Abstract
MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) act in complex with the Argonaute family of proteins to regulate target messenger RNAs (mRNAs) post transcriptionally. SiRNAs typically induce endonucleolytic cleavage of mRNA with near-perfect complementarity. For targets with less complementarity, both translational repression and mRNA destabilization mechanisms have been implicated in miRNA-mediated gene repression, although the timing, coupling, and relative importance of these events have not been determined. Here, we review gene-specific and global approaches that probe miRNA function and mechanism, looking for a unifying model. More systematic analyses of the molecular specificities of the core components coupled with analysis of the relative timing of the different events will ultimately shed light on the mechanism of miRNA-mediated repression.
Over the past decade, small RNAs emerged as a new class of key regulators of eukaryotic biology (reviewed in (1, 2)). This diverse class of RNAs includes small interfering RNAs (siRNA), micro RNAs (miRNAs) and PIWI-interacting RNAs (piRNA), all of which associate with multiple protein components within a complex to regulate partially or perfectly complementary transcripts. A key component of all such ribonucleoproteins (RNPs) is a member of the Argonaute family of proteins. Subdivided into two classes, the Argonautes interact primarily with siRNAs and miRNAs whereas PIWI proteins associate with piRNAs.
Structure, function and Allostery in the Argonaute superfamily
At the level of bioinformatics, Argonaute family members can be identified on the basis of the presence of the ubiquitous PAZ, MID and PIWI domains. Two major clades are identifiable when any of the three core domains or the ensemble (PAZ, MID and PIWI) are used for classification (3, 4). More detailed groupings are revealed when the isolated MID domain alone is used for classification; these groupings match known functional specificities that sometimes over-ride organismal boundaries (4).
The four domains of the Argonaute protein are structurally organized in a bilobal architecture (5), the N-terminal and PAZ domains comprising one lobe, and the MID and PIWI domains comprising the other (Fig. 1). Although the N-terminal domain is poorly conserved, it has been implicated in various biochemical functions of the Argonautes (6, 7). The PAZ domain structurally resembles a deviated oligosaccharide- or oligonucleotide-binding fold that interacts with the terminal (3′) two nucleotides of small oligonucleotides (RNA or DNA) (8, 9). The Argonaute MID domain adopts a Rossmann-like fold (5) and contains an oligonucleotide 5′ end binding pocket (10–14). The C-terminal, or PIWI, domain of Argonautes resembles the nuclease RNase H and is responsible for the endonucleolytic activity (5).
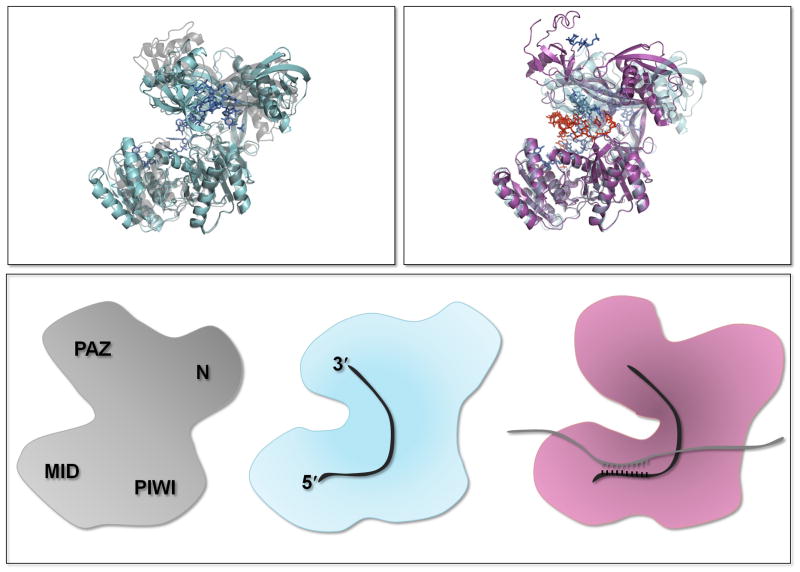
Figure 1.
Conformational changes in the Argonautes. (Top left) Comparison of apo-like (gray; PDB code 3DLB) and oligonucleotide-bound (blue; PDB code 3DLH) structures of bacterial Argonautes. (Top right) Comparison of the same oligonucleotide-bound (blue; PDB code 3DLH) and duplex-bound (pink; PDB code 3HXM) Argonautes. (Bottom) Conformational changes thought to reflect binding of miRNA and mRNA targets.
Duplex formation between the guide (the small RNA) and target (mRNA) oligonucleotides (where perfect complementarity extends to position 15 but not to position 12) brings about conformational changes in the bacterial Argonautes that preclude binding of the 3′ end of the guide strand to the PAZ binding pocket (6, 15). This observation are consistent with a two-step model wherein the 3′ end of the guide strand is released from the PAZ domain after target binding for the siRNA-loaded Argonautes, activating slicing (16). For miRNA-loaded Argonautes, target binding does not typically promote cleavage, likely because the interrupted helix permits continued binding of the 3′ end to the PAZ domain (6).
These structural observations converge with bioinformatic and biochemical insights into MID domain function. Allosteric behavior has been observed in the Argonaute proteins, with potential binding sites for small RNAs (and their target) and for m7G-cap like nucleotides (4). A recent MID domain structure from Neurospora crassa (14) reveals the 5′-end nucleotide binding site (10, 12) as well as an additional potential ligand-binding site; an equivalent second site was not observed in a recent structure of the hAgo2 MID domain (11). Association of the Argonaute protein with the miRNA alone (or duplexed with its target) may induce binding to the eukaryotic mRNA 5′ m7G cap structure as well as other proteins such as the GW182 family. Supporting this model, the loss of miRNA binding through mutations in the MID domain results in the loss of cap-binding activity, GW182 interaction, and the repression of gene expression (4, 17), although others argue that miRNA and GW182 binding are uncoupled (18). The GW182 class of proteins is likely a key mediator of cellular events for many members of the Argonaute family (19).
Insights into mechanism from gene-specific approaches
Analysis of the Caenorhabditis elegans heterochronic gene lin-14 and its regulation by the miRNA lin-4 provided early insights into the mechanism of miRNA-mediated gene regulation that have shaped the development of the field. These studies determined that miRNA-mediated regulation was posttranscriptional, because there were large effects on protein expression and no discernible effects on mRNA abundance (20, 21). Sucrose gradient analyses used to evaluate the translational status of the mRNA indicated that the lin-14 mRNA was found primarily in heavy polysomes and that its distribution in the profile was unaffected by the expression of its regulatory partner, miRNA lin-4. These data suggested that lin-4 regulates its target mRNA lin-14 through a process that affects translation at a postinitiation step (20).
These ideas were corroborated by results from other systems. Analysis both of authentic miRNA-mRNA partner interactions (in vivo) and of various engineered reporter genes fused to 3′ untranslated regions (UTRs) with miRNA-responsive elements (in cell culture systems, in vitro) revealed substantial degrees of regulation at the protein level, with more modest effects typically seen on the amounts of the mRNA target (7, 22–25). Other studies provided experimental support for the proposed postinitiation mechanism of translational repression (25, 26).
Concurrent studies, however, came to different conclusions. For example, in similar reporter systems, the migration of a miRNA-responsive mRNA reporter construct was shifted to lighter, nonpolysomal fractions in the presence of the miRNA (7); these data were consistent with models invoking the inhibition of translation initiation. There is now considerable evidence for this model. First, in human cells and C. elegans, endogenous mRNA targets are shifted to lighter polysomal fractions when actively repressed by miRNAs (27, 28). Second, in reporter-based cell culture systems, mRNAs with exogenously introduced internal ribosome entry sites fail to be repressed in a miRNA specific manner (7, 29). These data suggest that the miRNP complex specifically recognizes some feature of the cap-dependent initiation machinery; we note that other groups reported contradictory results (25, 30). Lastly, a number of in vitro reconstituted translation systems have implicated initiation as a likely target for miRNA-mediated repression (31–34). The likely importance of the m7G cap in miRNA-mediated repression is consistent with in vitro biochemical data showing that the Argonaute proteins can bind directly to the m7G cap (4), albeit not through a eukaryotic initiation factor 4E (eIF4E) motif as initially reported (35).
What can such experiments tell us about how translation might be targeted? Translation initiation in eukaryotes involves a large number of factors (eIFs) that deliver mRNA to the 40S ribosomal subunit. The initial encounter between the mRNA and the ribosome takes place at the m7G cap, “scanning” along the mRNA identifies the AUG start codon, the 60S subunit joins, and translation elongation begins. Any step along the way is a potential target for regulation (36). m7G binding by the Argonautes would be conceptually similar to the well-known role of 4E binding proteins in disrupting interactions between eIF4E and eIF4G (37). Alternatively, miRNAs could block initiation at a later stage, the subunit-joining step (38, 39), a model supported by in vivo and in vitro studies (40, 41). The data supporting miRNA-mediated repression of translation initiation could be satisfyingly explained by a model where the Argonaute-centered mRNP, bound to the 3′ UTR target site, directly binds to the m7G cap, diminishing the efficiency of translation initiation either by competing with eIF4E or by blocking subunit joining.
Despite initial indications that miRNAs repress translation of target genes without affecting the corresponding mRNA levels, many subsequent studies have challenged this simplistic view. Early microarray analyses showed that ectopic expression of a miRNA reduced the levels of target mRNAs carrying regions complementary to the seed (42). In zebrafish, deadenylation is triggered by miRNP interactions, suggesting that mRNAs are generally destabilized by the action of the miRNP complex (43). Endogenous targets of two C. elegans miRNAs (let-7 and lin-4) are degraded during the larval stage, when the relevant miRNAs are produced (44), contrary to earlier reports (20, 21). Lastly, numerous studies with reporter constructs show losses in mRNA abundance that correlate with the silencing of gene expression (23, 45). These connections between miRNAs and RNA degradation are supported by biochemical and genetic studies describing the GW182-dependent recruitment of poly (A)–binding protein (PABP) and the deadenylase (CCR4:NOT1) and decapping (DCP1:DCP2) machineries (23, 34, 46). These data make a compelling case for mRNA decay being an important component of miRNA-mediated gene silencing.
Insights into mechanism from genome-wide approaches
Global analysis of miRNA function began with microarray studies (42), which revealed that targets for a given miRNA could be identified directly through the analysis of mRNA amounts, although there was some concern that certain classes of target might be missed [for example, those regulated primarily at the level of translation (23, 24, 28)].
These concerns have been addressed by studies that evaluate in a global manner the effects of miRNAs on both mRNA abundance (the easy part) and translational efficiency (the harder part). Two groups used a proteomic approach (quantitative mass spectrometry) to discern the effects of specific miRNAs on protein output (and mRNA levels), both relying on miRNA knockdown or overexpression (47, 48). The Bartel study revealed a strong correlation between decreases in mRNA and protein abundance in response to miRNA silencing (for genes carrying potential target sites) at a single time point after the initiation of silencing. The Rajewsky study argued that the correlation was less strong overall and that, for certain genes, the protein levels were more significantly affected than those of mRNA (48), an idea consistent with the results of a recent more limited proteomic analysis (49).
Whereas proteomic approaches can measure protein levels as a direct readout for translation, the translational status of a gene can also be evaluated on the basis of the extent of its association with ribosomes. Two recent studies used somewhat different approaches to look at the ribosome association of targets regulated by ectopically or endogenously expressed miRNAs (50, 51). In one study, microarrays and a polysomal partitioning scheme were used to globally determine the occupancy and density of ribosomes on 8000 different mRNAs, thus reporting on the relative extent of translation of the various species (51). A more recent study used ribosome profiling (52) to derive translational efficiencies (50). In both studies, there were marked differences in the abundance of mRNAs (evaluated by microarrays or mRNA-Seq) that carry potential miRNA target sites in a predictable direction. Additionally, both studies found that ribosome association with the remaining miRNA-targeted mRNAs was modestly reduced, data consistent with a model where translation itself is targeted by the miRNAs.
These studies broadly extend the earlier observations of the field: miRNA-mediated silencing is typically manifested at the level of both protein and mRNA, in particular when steady state amounts are evaluated. At a minimum, these studies make it clear that mRNA analysis is able, given the selection of an appropriate time point, to rather comprehensively identify likely targets of a given miRNA (47, 50, 51). However, the global studies argue for regulation at the level of translation that cannot be accounted for by mRNA decay.
A parsimonious model for miRNA-mediated silencing
In this review, we are particularly interested in how miRNAs bound to Argonaute proteins bring about gene silencing. What are the relative contributions of various molecular events on repression (for example, translational inhibition, deadenylation, and mRNA decay); are the events coupled (and thus ordered) or uncoupled; is the pathway the same for all mRNA targets; and how is repression triggered through molecular interactions between the miRNP complex, the 3′ UTR, and other components of the cell? Answers to these questions will define the mechanism of silencing and will depend on experiments that incorporate kinetic and thermodynamic parameters into the evaluation of models. These questions are distinct, and more intractable, than the simpler question of which cellular components are affected, on some time scale, by the action of the miRNAs.
There are two basic models proposed to explain the effects of miRNAs on gene expression, one invoking initial mRNA decay (because when no mRNA is present, then of course there will be no protein) and the other initial translational repression (followed by mRNA decay). A complicating factor is that these processes are closely linked in the cell. Indeed, the overriding paradigm is that translational repression leads to mRNA decay unless an active mechanism is in place to protect the transcript from degradation (53). Indeed, many of the factors that facilitate mRNA degradation have been implicated in translational control (54).
We argue that essentially all of the existing data can be reconciled by a model for miRNA-mediated silencing that begins with initiation targeted translational repression, potentially triggered (or enhanced) by deadenylation, followed by general mRNA decay to consolidate the more transient translational repression (Fig. 2) (34, 55, 56). First, where miRNA-mediated silencing results in substantial inhibition of protein expression with more modest effects on mRNA abundance (7, 23–25, 49), the translation model is clearly indicated. The translation-triggered model also readily accounts for the observed concordant regulation of mRNA abundance and protein expression seen for certain reporters (45), certain in vivo targets (44), and the global analyses of gene expression (47, 50, 51). In these examples, although it is impossible to gauge what might be the triggering event, mRNA decay or translational inhibition, the data are certainly consistent with a model where the inhibition of translation leads to subsequent rapid mRNA decay. Lastly, we remember that, in the global analyses where generally concordant decreases in protein and mRNA abundance were observed, there was an additional contribution to repression that could most easily be accounted for at the level of translation (50, 51).
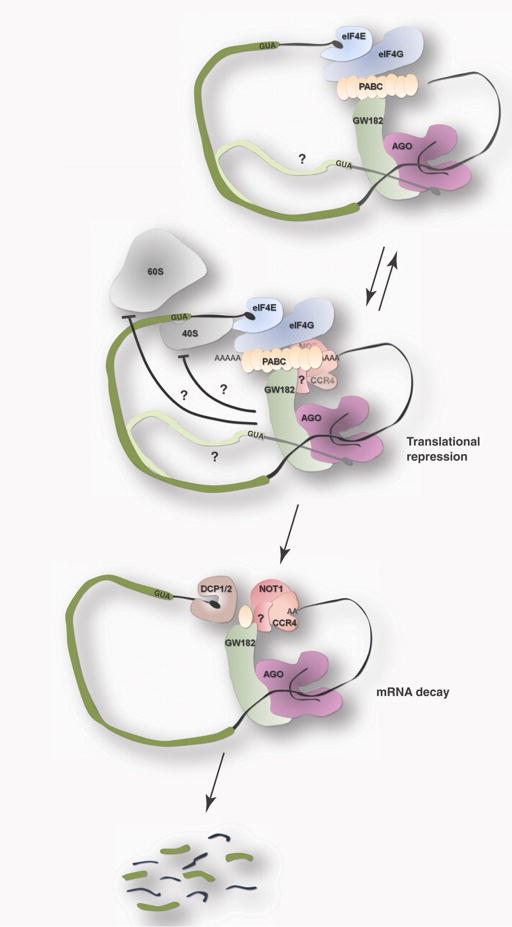
Figure 2.
Model for miRNA-mediated silencing. Cartoon outlines the proposed streamlined model, beginning with recruitment of Argonaute and associated GW182 and involving potential interactions with mRNA m7G cap. These binding events ultimately lead to translational repression by blocking some early step in translation initiation. Deadenylation by CCR4:NOT1 may contribute to translational repression at an initial step. Lastly, after deadenylation, miRNA targeted mRNA may be subjected to degradation (consolidation) through recruitment of decapping complex (DCP1/2).
Although the translation-triggered model can rationalize a vast majority of the data, what is generally missing from existing studies is a thorough analysis of the progression (or kinetics) of the repression events. For example, if the observed rates of inhibition of translation are more rapid than the observed rates of mRNA decay, then the translation triggered mechanism would be favored. Of course, if the rates are closely matched, it suggests that one process limits progression of the other, and it is impossible to establish order. Similarly, when the end points of the miRNA-mediated repression are the same (i.e., the effects on mRNA and protein or translation are matched) (47, 48, 50, 51), then the same uncertainty is associated with the conclusions.
Although most experiments to date have not included time as a variable, there are some notable exceptions. In one in vitro kinetic analysis of miRNA-mediated silencing (31), repression of protein synthesis was observed within 15 min of miRNA addition, whereas mRNA decay was not observed for another 45 min. Other studies, again using an in vitro system, more specifically looked at the rates of deadenylation and came to similar conclusions (33, 34). Together, these data support the translation-triggered model, although we acknowledge that others failed to observe such kinetic differences (50).
To fully decipher the mechanism of miRNA-mediated silencing, we must determine whether the different events (translational repression, deadenylation, and decay) are obligately connected or whether they occur independently. Although we argue for their linkage (on the basis of generally concordant reductions in mRNA and protein), a number of studies argue the contrary. Several studies argued that miRNA-mediated silencing occurs independent of events at the polyadenylated [poly(A)] tail because reporters lacking poly(A) tails are nevertheless repressed by miRNAs (7, 30, 45, 57). Other studies argued that miRNA-mediated mRNA decay takes place independent of translation because miRNA-dependent decay is observed even when translation is disrupted (33, 45, 57).
Despite these studies suggesting that translation and decay are uncoupled, we remain agnostic on this point. We highlight one careful study and in doing so point out the difficulties in designing a foolproof experiment. In this study, translation of the reporter mRNA was blocked through the incorporation of a stable stem loop structure in the 5′ UTR (a disruptor of scanning), and it was observed that miRNA-dependent decay of the reporter still took place (at the same rate and to the same extent) (57). This result was interpreted to mean that miRNA-mediated deadenylation and decay take place independent of translation. Yet, if the bound miRNP complex blocks translation at a stage before 5′ UTR scanning, then the additional translational inhibition that results from the inserted stem loop may be unimportant relative to the miRNA-mediated block that is already in place. This argument applies to most similar studies (29, 45).
Dramatic disruptions of a pathway often result in changes in the rate-limiting step of a multistep process, making it difficult to assess the implications of the observed outcome (58). For example, the incorporation of a stem loop in the 5′ UTR might result in scanning becoming rate-limiting, whereas the inclusion of cycloheximide might result in elongation becoming rate-limiting. If the normal rate-limiting step is cap recognition, the now rate-limiting later step may mask critical effects on cap recognition. The safest experiments involve minimal disruption of the natural process.
Although the interconnections between translation and decay are not wholly sorted out, there is abundant literature indicating that translational inhibition and mRNA decay are coupled throughout biology (53, 59). It stands to reason that a translational inhibition component of miRNA-mediated repression might directly lead to the mRNA decay that consolidates silencing. The generally concordant decreases in protein output and mRNA abundance are most easily explained by the proposed model invoking initial effects on translation, leading ultimately to mRNA decay in a coupled process, with differences in mRNA stability rationalizing the documented discordant examples. We argue that the proposal of a relatively streamlined model that accounts for a majority of the data will serve to clarify the gaps in our knowledge and set the stage for further critical experimentation.
Acknowledgments
We thank Jeff Coller, Geraldine Seydoux, Rita Strack and Karen Wehner for critical comments. Supported by HHMI and Life Sciences Research Foundation (A.N.).
References
- 1.Farazi TA, Juranek SA, Tuschl T. Development. 2008;135:1201. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 2.Jinek M, Doudna JA. Nature. 2009;457:405. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 3.Parker JS, Barford D. Trends Biochem Sci. 2006;31:622. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Djuranovic S, et al. Nat Struct Mol Biol. 2010;17:144. doi: 10.1038/nsmb.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Science. 2004;305:1434. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Nature. 2009;461:754. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai RS, et al. Science. 2005;309:1573. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 8.Lingel A, Simon B, Izaurralde E, Sattler M. Nat Struct Mol Biol. 2004;11:576. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 9.Ma JB, Ye K, Patel DJ. Nature. 2004;429:318. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma JB, et al. Nature. 2005;434:666. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank F, Sonenberg N, Nagar B. Nature. 2010;465:818. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 12.Parker JS, Roe SM, Barford D. Nature. 2005;434:663. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mi S, et al. Cell. 2008;133:116. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boland A, Tritschler F, Heimstadt S, Izaurralde E, Weichenrieder O. EMBO Rep. 2010 doi: 10.1038/embor.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Nature. 2008;456:921. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomari Y, Zamore PD. Genes Dev. 2005;19:517. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 17.Eulalio A, Huntzinger E, Izaurralde E. Nat Struct Mol Biol. 2008;15:346. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 18.Mioyoshi K, Okada TN, Siomi H, Siomi MC. RNA. 2009;15:1282. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tritschler F, Huntzinger E, Izaurralde E. Nat Rev Mol Cell Biol. 2010;11:379. doi: 10.1038/nrm2885. [DOI] [PubMed] [Google Scholar]
- 20.Olsen PH, Ambros V. Dev Biol. 1999;216:671. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 21.Wightman B, Ha I, Ruvkun G. Cell. 1993;75:855. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 22.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Cell. 2003;113:25. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 23.Behm-Ansmant I, et al. Genes Dev. 2006;20:1885. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nahvi A, Shoemaker CJ, Green R. RNA. 2009;15:814. doi: 10.1261/rna.1565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Mol Cell. 2006;21:533. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 26.Nottrott S, Simard MJ, Richter JD. Nat Struct Mol Biol. 2006;13:1108. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Cell. 2006;125:1111. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 28.Ding XC, Grosshans H. EMBO J. 2009;28:213. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Genes Dev. 2007;21:1857. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwasaki S, Kawamata T, Tomari Y. Mol Cell. 2009;34:58. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Mathonnet G, et al. Science. 2007;317:1764. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 32.Thermann R, Hentze MW. Nature. 2007;447:875. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 33.Zdanowicz A, et al. Mol Cell. 2009;35:881. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Fabian MR, et al. Mol Cell. 2009;35:868. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiriakidou M, et al. Cell. 2007;129:1141. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Sonenberg N, Hinnebusch AG. Cell. 2009;136:731. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter JD, Sonenberg N. Nature. 2005;433:477. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 38.Huttelmaier S, et al. Nature. 2005;438:512. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 39.Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Cell. 2001;104:281. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 40.Chendrimada TP, et al. Nature. 2007;447:823. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 41.Wang B, Yanez A, Novina CD. Proc Natl Acad Sci U S A. 2008;105:5343. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim LP, et al. Nature. 2005;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 43.Giraldez AJ, et al. Science. 2006;312:75. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 44.Bagga S, et al. Cell. 2005;122:553. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 45.Eulalio A, et al. RNA. 2009;15:21. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. Mol Cell Biol. 2009;29:6220. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baek D, et al. Nature. 2008;455:64. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selbach M, et al. Nature. 2008;455:58. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y, et al. Mol Biosyst. 2010 [Google Scholar]
- 50.Guo H, Ingolia NT, Weissman JS, Bartel DP. Nature. 2010;466:835. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendrickson DG, et al. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Science. 2009;324:218. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz DC, Parker R. Mol Cell Biol. 1999;19:5247. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coller J, Parker R. Cell. 2005;122:875. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fabian MR, Sonenberg N, Filipowicz W. Annu Rev Biochem. 2010;79:351. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 56.Eulalio A, Huntzinger E, Izaurralde E. Cell. 2008;132:9. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 57.Wu L, Fan J, Belasco JG. Proc Natl Acad Sci U S A. 2006;103:4034. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nissan T, Parker R. RNA. 2008;14:1480. doi: 10.1261/rna.1072808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isken O, Maquat LE. Genes Dev. 2007;21:1833. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]