmC IR
Besides DNA methylation marker 5-mC, which reflects a wellknown epigenetic modification, the present study also addresses 5-hmC, which represents a recently described epigenetic modification. To our knowledge, this is the first study to investigate changes of 5-hmC in human AD. Pooled analyses showed a decrease of 5-hmC IR in the hippocampus of AD patients compared to ND controls, and stratified analyses showed that the observed changes were more pronounced in the CA3 and the DG regions. As with 5-mC IR, an AD associated decrease was the main effect, in both neurons and glial cells. Because 5-hmC is an exclusive product of 5-mC, it could be speculated that the observed changes in 5-hmC are closely associated with the decrease of 5-mC. Interestingly, although age associated increases of hippocampal 5-hmC have been reported (Chouliaras et al., 2012; Song et al., 2011) in normal aging, our recent work in transgenic APPswe/PS1ΔE9 mice suggests a imbalance of hippocampal 5-mC and 5-hmC in APPswe/Ps1ΔE9 when compared to that in wild-type mice (Chouliaras et al., submitted), In parallel, another study reported that a genetic variant of TET1, 1 of the enzymes that catalyze DNA hydroxymethylation, is associated with an increased risk of developing AD (Morgan et al., 2008). Such findings suggest that 5-hmC and an altered methylation/hydroxymethylation potential might be linked with AD pathology (Van den Hove et al., 2012). The exact consequences of the observed changes of 5-hmC are unknown, but as with 5-mC, could be associated with gene expression alterations and cell-cycle reprogramming of cells in AD (Blalock et al., 2004; Busser et al., 1998; Nagy et al., 1998), considering that 5-hmC is known to play an important role in regulating stem cell function (Haffner et al., 2011; Pastor et al., 2011; Szulwach et al., 2011a).
4.3. Twins
Similar to the comparison between AD cases and controls, 5-hmC IR was diminished in the hippocampus of the AD twin compared to the ND sibling. Unfortunately, because of limitations associated with the availability of brain materials from monozygotic twins discordant for AD, we were able to examine 5-hmC IR only in the CA1 hippocampal subregion. Previous reports on the same cases have already shown decrements of DNA methylation and associated markers in the temporal neocortex of this twin pair (Mastroeni et al., 2009). We now report that the hippocampal 5-hmC changes are comparable to the general cohort, with an AD-associated decrease of hippocampal 5-hmC IR. Notably, the occupation-related environmental exposures of the AD twin were different from those of the ND twin, and this could be associated with the development of AD (Mastroeni et al., 2009).
4.4. Negative correlations between hippocampal AD pathology and DNA (hydroxy)methylation markers
There were negative correlations between hippocampal 5-mC and 5-hmC IR and amyloid plaque pathology. Moreover, a trend toward significance pointed to a negative correlation between 5 mC IR and hippocampal neurofibrillary tangle load Such findings suggest that AD pathology is associated with epigenetic changes. However, it is not yet clear whether the observed epigenetic changes are the cause or the consequence of AD pathology, and future studies should therefore assess whether the observed epigenetic changes precede AD pathology.
4.5. Conclusions and future directions
Future studies should identify the gene-specific epigenetic changes that are associated with AD. Most of the techniques developed to date require large amounts of input DNA, and whole-tissue homogenates are mainly used for that purpose. However, cell-type- and subregion-specific changes, and neuronal loss occur in AD, which might alter the neurons/glia ratio when comparing AD brains to controls (Blalock et al., 2011; Ginsberg et al., 2012 ; Liang et al., 2008; Morrison and Hof, 2002; Torres-Munoz et al., 2004). Thus, novel epigenetic approaches using cell-sorting or laser-capture microd issected tissues and requiring less input material should be further developed.
Interestingly, as epigenetic changes are responsive to environmental stimuli, it should be examined whether brain-related profiles correlate with peripheral profiles, and whether changes of epigenetic markers in accessible tissue types may be used as a biomarker for AD. Epigenetic mechanisms are influenced by environmental exposures, and the data on discordant monozygotic twins further supports this notion. Specific environmental exposures that might be associated with AD, such as previous reports on metal/lead exposure, related to the latent early-life associated regulation (LEARn) model hypothesis, appear to be promising in this aspect (Bihaqi and Zawia, 2012; Lahiri et al., 2009). Furthermore, specific gene—environment interactions should be investigated, as it is known that the effect of certain environmental exposures is highly dependent on individual genetic susceptibility (Chouliaras et al., 2010b).
Future research needs also to investigate the exact genomic localization and downstream effects of the observed AD-associated epigenetic changes, such as the effects on histone modifications, and gene and protein expression. Although it is not yet clear whether epigenetic dysregulation is a cause or a consequence of the disease, the fact that similar effects have also been observed in transgenic APPswe/PS1ΔE9 mice (Chouliaras et al., submitted) suggest that the observed DNA methylation and hydroxymethylation changes might be triggered by the Aβ pathology and may be associated with disturbances of nuclear-cytoplasmic transport of epigenetic molecules (Mastroeni et al., 2013). Given the limitations of the current technologies and the limited availability of brain tissue, examination of peripheral blood in cohort studies or animal studies could possibly answer these specific and related questions. Furthermore, examination of correlations with markers associated with the disease, such as cognitive decline, synaptic deficits, and neuronal loss, is of high importance (Coleman and Yao, 2003; Rutten et al., 2005; Selkoe, 2002). Meanwhile, the exact consequences of the observed epigenetic changes are unknown; however, considering the role of epigenetic programming in the development and differentiation of cells, it could be speculated that the observed epigenetic changes induce gene expression changes, the accumulation of which may result in neuronal loss. As such, it will be crucial to clarify the involvement of epigenetic changes in the pathophysiology of AD. The present findings suggest that AD is associated with global changes of DNA methylation and hydroxymethylation, and further show negative correlations between these epigenetic markers and hippocampal AD pathology.
Acknowledgements
Funds have been provided by the Internationale Stichting Alzheimer Onderzoek (ISAO), grant number 07551 and 11532 (D.L.A. vdH.), by the ISAO grant number 09552, and the Netherlands Organization for Scientific Research (NWO, Veni Award 916.11.086) (B.P.F.R.), and by a Marie Curie Host Fellowship Grant MC-EST 020589 EURON (L.C). P.R.H. is supported by National Institutes of Health (NIH) grant P50 AG05138. P.D.C. is supported by NIH RO1 AG036400 grant and Arizona Alzheimer’s Research Center DHS Award FY 2012.
Footnotes
L.C, D.M., B.P.F.R., and D.L.A. vdH. contributed equally to this work.
Disclosure statement
None of the authors has any conflicts of interest with the present work.
References
- Bakulski KM, Dolinoy DC, Sartor MA, Paulson HL, Konen JR, Lieberman AP, Albin RL, Hu H, Rozek LS. Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J. Alzheimers Dis. 2012;29:571–588. doi: 10.3233/JAD-2012-111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach TG, Sue LI, Walker DG, Sabbagh MN, Serrano G, Dugger BN, Mariner M, Yantos K, Henry-Watson J, Chiarolanza G, Hidalgo JA, Souders L. Striatal amyloid plaque density predicts Braak neurofibrillary stage and clinicopathological Alzheimer’s disease: implications for amyloid imaging. J. Alzheimers Dis. 2012;28:869–876. doi: 10.3233/JAD-2011-111340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi SW, Huang H, Wu J, Zawia NH. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer’s disease. J. Alzheimers Dis. 2011;27:819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- Bihaqi SW, Zawia NH. Alzheimer’s disease biomarkers and epigenetic intermediates following exposure to Pb in vitro. Curr. Alzheimer Res. 2012;9:555–562. doi: 10.2174/156720512800617964. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW. Micro-array analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J. Chem. Neuroanat. 2011;42:118–126. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Galimberti D, Pergoli L, Dalla Valle E, Barretta F, Cortini F, Scarpini E, Bertazzi PA, Baccarelli A. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav. Immun. 2011;25:1078–1083. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer’s disease brain. J. Neurosci. 1998;18:2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Rutten BP, Kenis G, Peerbooms O, Visser PJ, Verhey F, van Os J, Steinbusch HW, van den Hove DL. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog. Neurobiol. 2010a;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Sierksma AS, Kenis G, Prickaerts J, Lemmens MA, Brasnjevic I, van Donkelaar EL, Martinez-Martinez P, Losen M, De Baets MH, Kholod N, van Leeuwen F, Hof PR, van Os J, Steinbusch HW, van den Hove DL, Rutten BP. Gene-environment interaction research and transgenic mouse models of Alzheimer’s disease. Int. J. Alzheimers Dis. 2010b doi: 10.4061/2010/859101. 2010. http://dx.doi.org/10.4061/2010/859101. [DOI] [PMC free article] [PubMed]
- Chouliaras L, van den Hove DL, Kenis G, Keitel S, Hof PR, van Os J, Steinbusch HW, Schmitz C, Rutten BP. Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr. Alzheimer Res. 2012;9:536–544. doi: 10.2174/156720512800618035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, Rutten BP, Mastroeni D, Kenis G, van Os J, Steinbusch HW, Coleman P, van den Hove DL. Age-related disturbances in DNA methylation and hydroxymethylation in hippocampus of transgenic APPswe/PS1dE9 mice. Submitted. [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman PD, Yao PJ. Synaptic slaughter in Alzheimer’s disease. Neurobiol. Aging. 2003;24:1023–1027. doi: 10.1016/j.neurobiolaging.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Fuso A, Nicolia V, Pasqualato A, Fiorenza MT, Cavallaro RA, Scarpa S. Changes in Presenilin 1 gene methylation pattern in diet-induced B vitamin deficiency. Neurobiol. Aging. 2011;32:187–199. doi: 10.1016/j.neurobiolaging.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Fuso A, Nicolia V, Ricceri L, Cavallaro RA, Isopi E, Mangia F, Fiorenza MT, Scarpa S. S-adenosylmethionine reduces the progress of the Alzheimer-like features induced by B-vitamin deficiency in mice. Neurobiol. Aging. 2012;33:1482. doi: 10.1016/j.neurobiolaging.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Che S. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer’s disease. Neurobiol. Dis. 2012;45:99–107. doi: 10.1016/j.nbd.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, Netto GJ, De Marzo AM, Yegnasubramanian S. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, Singleton AB. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;(33 Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Zawia NH. The LEARn model: an epigenetic explanation for idiopathic neurobiological diseases. Mol. Psychiatry. 2009;14:992–1003. doi: 10.1038/mp.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Ramsey K, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette CM, Schmechel D, Reiman EM, Rogers J, Stephan DA. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol. Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Chouliaras L, Grover A, Liang WS, Hauns K, Rogers J, Coleman PD. Reduced RAN expression and disrupted transport between cytoplasm and nucleus: a key event in Alzheimer’s disease pathophysiology. PLoS One. 2013;8:e53349. doi: 10.1371/journal.pone.0053349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol. Aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer’s disease. Neurobiol. Aging. 2011;32:1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AR, Hamilton G, Turic D, Jehu L, Harold D, Abraham R, Hollingworth P, Moskvina V, Brayne C, Rubinsztein DC, Lynch A, Lawlor B, Gill M, O’Donovan M, Powell J, Lovestone S, Williams J, Owen MJ. Association analysis of 528 intra-genic SNPs in a region of chromosome 10 linked to late onset Alzheimer’s disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:727–731. doi: 10.1002/ajmg.b.30670. [DOI] [PubMed] [Google Scholar]
- Morrison JH, Hof PR. Selective vulnerability of corticocortical and hippocampal circuits in aging and Alzheimer’s disease. Prog. Brain Res. 2002;136:467–486. doi: 10.1016/s0079-6123(02)36039-4. [DOI] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem. Int. Ed. Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Smith AD. The cell division cycle and the pathophysiology of Alzheimer’s disease. Neuroscience. 1998;87:731–739. doi: 10.1016/s0306-4522(98)00293-0. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten BP, Van der Kolk NM, Schafer S, van Zandvoort MA, Bayer TA, Steinbusch HW, Schmitz C. Age-related loss of synaptophysin immunoreactive presynaptic boutons within the hippocampus of APP751SL, PS1M146L, and APP751SL/PS1M146L transgenic mice. Am. J. Pathol. 2005;167:161–173. doi: 10.1016/S0002-9440(10)62963-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirnjak C, Vissel B, Bollinger J, Faulstich M, du Lac S. Purkinje cell synapses target physiologically unique brainstem neurons. J. Neurosci. 2003;23:6392–6398. doi: 10.1523/JNEUROSCI.23-15-06392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, Yoon YS, Ren B, He C, Jin P. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011a;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-Mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011b;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Munoz JE, Van Waveren C, Keegan MG, Bookman RJ, Petito CK. Gene expression profiles in microdissected neurons from human hippocampal subregions. Brain Res. Mol. Brain Res. 2004;127:105–114. doi: 10.1016/j.molbrainres.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hove DL, Chouliaras L, Rutten BP. The role of 5-hydroxymethylcytosine in aging and Alzheimer’s disease: current status and prospects for future studies. Curr. Alzheimer Res. 2012;9:545–549. doi: 10.2174/156720512800618008. [DOI] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]


