Abstract
Protein misfolding and aggregation are widely implicated in an increasing number of human diseases providing for new therapeutic opportunities targeting protein homeostasis (proteostasis). The cellular response to proteotoxicity is highly regulated by stress signaling pathways, molecular chaperones, transport and clearance machineries that function as a proteostasis network (PN) to protect the stability and functional properties of the proteome. Consequently, the PN is essential at the cellular and organismal level for development and lifespan. However, when challenged during aging, stress, and disease, the folding and clearance machineries can become compromised leading to both gain-of-function and loss-of-function proteinopathies. Here, we assess the role of small molecules that activate the heat shock response, the unfolded protein response, and clearance mechanisms to increase PN capacity and protect cellular proteostasis against proteotoxicity. We propose that this strategy to enhance cell stress pathways and chaperone activity establishes a cytoprotective state against misfolding and/or aggregation and represents a promising therapeutic avenue to prevent the cellular damage associated with the variety of protein conformational diseases.
Keywords: Protein conformational diseases, proteostasis network, proteostasis regulators, stress responses
INTRODUCTION
All proteins must achieve their native conformation to ensure optimal cellular function. This is achieved by the protein homeostasis (proteostasis) network (PN), comprised of conserved stress response pathways that control protein synthesis, folding, trafficking, and degradation, to collectively achieve the stability and functional properties of the proteome [1]. The PN integrates the environmental state of the cell through stress signaling pathways including the heat shock response (HSR) and the endoplasmic reticulum [2] and mitochondrial unfolded protein responses (UPR) [3] that sense an imbalance in compartmental proteostasis in the face of extrinsic and intrinsic challenges [4]. Other components of the PN include the ubiquitin-proteasome and the autophagic-lysosomal systems that provide the major pathways for degradation and clearance; pathways that regulate the Ca2+ gradient between the cytoplasm and the endoplasmic reticulum; the inflammatory and oxidative-stress defense pathways; and histone deacetylases (HDAC) that regulate epigenetic chromatin remodeling [4]. These mechanisms are highly orchestrated for optimal proteostasis throughout development and differentiation, and compromised during aging to affect lifespan. The balance of folding stability therefore is tightly linked to the robustness of the HSR and UPR to ensure that the PN is optimized to minimize the stress of misfolded proteins and the incorporation of damaged molecules into the cellular machinery. The significance of this cannot be overstated for the chronic expression of a single misfolded protein has the potential to imbalance the proteostatic machinery leading to collateral damage on multiple biochemical pathways due to the subsequent misfolding and aggregation of other metastable proteins [5–7].
Misfolding and aggregation is the consequence of physical, chemical, environmental, and metabolic stress on protein stability and the cellular environment (Fig. 1). Protein damage is further amplified by mutations and polymorphisms, error-prone synthesis, and post-translational modifications, that in humans are associated with a wide range of proteinopathies leading to both loss- or gain-of-function diseases [8]. Loss-of-function diseases, including cystic fibrosis and the lysosomal storage disorders typically result from inherited mutations that affect folding leading to premature degradation [9, 10]. Likewise, gain-of-toxic function diseases such as cancer and the multitude of neurodegenerative diseases including Alzheimer’s, Parkinson’s, Huntington’s, and amyotrophic lateral sclerosis (ALS) associated with aging are characterized by the expression of highly aggregation-prone proteins that interfere with proteostasis with deleterious consequences on a multitude of downstream cellular processes [1, 5, 8, 11].
Fig. 1.
Pathways and modifiers influencing proteostasis. Genetic, epigenetic, physiological and environmental stressors affect proteostasis and cause the accumulation of misfunctional proteins. Small molecule modulators of the activities of the proteostasis network pathways (small molecule proteostasis regulators) facilitate chaperone-mediate refolding and/or induce the degradation of misfolded and damaged proteins therefore rebalancing cellular proteostasis. In parenthesis are indicated some of the genes responsible for proteostasis maintenance.
Currently there are no treatments for protein conformational diseases directed to restoring or enhancing cellular proteostasis. For some loss-of-function diseases such as type 1 Gaucher disease, enzyme replacement therapy is an effective treatment, though alternative approaches for patients with neuropathic symptoms are required, since the recombinant proteins are not blood-brain barrier permeable [12]. Perhaps the most urgent need is for the development of therapeutic approaches to arrest, stabilize, or reverse the progression of neurodegeneration. The current use of L-dopamine (L-dopa), a precursor of dopamine, for Parkinson’s disease, to improve motor function and patient quality of life, has a limited long-term efficacy due to motor complications and drug-induced dyskinesia [13]. Likewise riluzole, the only drug approved for ALS, and the antipsychotics and neuroleptics used in Huntington’s disease only have modest beneficial effects to extend patient survival [14, 15].
The urgency is substantial, therefore, to develop novel therapeutic strategies to treat diseases associated with protein misfolding. Even though our understanding of the PN is incomplete, there is now sufficient evidence that modulation of the PN, either by genetic modifiers or through small molecules can restore proteostasis and suppress misfolding and/or aggregation-associated toxicity in various cell based and animal model systems of disease [1, 4, 16]. This complements other approaches including pharmacological chaperones and kinetic stabilizers that lower the folded free energy of a specific misfolding-prone protein thus increasing its stability [16]. Compounds in this category are currently in clinical trials for the treatment of some loss- and gain-of-function disorders, respectively. Another approach is represented by small molecules that modulate the activity of specific chaperones such as Hsp90 and Hsp70 that both have essential roles in proteostasis [17, 18].
A general strategy to restore proteostasis relies on reprogramming stress response pathways using small molecules or biologicals (i.e. siRNAs). The appearance of misfolded, oxidized and aggregated proteins, as occurs in inherited and sporadic misfolding-prone protein diseases and during aging, indicates that the quality control pathways of the PN become compromised, leading to cellular dysfunction and death [5, 11, 19–23]. Therefore, small molecules that can transiently expand the PN and therefore the folding capacity of the cell should be cytoprotective against proteotoxic stress, as is observed for the inducible stress responses such as the HSR and UPR (Fig. 1) [4]. In this article, we will review the most relevant findings on the regulation of the PN pathways by small molecule proteostasis regulators, and propose that pharmacological modulation of the proteostasis boundary could provide a new avenue to ameliorate the numerous diseases of protein conformation.
MODULATION OF THE HEAT SHOCK RESPONSE BY PROTEOSTASIS REGULATORS
The HSR is rapidly induced by heat shock and other environmental and physiological stress conditions that results in the appearance of misfolded and damaged proteins in the cytoplasm and nucleus. The master regulator of the HSR is the heat shock transcription factor HSF-1, a member of the HSF gene family (HSF-1–4) [24]. HSF-1 activity is a multistep process that is finely tuned by both positive and negative regulators to respond to diverse forms of physiological, environmental, and metabolic stress conditions (Fig. 2). In the control unactivated state, HSF-1 resides in the cytoplasmic or nuclear compartments and is maintained as an inert monomer transiently bound to molecular chaperones (Hsp90, Hsp70, and other co-chaperones, including Hsp40) (Fig. 2) [24]. Stress activation of HSF-1 is associated with the dissociation from heat shock proteins (hsps), leading to the formation of HSF-1 trimers that bind with high affinity in the nucleus to cis-acting heat shock elements (HSE) comprised of multiple adjacent and inverted nGAAn repeats located in the promoter regions of target genes, including those encoding molecular chaperones and other hsps (Fig. 2) [24].
Fig. 2.
The heat shock response (HSR) pathway. The mammalian HSR is governed by the transcription factor HSF-1. In absence of stress, chaperones maintain HSF-1 as an inert monomer. With the appearance of misfolded proteins in the cytoplasm, chaperones are titrated away from HSF-1 (1) to deal with misfolding and aggregation. This allows HSF-1 to trimerize (2), bind to its consensus sequence (3) and be post-translational modified (4) therefore inducing the transcription of chaperones and heat shock proteins (hsps) (5). HSF-1 activation pathway is negatively regulated by the newly expressed chaperones and hsps, which rebind HSF-1 (6), and by acetyl transferases (HATs) (7). SIRT1 positively regulates HSF1 DNA-binding activity (7).
Positive and negative regulation of HSF-1 activity is achieved through multiple mechanisms involving a combination of chaperone interactions and post-translational modifications (Fig. 2) [24]. Transient interactions of molecular chaperones with the control form of HSF-1 maintain the inert monomer in a stress responsive state primed for activation. Likewise, chaperone interactions with the DNA-bound stress-inducible HSF-1 trimer negatively regulate transcriptional activity. In addition to the conformational regulation involving chaperone interactions, HSF-1 is extensively post-translationally modified by phosphorylation, sumoylation, and acetylation. Inducible phosphorylation at serines 230, 326, and 419 enhances HSF-1 transcriptional activity, whereas constitutive phosphorylation at serines 303, 307, and 308, and sumoylation at lysine 298, has negative effects on HSF-1-activity [25]. Likewise, sumoylation of HSF-1 requires phosphorylation at Ser303 and Ser307 [26, 27], but the mechanism by which sumoylation mediates HSF-1 repression remains to be elucidated. Finally, a key regulatory step in attenuation of the heat shock transcriptional response involves acetylation of HSF-1 at lysine 80 in the DNA binding domain that prevents binding to DNA [24].
The multistep process of HSF-1 regulation enhances the dynamic range for controlling the HSR and reveals a number of potential targets for therapeutic opportunities in protein misfolding disease. Although most efforts to date have been in the development of compounds that activate HSF-1, the other members of the HSF family should also be investigated, in particular HSF-2, that has been shown to contribute to the expression of chaperone genes by forming a hetero-complex with HSF-1 [28].
Small Molecule Activators of the HSR
Non-steroidal Anti-inflammatory Drugs (NSAIDS)
NSAIDS including sodium salicylate have multiple properties; at higher concentrations they partially activate HSF-1 and at lower concentrations they can synergize with other stress conditions to fully induce the HSR [29]. Exposure of human tissue culture cells to sodium salicylate activates HSF-1 trimers that bind in vivo to the HSEs of the Hsp70 gene, yet do not induce Hsp70 transcription. Salicylate-treated cells, however, are sensitized to stress and readily activate heat shock genes upon exposure to a subsequent or co-exposure to mild stress conditions. Likewise, indomethacin induces HSF-1 DNA binding with full Hsp70 transcription requiring a secondary stress [30]. Among the inflammatory modulators, arachidonic acid and the cyclopentenone prostaglandins, including PGA1, PGA2 and PGJ2, all induce HSF-1 [31, 32]. Of importance, NSAIDS do not have associated toxicity thus providing a means to activate the HSR without long-term deleterious effects.
Proteasome inhibitors and small molecule inducers of protein misfolding
A number of small molecule activators of the HSR have been reported, and for each of these compounds HSF-1 is activated indirectly, either by promoting protein aggregation or by inhibiting HSF-1 negative regulators such as chaperones, protein kinases and acetyltransferases [25, 33]. Among the compounds that cause the accumulation of misfolded proteins targeted for degradation are proteasome inhibitors such as Velcade® (bortezomib) [34], MG132 and lactacystin [35], and the serine protease inhibitors dichloroisocoumarin (DCIC), N-tosyl-L-phenylalanyl chloromethyl ketone (TPCK), and N-alpha-tosyl-L-lysinyl-chloromethylketone (TLCK) [36]. Similarly, the proline analogue azetidine and the protein synthesis inhibitor puromycin result in the expression of damaged proteins with increased propensity to misfold [25]; azetidine alters protein tertiary structure, therefore affecting folding stability, and puromycin causes the premature release of truncated protein chains resulting in folding-incompetent peptides.
Celastrol
Another class of HSR activators is represented by the quinone methide triterpene celastrol that is a natural product isolated from the root barks of the Celastraceae family of plants and is commonly used in traditional Chinese medicine for its anti-inflammatory properties. Consistent with this, celastrol has been identified as an inhibitor of nuclear factor-κB [37, 38]. Celastrol has additional pharmacological properties including anti-neurodegenerative properties [39] that have been attributed to induction of the HSR through activation of HSF-1 and the expression of molecular chaperones [40]. Several hypotheses have been proposed for the chemical activity of celastrol, although the mechanism by which it activates the HSR is not fully understood. Celastrol has been suggested to covalently react with protein thiol groups thus affecting protein conformation [41]; therefore activation of HSF-1 could be due to celastrol-induced oxidative damage of cellular proteins. Celastrol has also been suggested to inhibit Hsp90, a chaperone that also functions as an HSF-1-repressor [42–44]. Binding of celastrol to the C-terminal domain of Hsp90 has been proposed to promote the degradation of the co-chaperone Cdc37 [42, 43], which is essential for Hsp90 client interactions. Other studies suggest however that the mechanism of Hsp90 inhibition is through modification of Hsp90 co-chaperones Cdc37 and p23 [45, 46]. Consistent with this, Hsp90 was not identified as a cellular target of celastrol [47] and rather annexin II, eEF1A and β-tubulin were identified as molecular targets of celastrol from in vitro pull-down experiments using biotinylated conjugates of celastrol [47]. Finally, celastrol was shown to inhibit proteasome chymotrypsin-like activity and to promote the accumulation of polyubiquitinated proteins [48].
Of interest, celastrol and MG132 have been shown to activate the UPR and partially restore the folding, trafficking and function of mutations in proteins responsible for lysosomal storage diseases [49]. Further enhancement was obtained when either celastrol or MG132 were used together with a pharmacological chaperone. Whether this approach can be generalized remains to be shown, nevertheless the ability of various combinations of small molecules to restore mutant protein folding and function in different loss-of-function diseases shows promise.
Hsp90 inhibitors
Hsp90 is a ubiquitous molecular chaperone that is essential for the function of a large and diverse array of client proteins including steroid hormone receptors, kinases, phosphatases, and transcription factors [50]. For example, Hsp90 interacts with HSF-1 to maintain a repressed, inactive state, consequently small molecule inhibitors of Hsp90 activate the HSR [51].
Among the inhibitors of Hsp90 are the macrocyclic anti-fungal antibiotic radicicol and the benzoquinone ansamycin antibiotics geldanamycin and its derivatives 17-allylamino-17-demethoxy-geldanamycin (17-AAG) and 17-dimethyl-aminoethylamino-17-demethoxygeldanamycin (17-DMAG). These compounds inhibit Hsp90 ATPase activity by binding to the Hsp90 N-terminal ATP-binding pocket leading to the release of client proteins [52].
Geldanamycin, radicicol and 17-AAG reduce the toxicity associated with protein aggregation in several models of neurodegenerative diseases. For example, geldanamycin treatment induces expression of molecular chaperones and suppresses aggregation-mediated toxicity of mutant huntingtin, α-synuclein and mutant SOD1 in mammalian cell lines [53–55]. Geldanamycin was also shown to induce chaperone expression and reduce tau aggregation [56], and in a Drosophila model of Parkinson’s disease protected dopaminergic neurons from α-synuclein toxicity [57]. In this study, geldanamycin upregulated Hsp70 expression only in α-synuclein-expressing flies, suggesting selectivity for ‘stressed’ cells. Geldanamycin was also protective in a mouse model of Parkinson’s disease in which dopaminergic neurotoxicity induced by 1-methyl-4-pheny-1,2,3,6-tetrahydropyridine (MPTP) was reduced by pretreatment with geldanamycin [58]. Likewise, geldanamycin and radicicol treatment delayed disease progression in a mouse model of Huntington’s disease [59].
The small molecule inhibitor of Hsp90, 17-AAG, also induces expression of hsps and rescued eye degeneration and lethality in a fly model of spinocerebellar ataxias [60]. Likewise, 17-AAG suppressed neurodegeneration in an HSF-1-dependent manner in a Drosophila model of Huntington’s disease [60] and in a mouse model of spino bulbar muscular atrophy caused by expression of the androgen receptor (mAR) with a polyQ-expansion [61].
Another role for Hsp90 in disease progression is associated with effects on the stability and function of Hsp90 client proteins. For example, huntingtin has been shown to interact with Hsp90 and consequently Hsp90 inhibitors induced clearance of huntingtin aggregates through the ubiquitin-proteasome system [62]. Similarly, Hsp90 interaction with the leucine-rich repeat kinase 2 [63], PTEN-induced kinase 1 [64] and α-synuclein [65] is affected by the Hsp90 inhibitors PU-H71 [63] and geldanamycin [64] leading to client protein degradation by the proteasome. Accordingly, in mice treated with the brain barrier-permeable Hsp90 inhibitor PU-DZ8, the levels of aggregated tau were reduced [66] and likewise inhibition of Hsp90 activity led to a reduction in phosphorylated tau in a mouse model of Alzheimer’s disease [67]. This effect appears to be due to the ability of Hsp90 inhibitors to convert the Hsp90-cochaperone complex from protein folding to protein degradation. Another client of Hsp90 is hyperphosphorylated tau; consequently tau clearance by the proteasome through interaction with Hsp90, Hsp70 and Hsp40 involves recruitment of the ubiquitin ligase, CHIP.
Collectively, the Hsp90 inhibitors reveal potential opportunities for the treatment of proteinopathies. However, for long-term therapeutic use in neurodegenerative diseases, some concerns of many Hsp90 inhibitors include cytotoxicity due to clearance of Hsp90 client proteins and activation of the HSR, together with the low blood brain barrier permeability. A new class of Hsp90 inhibitors is represented by the 2-amino-7,8-dihydro-6H-pyrido[4,3-D]pyrimidin-5-one compound NVP-HSP990 that binds at the Hsp90 N-terminal domain, is orally available, brain penetrant, and was shown to induce the HSR in the brain of Huntington’s disease mouse models, reducing aggregation and improving survival [68]. As a primary indication, NVP-HSP990 is currently in phase 1 clinical trials for treatment of solid and hematological tumors [69].
Compounds that target the Hsp90 C-terminal ATP-binding site have also been developed using the core structure of the C-terminal domain inhibitor, novobiocin. The novobiocin analogue A4 exhibits higher inhibitory potency compared to the parent compound, and induces chaperone expression and Hsp90 client protein degradation at nanomolar concentrations. A4 was also shown to confer neuroprotection against Aβ-induced toxicity [70, 71]. Distinct to novobiocin and its analogues, compounds AEG3482 and ITZ-1 bind to Hsp90 and promote HSF-1 activation without causing degradation of Hsp90 client proteins [72, 73]. This effect may account for the low toxicity displayed by these two compounds. Whereas ITZ-1 was still found to bind to the C-terminal binding site of Hsp90, AEG3482 instead appeared to bind to a portion of the Hsp90 peptide-binding domain this way facilitating the dissociation of HSF-1 from Hsp90.
Although the range of biological functions described above supports inhibitors of Hsp90 as a promising therapeutic approach, the likelihood that Hsp90 inhibitors can be used for the chronic treatment of neurodegenerative diseases remains to be demonstrated. Due to the variety and essential nature of Hsp90 client proteins, that include proteins involved in cell growth, proliferation, differentiation, and survival, inhibition of this molecular chaperone has complicating side-effects and cytotoxicity. For these reasons the current emphasis of Hsp90 therapeutics has concentrated on acute treatment of cancer.
HSF1A
The benzyl pyrazole derivative HSF1A, a small molecule activator of HSF-1, was identified in a yeast-based high-throughput screen [74]. Induction of chaperones by HSF1A was shown to reduce protein misfolding and aggregation-mediated toxicity in cellular and fly models of polyQ-related diseases, and to activate HSF-1 in Drosophila and mammalian cells without inhibition of Hsp90 activity or causing proteotoxicity. Rather, HSF1A was suggested to interact with the cytosolic TCP-1 ring complex (TRiC). This proposed mechanism of action is of interest as TRiC binds to polyglutamine-expanded variants of huntingtin to inhibit their aggregation [75, 76]. Whether HSF1A can also ameliorate cytotoxicity in other models of misfolding remains to be demonstrated [77].
Small molecule proteostasis regulators that regulate distinct stress pathways
A recent high-throughput screen of ~ 1 million small molecules for novel HSR activators identified nearly 300 compounds that induce expression of human Hsp70 in a human cell-based assay [78]. These chemical hits were classified into seven scaffold clusters, β-aryl-α,β-unsaturated-carbonyls (cluster A), β–nitrostyrenes (cluster B), β-Cl-α,β-unsaturated-carbonyls (cluster C), nitrobenzofurazans (cluster D), nitrofuranylamides (cluster E), unsaturated barbituric acids (cluster F) and 2-cyanopentadienamide (cluster G), of which a representative subset were characterized for induction of HSF-1 and HSF-1-dependent expression of chaperones in human cell lines and C. elegans. Of these, compounds A1, D1, and F1 suppressed aggregation and toxicity in cellular and animal models of polyQ-related diseases, whereas A3, C1, and F1 induced the HSR and the UPR and rescued misfolding and promoted trafficking of the mutated CFTR protein (ΔF508CFTR) in a cellular model of cystic fibrosis. While the molecular targets of these compounds and their mechanisms of action remains to be elucidated, these compounds do not appear to activate the HSR by causing protein misfolding, inhibition of Hsp90, or proteasome activity. Of these compounds, the barbiturate-analog F1 represents a new class of small molecules that rescues the folding stability of proteins associated with both gain- and loss-of-function diseases, presumably through poly-pharmacological activation of multiple cell stress responses to establish a cytoprotective state against protein damage.
MODULATION OF THE UNFOLDED PROTEIN RESPONSE AND ER-ASSOCIATED DEGRADATION BY PROTEOSTASIS REGULATORS
Protein folding in the endoplasmic reticulum (ER) is dependent upon lumen-localized chaperones for the function of secreted proteins [79]. Consequently, perturbation in the ER folding environment requires an adjustment in the folding capacity that is mediated by the UPR comprised of three intracellular signal transduction pathways (Fig. 3) [80]. These pathways are each identified by a different class of stress-response transmembrane protein: the activating transcription factor 6 (ATF6), the protein kinase RNA (PKR)-like ER kinase (PERK), and the inositol-requiring protein 1 (IRE1) (Fig. 3). These integral membrane proteins act as ER stress transducers and respond to the accumulation of misfolded proteins in the ER lumen by leading to the activation of transcription factors that regulate the expression of UPR target genes (Fig. 3) [80].
Fig. 3.

The unfolded protein response (UPR) pathway and its chemical modulators. Accumulation of misfolded proteins in the ER lumen activate the three UPR signal transducers ATF6, PERK and IRE1. This results in the production of transcription factors that migrate into the nucleus and activate UPR target genes that cause attenuation of protein synthesis and increase both the ER folding-capacity and ER-associated degradation.
The transcription factor ATF6 is expressed as an inactive precursor bound to the ER membrane that is delivered upon ER stress to the Golgi where it is cleaved by the proteases, S1P and S2P. The released N-terminal cytosolic DNA-binding domain, ATF6(N), migrates to the nucleus to activate the transcription of genes involved in protein folding (i.e. the molecular chaperones BiP and GRP94 that correspond to the ER Hsp70 and Hsp90 homologs, and protein disulfide isomerases). The second branch of the UPR is mediated by PERK, a transmembrane protein kinase that undergoes oligomerization and autophosphorylation, and phosphorylates the translation initiation factor eIF2α upon ER stress resulting in the inhibition of mRNA translation, thus decreasing the load of damaged proteins in the ER. PERK also contributes to the translational upregulation of the transcription factor ATF4, that induces the expression of the target genes CHOP (transcription factor C/EBP homologous protein) and PPPR15A/GADD34 (growth arrest and DNA damage-inducible 34). CHOP, in turn, regulates the transcription of genes involved in apoptosis and upon persistent activation can lead to cell death. Activation of PERK, at modest levels of signaling, however is protective. ATF4 expression also upregulates ERAD retrograde clearance machinery to further relieve stress. Restoration of ER homeostasis involves the phosphatase PPPR15A/GADD34 that can quickly dephosphorylate eIF2α, thus providing a negative feedback loop in the PERK signaling pathway. The third arm of the UPR corresponds to IRE1; active IRE1 displays ribonuclease activity and cleaves the mRNA encoding an UPR-specific transcription factor, the X-box binding protein 1 (XBP1). XBP1 has a role in regulating lipid biosynthetic enzymes and ER-associated degradation components (ERAD) (Fig. 3).
It has become increasingly evident that UPR dysfunction has an important role in human disease, in particular those involving tissues with increased requirement for protein synthesis, such as cancer cells, that need extensive protein synthesis to sustain growth [79], and pancreatic β-cells, that express high levels of insulin [81]. Therefore targeting the UPR with small molecule modulators could have beneficial therapeutic implications. However, the presence of opposing activities (cytoprotective and pro-apoptotic) resulting from UPR activation suggests that success in modulation of the UPR should focus on small molecules that are selective for individual components of this stress response pathway [79].
Guanabenz
Guanabenz, an agonist of the α2-adrenergic receptor used for treatment of high blood pressure, was recently shown to protect the cell from the toxicity associated with the expression of misfolding-prone insulin in the ER of pancreatic β-cells [82] by selectively inhibiting the phosphatase activity of the stress-inducible PPPR15A/GADD34 protein (Fig. 3). Guanabenz was shown to prolong eIF2a phoshporylation in stressed cells and translation attenuation, thus increasing the balance of chaperones to damaged substrates. Guanabenz was not cytotoxic to unstressed cells since they do not express PPPR15A/GADD34 and therefore are not subjected to inhibition of protein synthesis. The guanabenz effect is reminiscent of salubrinal, a small molecule inhibitor of eIF2a dephoshporylation with cytoprotective properties against ER-stress inducers and viral assault by herpes simplex virus [83]. It would be of interest to determine whether the effect of guanabenz is selective for mutant insulin or could be applied to other diseases associated with ER protein misfolding.
Small molecule inhibitors of IRE1
4-methyl umbelliferone 8-carbaldehyde (4μ8C) [84] and other aldehydes, including MK0186893 [85] and STF-083010 [86], have been identified as inhibitors of the RNase activity of IRE1 (Fig. 3). Inhibition of IRE1 affects the integrity of secretory tissues [87–91], therefore IRE1 inhibitors have been proposed for pathologies associated with elevated ER secretory load, such as solid tumors and hematologic malignancies that possess a robust secretory apparatus. In support of this, 4μ8C and STF-083010 have antimyeloma activity in both in vitro and in vivo models of multiple myeloma [86]. Modulation of protein synthesis by small molecules inhibitors of IRE1 endonuclease activity could therefore be used for conditions such as type-2 diabetes and viral infection.
Small molecule inhibitors of ER-associated degradation (ERAD)
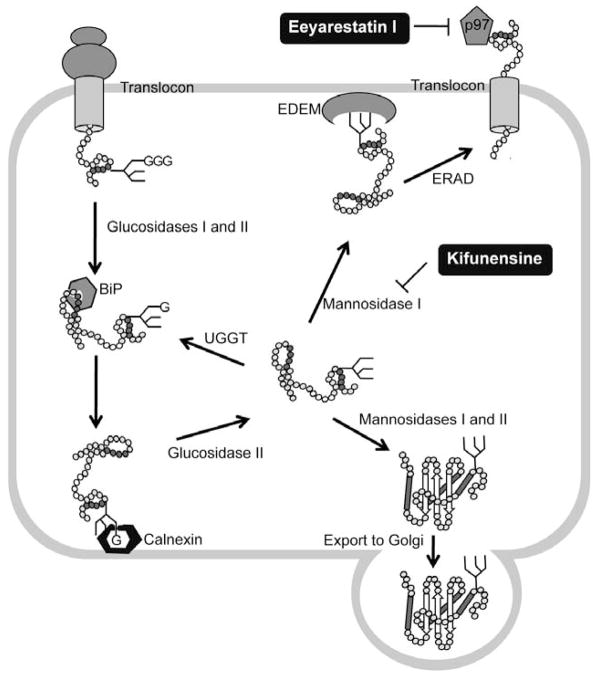
The folding and activity of destabilized lysosomal enzymes prone to degradation can be restored by inhibition of ERAD using kifunensine (Kif) and eeyarestatin I (EerI), that interfere respectively with the recognition and retrotraslocation of misfolded proteins in the ER (Fig. 4) [92]. The rescue of activity by Eerl was suggested to be promoted by upregulation of BiP expression, whereas Kif treatment did not induce the UPR or result in CHOP activation and apoptosis. Although Kif is apotent inhibitor of the ER-mannosidase I enzyme and therefore could cause major defects in many cellular essential glycoproteins, associated cytotoxicity was not observed. Thus, the identification of the steps in the ERAD pathway that can be modulated by small molecules without disruption of the ER quality-control system offers potential targets for the therapeutic rescue of misfolded proteins targeted for degradation [92].
Fig. 4.
ER-associated degradation (ERAD) and its chemical modulators. Proteins entering the endoplasmic reticulum (ER) are immediately recognized by BiP and often modified by the addition of a GlcNAc2-Man9-Glc3 glycan. Glucosidases I and II sequentially remove two terminal glucoses (G) from the glycan and generate monoglucosylated substrates that are recognized by calnexin and calreticulin (calreticulin is a soluble protein and is not shown), which facilitate substrate folding. Once the substrate is released from the calnexin–calreticulin cycle, glucosidase II trims the last glucose. Proteins that have adopted their native conformation are demannosylated by mannosidases I and II and exit the ER. If proteins have not been folded properly, they re-enter the calnexin–calreticulin cycle. Such proteins are reglucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT), which promotes re-entry into the folding cycle. Terminally misfolded proteins are processed by mannosidase I and then targeted for ERAD with the participation, through an undetermined mechanism, of the ER degradation-enhancing -mannosidase-like lectins (EDEM). Retrotranslocation of the misfolding-prone substrate to the cytoplasm is mediated by p97 complex. The small molecule ERAD inhibitors kifunesine and Eeyarestatin I block different steps of the ERAD pathways.
MODULATION OF AUTOPHAGY AND UBIQUITIN PROTEASOME SYSTEM BY PROTEOSTASIS REGULATORS
Autophagy [93] and the ubiquitin proteasome system (UPS) [94–96] are important components of the PN that regulate protein degradation in the eukaryotic cell. These pathways are essential for normal physiology and development, and are also involved in numerous diseases including cancer and neurodegeneration (Fig. 5). The UPS and autophagy are proposed to have a key role in proteostasis by clearance of proteins that control the cell cycle, signaling events, transcription and translation, and removal of damaged and dysfunctional cellular components. Therefore, it is not unexpected that the activities of these two proteostatic machines are tightly regulated.
Fig. 5.
Autophagy, the ubiquitin proteasome system (UPS) and their chemical modulators. Both mTOR-dependent and -independent pathways induce autophagy, the main degradation pathway for aggregate-prone proteins. One of the major pathways that regulate mTOR in mammalian cells is the PI3K–Akt pathway, which is triggered by the binding of insulin growth factors to its receptor (IR). An mTOR-independent mechanism instead involves G protein-coupled receptors, which regulate intracellular inositol and inositol 1,4,5-trisphosphate (IP3) levels. Inositol and IP3 are negative regulators of autophagy. Induction of autophagy involves the formation of a phagophore, a double-membrane structure that sequesters aggregated proteins, thus creating an autophagosome. Autophagosomes fuse with lysosome to form autolysosomes in which lysosomal hydrolases degrade their content. Chaperone-mediated autophagy (CMA) instead targets cytosolic proteins to the lysosome surface where they bind to the transmembrane protein lysosome-associated membrane protein type 2A (LAMP-2A). LAMP-2A mediates the protein translocation across the lysosomal membrane for degradation. Unfolded and misfolded proteins that cannot be re-folded correctly by molecular chaperones are polyubiquitinated and then targeted for degradation by the proteasome. SV, sodium valproate; CA, carbamazepine.
Autophagy substrates include long-lived cytosolic proteins, protein complexes and organelles that are sequestered into double-membrane cytosolic vescicles, the autophagosomes (Fig. 5). Fusion of autophagosomes with lysosomes results in degradation of the vescicular contents. Basal levels of autophagy are important for proteostasis and confer cytoprotection, whereas an imbalance in degradation can lead to cell death. Autophagy is regulated by mammalian target of rapamycin(mTOR)-dependent and–independent pathways (Fig. 5) [97] and is affected by growth factor and nutrient signals. Rapamycin is an inhibitor of mTOR and induces autophagy [97], whereas other small molecules that activate autophagy in an mTOR-independent pathway decrease the levels of inositol or inositol-triphosphate (IP3) [97].
The proteasome is the proteolytic arm of the UPS (Fig. 5) and is comprised of a regulatory and a core particle into which substrates are translocated and degraded [98]. Most often, substrates are targeted to the proteasome through polyubiquitination (Fig. 5). Protein complexes and aggregates are poor proteasome substrates due to the size of the proteasomal catalytic pore, and require partial unfolding and interactions with ubiquitin ligases and deubiquitinating enzymes (DUBs). These concerted events alter the ubiquitin chain length on misfolded substrates and enhance substrate affinity, and rate of degradation by the proteasome (Fig. 5) [99].
Small Molecule Enhancers of Autophagy
Rapamycin
Rapamycin is a lipophilic macrolide antibiotic that inhibits the kinase activity of mTOR (Fig. 5) [100] and can upregulate autophagy in mammalian brains. Rapamycin-induced autophagy enhances the degradation of several misfolded proteins, including mutant huntingtin, α-synuclein, ataxin 3 and tau [97] and is protective in fly and mouse models of Huntington’s disease and tauopathy. However, because TOR proteins are involved in other processes, including ribosome biogenesis and protein translation, long-term use of rapamycin has toxic side effects, such as immunosuppression, that may limit use for chronic diseases. Therefore the search for compounds that either work through mTOR-independent pathways or that increase the activity of rapamycin has been pursued.
Inositol-lowering compounds
Lithium, carbamazepine and valproic acid are used as mood-stabilizers drugs with effects on autophagy by reducing the levels of inositol and IP3 (Fig. 5) [97]. Lithium, a drug widely used in the treatment of bipolar disorders [101], inhibits the inositol monophosphatase (IMPase) enzyme that catalyzes the hydrolysis of inositol monophosphate into free inositol. These compounds have been shown to accelerate the clearance of mutant huntingtin and α-synuclein and have synergistic effects with rapamycin in clearance of toxic protein species [97].
Small molecule enhancers of rapamycin (SMERs)
A yeast-based high-throughput screen for small molecule enhancers of rapamycin (SMERs) identified SMER10, 18 and 28 that induced mTOR-independent autophagy and enhanced clearance of α-synuclein and mutant huntingtin in mammalian cells and reduced polyglutamine toxicity in a Drosophila model of Huntington’s disease [102]. In addition, these compounds enhanced the clearance of mycobacteria in primary human macrophages [103]. SMER10 is an aminopyrimidone, SMER18 a vinylogous amide and SMER28 a bromo-substitued quinazoline, and limited SAR analysis has indicated that the pyrimidone functionality of SMER10 is critical for activity, and the hydroxyl group at the meta position of SMER18 is important for the autophagy-inducing effect. SMER28 also enhances degradation of Aβ peptide and amyloid precursor protein-derived fragment in cell lines and primary neuronal cultures. These effects were proposed to be mediated by the autophagy-related protein Atg5 and the autophagy pathway [104].
Bioactive compounds
Autophagy-inducing compounds were identified in a high-throughput screen using human glioblastoma cells expressing the autophagy marker LC3-GFP [105]. Eight compounds (fluspirilene, trifluoperazine, pimozide, niguldipine, nicardipine, amiodarone, loperamide and penitrem A) were shown to induce mTOR-independent autophagic degradation of long-lived proteins without apparent toxicity. With the exception of nicardipine, these compounds also reduced the accumulation of polyQ in a dose-dependent manner. Nearly all of these compounds are FDA-approved drugs for different indications, and of these, verapamil, had been identified in a screen for drugs that induced autophagy [106] with beneficial effects in cell, fly and zebrafish models of Huntington’s disease. Verapamil is a Ca2+-channel blocker drug commonly used to treat hypertension and other cardiac conditions, such as angina and cardiac arrythmia [101]. Verapamil is also an inhibitor of the multi-drug resistance transporter 1 (MDR1), an efflux pump that serves to limit the absorption of xenobiotics and also mediates their elimination.
Trehalose
The disaccaride trehalose has properties as a chemical chaperone that can reduce aggregation of misfolding-prone proteins, including expanded polyQ, α-synuclein, and β-amyloid [97]. Trehalose was also shown to induce mTOR-independent autophagy and consequently to reduce mutant huntingtin and α-synuclein aggregation and toxicity [97]. In cells, the trehalose concentration required to reduce mutant huntingtin and α-synuclein aggregates was 100 mM [107]. These properties, together with the lack of toxicity, makes trehalose and other compounds with similar properties an intriguing general class of proteostasis regulators.
N10-Substituted Phenoxazine
Neuronal cells have been suggested to be less responsive to autophagy regulation as compared to other cell types. A reduced capacity of autophagy in the central nervous system has been linked to neurodegeneration [108, 109], therefore justifying a search for compounds that enhance neuronal autophagy. The Akt inhibitor 10-[4-(N-diethylamino)butyl]-2-chlorophenoxazine (10-NCP) was shown to induce autophagy in primary neurons in an Akt- and mTOR-independent way [110]. Although 10-NCP was protective in a neuronal model of Huntington’s disease without causing cell toxicity, the long-term inhibition of Akt could compromise neuronal survival.
Carbamazepine
The accumulation of aggregated Z-variant (Glu342Lys) α1-antitrypsin, a hepatic secretory glycoprotein, in the ER leads to liver degeneration [111]. Carbamazepine, an autophagy-enhancing drug, has been shown to decrease the hepatic load of the mutated α1-antitrypsin with beneficial effects on hepatic fibrosis in a mouse model [112]. Carbamazepine likely functions through a mTOR-independent pathway, since rapamycin was not able to increase the degradation of the mutated protein, and is currently in clinical trials for severe liver disease (ClinicalTrials.gov)
Small Molecule Enhancers of Proteasomal Activity
Small molecule inhibitor of USP14
A highly promising approach to restore proteostatic imblance is based on USP14, a DUBs that inhibits proteasome-mediated degradation by trimming the polyubiquitin chain on the client proteins (Fig. 5) [113]. IU1 (1-[1-(4-fluorophenyl)-2,5-dimethylpyrrol-3-yl]-2-pyrrolidin-1-ylethanone), an active-site-directed thiol protease inhibitor of USP14, was identified by a high-throughput screen and was shown to enhance degradation of tau, ataxin 3, and glial fibrillary acidic protein, independent of autophagy. In addition, IU1 reduced the accumulation of oxidized proteins, suggesting that IU1 can enhance cell survival during proteotoxic stress. As a therapeutic strategy, the induction of proteasome activity could be beneficial in diseases of protein aggregation, during aging when the proteasome function is impaired, and also in diseases resulting from loss-of-function mutations in the components of the ubiquitin pathway. Among the critical questions for the further development of this class of small molecules will be client selectivity to ensure that activation of the proteasome does not have deleterious consequences on normal cell function.
MODULATION OF CALCIUM SIGNALING BY PROTEOSTASIS REGULATORS
Ca2+ ions function as second messengers, and therefore have a fundamental roles in cell signaling through a range of cellular processes from gene expression to hormone secretion and ER function [114]. ER Ca2+ homeostasis is fundamental in the biogenesis of secretory proteins and regulates the activities of ER-resident Ca2+-dependent chaperones, such as the glucose-regulated protein GRP78/BiP, GRP94, calnexin and calreticulin, that stabilize folding intermediates [115]. Alteration of Ca2+ homeostasis by small molecules therefore has substantial appeal as an approach to enhance the proteostasis network capacity and restore the folding, trafficking and activity of several misfolded-prone proteins in the cell (Fig. 6).
Fig. 6.
Modulation of ER calcium levels by small molecules alters mutant protein-chaperone interaction and restores proteostasis. The activity of ER-resident chaperones are regulated by calcium. Increased ER calcium concentration is beneficial for those misfolded-prone proteins (such as those involved in different lysosomal storage diseases), which require chaperone-assisted folding (left panel). On the contrary, a reduction in ER calcium levels allows ΔF508CFTR to escape from chaperone-mediated proteasomal degradation (right panel). The question mark indicates that the original findings on thapsigargin and curcumin were not reproducible.
Thapsigargin
Among the first indications that modulation of Ca2+ signaling could have benefical effects on protein homeostasis and disease was the demonstration that thapsigargin, an inhibitor of the ER Ca2+-ATPase (SERCA), enhanced channel activity and the trafficking of ΔF508 CFTR from the ER to the apical membrane in cystic fibrosis cell lines (Fig. 6) [116]. Similar effects were also observed in a mouse model of cystic fibrosis. Thapsigargin reduced intraluminal levels of Ca2+ in the ER without upregulating the expression of stress-related ER chaperones or causing an alteration of the ER ultrastructure. Rather, thapsigargin treatment appears to alter the association among ER chaperones and the newly synthesized ΔF508 CFTR, therefore reducing ΔF508 CFTR ER retention and degradation. Although this study suggests that thpsigargin and possibly other Ca2+ pump inhibitors could have a potential therapeutic effect in cystic fribrosis-affected patients, several other investigators were not able to reproduce these original findings [117, 118]
Curcumin
Curcumin has effects as a low-affinity inhibitor of the SERCA pump and has been shown to correct ΔF508 CFTR defects in cell lines and a mouse model of cystic fibrosis (Fig. 6) [119]. By reducing ER Ca2+ levels, curcumin interfered with calnexin function and therefore reduced calnexin-ΔF508 CFTR interaction, thus allowing ΔF508 CFTR to escape proteasomal degradation. Similar to the previous study on thapsigargin, the results on curcumin have also been inconsistent [117, 118, 120–122].
Dltiazem, verapamil, ryanodine, dantrolene and lacidipine
Manipulation of Ca2+ homeostasis has been shown to improve the folding, trafficking and activity of mutant enzymes responsible for multiple lysosomal storage diseases (LSDs) (Fig. 6) [123–125]. Diltiazem and verapamil are FDA approved hypertension drugs that inhibit the L-type voltage-gated Ca2+ channels and lead to an increase in ER Ca2+ levels. These compounds enhance the folding and activity of a number of lysosomal mutant enzymes including L444P glucocerebrosidase (GC), P356R α-mannosidase, and S66W sulfamidase. Diltiazem and verapamil have been suggested to upregulate a subset of cytosolic (Hsp40 and Hsp90) and ER chaperones (BiP). Likewise, by increasing the ER Ca2+ store by either blocking the ryanodine receptors (RyRs) or by overexpressing ER Ca2+ influx ATPase pumps, mutant GC proteostasis was enhanced [123, 126]. Consistent with these observations, the siRNA knockdown of RyRs or treatment of L444P GC fibroblasts with small molecule inhibitors of RyR activity (diltiazem, verapamil, dantrolene and ryanodine) modestly restored L444P GC folding, trafficking and function. In contrast with inhibitors of plasma membrane L-type Ca2+ channels, antagonism at the RyRs did not induce a stress response [125], but rather resulted in the elevated expression of the Ca2+-regulated chaperone calnexin, which in turn enhanced folding of mutant GC. Treatment of L444P GC fibroblasts with ryanodine reduced MG-132-induced cytotoxicity and apoptosis, suggesting that ryanodine can re-restore the ER folding environment.
Lacidipine, a more selective inhibitor of the L-type Ca2+ channel and RyRs, was shown to rescue L444P GC folding with a greater efficiency than other Ca2+ channel blockers previously reported (Fig. 6) [124]. The marked increase in mutated GC activity was attributed to the more hydrophobic nature of lacidipine, which allows the compound to easily diffuse into the cell compared to the charged diltiazem and verapamil. Lacidipine enhances the expression of BiP and protects against apoptosis associated with sustained activation of the UPR.
INHIBITION OF HISTONE DEACETYLASES BY PROTEOSTASIS REGULATORS
Histone deacetylases (HDACs) and histone acetylases (HATs) have wide-ranging effects on gene expression by affecting the acetylation of histones and other regulatory proteins including transcription factors (i.e. HSF-1) [127], nuclear hormone receptors and signal-transduction proteins [128]. HDACs have been recognized as potential targets for the treatment of numerous disorders, including cancer [129] and neurodegenerative diseases [128, 130].
HDAC Inhibitors
HDAC inhibition by small molecules is neuroprotective in various models of neurodegenerative disease associated with protein misfolding [131]. Administration of the short fatty acid 4-phenylbutyrate (4-PBA) to a mouse model of Alzheimer’s disease restored learning behavior [132] that was dependent upon elevated levels of H4 acetylation and increased synthesis of proteins involved in synaptic function. Similar beneficial effects also associated with elevated acetylation of histones [133] were obtained in mouse models of Alzheimer’s disease treated with the pan-HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) [134]. The neuroprotective effects of HDAC inhibitors extend to invertebrate and mouse models of several polyQ-related diseases. Treatment of a mouse model of spinal bulbar muscular atrophy (SBMA) with the HDAC inhibitor sodium butyrate increased histone acetylation and partially ameliorated pathological phenotypes, and likewise treatment of mouse models of ALS by phenylbutyrate and sodium valproate promoted motor-neuron survival [130].
The beneficial effects of HDAC inhibition on proteostasis has implications for other protein conformational diseases, including lysosomal storage disorders [135], cystic fibrosis [136], and type 2 diabetes [137]. For cystic fibrosis, inhibition of HDAC7 by SAHA restores folding of mutant CFTR, trafficking, and chloride channel activity in patient-derived bronchial epithelial cells [136]. These beneficial effects are not mediated by the HSR or UPR, but rather due to inhibition of Hsp90 activity by acetylation [138], resulting in altered expression of CFTR proteostasis network genes [136]. These studies show that chronic low-dose treatment with SAHA maintained channel activity even after the compound had been removed from the culture medium. SAHA has been approved by the FDA for cancer treatment [139], suggesting that chronic treatment with low-doses of SAHA could also be benefical for cystic fibrosis [140]. However, concerns with the pleiotropic effects of pharmacological modulation of HDACs and HATs on acetylation homeostasis and activation of inflammation and suppression of immune system function, could result in severe side effects. Therefore the identification of compounds that target specific HDACs and HATs and also specific cell types could lead to the desired therapeutic outcome.
SMALL MOLECULE REGULATORS OF Hsp70
Hsp70 is a highly conserved ubiquitous molecular chaperone that is essential for protein synthesis, folding, assembly of macromolecular complexes, trafficking to subcellular compartments, and clearance [141]. Hsp70 also prevents protein misfolding and aggregation, thus shifting the equilibrium to the folded state [142]. Consequently, Hsp70 has diverse functions in multiple steps of protein biogenesis affecting, for example, DNA replication, transcription, cell division and apoptosis [18].
Hsp70 regulates the protein folding cycle through rounds of ATP-dependent binding of non-native substrates and release of folded proteins (Fig. 7) [141]. This requires the coordinated activities of three structural domains: an N-terminal ATPase domain, a hydrophobic peptide binding domain, and the C-terminal helical lid domain. Binding of the non-native substrate favors ATP hydrolysis, and induces a conformational change in the chaperone that leads to the lid closure and trapping of the substrate. The rate of the Hsp70 cycle is limited by the low ability of Hsp70 to hydrolyze ATP and release ADP. The J-domain co-chaperone Hsp40 stimulates the Hsp70 ATPase, and the co-chaperone BAG-1 functions as the nucleotide exchange factor to regulate the nucleotide state, and the substrate binding and release cycle of Hsp70 [143–149]. Due to the central role of Hsp70 and its co-chaperones in regulation of diverse cellular processes, modulation of chaperone activity by small molecules could be highly beneficial for the treatment of multiple human diseases including cancer, metabolic diseases, and degenerative diseases.
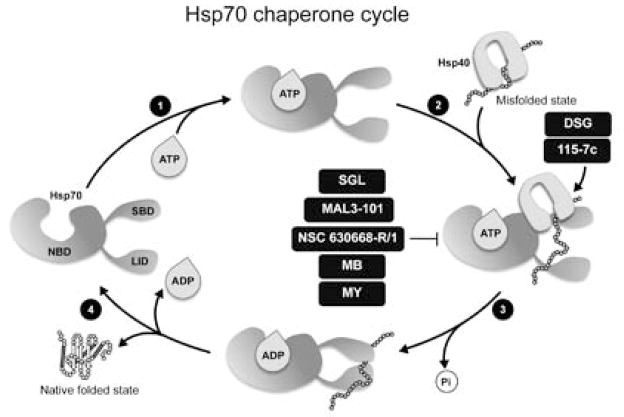
Fig. 7.
The Hsp70 chaperone cycle and chemical modulators of its activity. (1) Binding of ATP to the nucleotide binding domain (NBD) of Hsp70 causes the lid to be in an open state, which has a weak affinity for peptide substrates. When a peptide binds to the Hsp70 substrate binding domain (SBD) (2), ATP is hydrolyzed and this event leads to a conformational change that causes the lid closure (3) and increases the affinity of Hsp70 for the substrate. J-domain containing co-chaperones, such as Hsp40, increase the ATPase activity of Hsp70. Replacement of ADP with ATP is required for the release of the folded substrate (4). Small molecules alter Hsp70 functional activity by interfering with different regions of the chaperone. DSG, 15-deoxyspergualin; SGL, 3′-sulfogalactolipid; MB, methylene blue; MY, myricetin.
Small molecule regulators of Hsp70 folding and ATPase activity
The potent immunosuppressant and cytostatic agent, 15-deoxyspergualin (DSG), interacts with micromolar affinity to Hsp70 to slightly enhance the steady-state ATPase activity, and compete for binding of substrates (Fig. 7) [150, 151]. DSG was shown to have effects on chloride conductance of ΔF508 CFTR expressed in epithelial cells, similar to sodium butyrate or low temperature incubation [152]. However, whether this effect is due to DSG interaction with Hsp70 remains to be determined as other higher affinity targets of DSG have also been implicated in ΔF508 CFTR rescue.
Efforts to identify other compounds structurally related to DSG using in silico methods have led to the dihydropyrimidine NSC-630668-R/1 (R/1), reported to inhibit the Hsp70 ATPase activity (Fig. 7). R/1 affects the activity of both the cytosolic and the ER Hsp70 chaperones [153] by mimicking a peptide substrate and therefore competing with the Hsp70 client protein for binding to the substrate-binding pocket.
Another Hsp70 interacting small molecule, 3′l-sulfo-galactolipid (SGL), was shown to bind the N-terminal AT-Pase domain of Hsp70 [154] and inhibit the Hsp40-mediated stimulation of the Hsp70 ATPase cycle (Fig. 7) [155]. In another study, examining a series of DSG and R/1 analogs, two compounds, MAL3-39 and MAL3-101, were identified that inhibit the Hsp40-dependent simulation of the Hsp70 ATPase cycle (Fig. 7) [153] with inhibitory effects on protein post-translational translocation. Finally, a HTS performed on a library of 204 dihydropyrimidines identified 7 compounds that inhibit the ATPase activity of Hsp70 [156]. Although the binding site for these inhibitors has not been established, they appear to act at a site distinct from the substrate-binding pocket of Hsp70. Due to the central role of Hsp70 in proteostasis, small molecules that target the activity of this chaperone have value as research tools and, potentially, as therapeutic leads [156].
A recent HTS of 2,800 bioactive compounds for modulators of Hsp70 activity identified five active compounds belonging to three chemical scaffolds [156, 157]. The benzothiazine methylene blue (MB), the demethylated analog azure C (AC), and the flavonol myricetin (MY) decreased the ATPase activity of Hsp70 by >80%, whereas dihydropyrimidine compounds (115-7c and SW02) increased Hsp70 activity by ~45% (Fig. 7). The potential disease relevance of these compounds was demonstrated by western blot analysis of HeLa cells overexpressing tau showing that MB and AC caused a significant reduction of both total tau and phosphorylated tau levels, whereas treatment with 115-7c or SW02 led to accumulation of these tau species. These inhibitors do not affect the levels of other aggregation-prone proteins, α-synuclein and TAR-DNA binding protein (TARDBP or TDP-43), suggesting a level of selectivity. These studies indicate that MB and AC stimulate the clearance of both normal and abnormal tau from mice brain tissue and suggest their potential for treatment of Alzheimer’s disease and other tauopathies. In support of this, MB has reached phase III of clinical trials for the treatment of Alzheimer’s disease [158]. In addition to neurodegeneration, these inhibitors can also be used for cancer therapy, as inhibition of Hsp70 ATPase activity by MB, MY and AC reduces Akt levels. Further reduction in Akt levels can be obtained by combining Hsp70 overexpression with its inhibition, taking advantage of cancer cells reliance on high levels of chaperones and Akt for survival. Structure/function studies on MY indicate binding to Hsp70 between the IB and IIB subdomains, thus allosterically blocking the binding of Hsp40 [159].
Despite being ineffective in reducing tau levels [157], compound 115-7c (Fig. 7), a small molecule inducer of Hsp70 ATPase activity that stimulates refolding of denatured luciferase, enhanced the growth of a ydj1Δ mutant yeast that is normally severely compromised when exposed to elevated temperatures. This suggests that 115-7c functions as a chemical co-chaperone, at least at the high doses employed in this study [160]. In addition, treatment of yeast cells expressing either Q72 or Q103 with 115-7c resulted in the formation of smaller puncta. Since 115-7c failed to induce a HSR or a UPR, these results suggest that this compound could modulate polyQ aggregation by affecting the Hsp70 ATPase activity [160]. The binding site of 115-7c resides in the IIA subdomain of Hsp70 required for binding to the J-domains of Hsp40 [160]. Binding of 115-7c to Hsp70 was more favorable in the presence of Hsp40, indicating an increased affinity to the Hsp70-Hsp40 complex. Consequently, addition of a bulkier substituent (a dichlorobenzyl moiety was substituted by a diphenyl group) resulted in compound 116-9e [156], that suppressed J-domain-induced stimulation of the Hsp70 ATPase activity and inhibited Hsp70-mediated chaperone activity. As a result, compound 116-9c was unable to alter polyQ aggregation in cells, suggesting that the ATPase stimulatory activity is required for the suppression of aggregation [160]. The development of these novel classes of chemical probes that disrupt Hsp70 function offers an important complement to dissect proteostasis and alter the properties of specific chaperone client proteins.
OUTLOOK
Novel potential therapeutic strategies that target specific biological pathways of the PN now emerge as promising avenues for the treatment of diverse diseases of protein conformation [4, 16]. These strategies are based on the concept that modulation of the PN is more likely to ameliorate diseases of proteostasis deficiency compared to the more standard single-target approach due to the properties of the PN to achieve and maintain balance in the folding, activity, and clearance of specific protein substrates affected in disease. Regulation of the PN also offers the advantage that a single PR can restore proteostasis in multiple diseases of protein conformations in which the compromised proteins utilize the same PN components. Several small molecule PRs that modulate the PN pathways have been developed and have shown efficacy in a variety of cellular and animal models of conformational disorders. However despite their beneficial effects, several factors need to be considered before these small molecules are optimized for their clinical efficacy.
First, for conformational diseases involving the central nervous system, the small molecule PRs will need to permeate the blood brain barrier. Second, owing to the dichotomy in outcomes of activation of PN pathways such as the HSR and the UPR, a fine-tuning of these pathways by the small molecule PRs should be achieved for the chronic low-level activation of these stress responses to achieve correction of proteostatic deficiencies. In addition, the high degree of integration of the PN makes manipulation of the PN pathways by PRs a challenging task for which toxic-side effects will need to be addressed. Accordingly, the risk of tilting the proteostasis balance in favor of re-establishing a better environment for a misfolded-prone protein could also compromise the stability of other proteins with potential deleterious consequences. In this regard, pharmacological chaperones and kinetic stabilizers, small molecules that bind selectively to the native state of mutant proteins and stabilize their folding, could also be effective in combination for the treatment of conformational diseases such as lysosomal storage diseases [16]. Indeed, the active-site-directed pharmacological chaperone migalastat hydrochloride (1-deoxygalactonojirimycin, Amigal®) and the kinetic stabilizer tafamidis have reached phase III clinical trials for the treatment of Fabry disease (www.clinicaltrials.gov) and transthyretin amyloidosis [161], respectively. Migalastat hydrochloride is a potent competitive inhibitor of the enzyme α-D-galactosidase, but at subinhibitory concentrations it is thought to act as a pharmacological chaperone [9, 162]. Analogously, the pharmacological chaperone miglustat (N-butyl-deoxynojirimycin), an inhibitor of the enzyme glucosylceramide synthase, is used to treat adults with type 1 Gaucher disease and it is the first treatment to be approved for patients with Niemann-Pick type C disease. Third, at least for the HSR, development of small molecules that activate this cytoprotective response by direct activation of the transcription factor HSF-1 could result in a new class of compounds with a reduced toxicity and an improved therapeutic window. In addition, although activation of the HSR has not been associated to cancer promotion, the observation that HSF-1 can maintain cancer proliferation and survival in oncogenic cells by engaging a transcriptional program in malignant cells that is distinct from the HSR [163, 164] suggests that small molecule that induce high level activation of this transcription factor could tilt the balance towards the cancer phenotype. Finally, a comprehensive understanding of the cell- and tissue-specific composition of the PN in aging and diseases will be necessary so that small molecule PRs enhance and restore the cellular and organismal phenotype.
In conclusion we propose that the regulation of the PN by small molecule PRs offers a promising new avenue for the small molecule treatment of many human diseases associated with altered protein conformation.
Acknowledgments
This work was supported by grants to R.I.M. from the NIH (NIGMS, NIA, and NINDS), DOD, the Ellison Medical Foundation, and the Daniel F. and Ada L. Rice Foundation.
Footnotes
This article is published as part of a themed issue on Protein Misfolding in Conformational Disorders, Guest Edited by Cláudio M. Gomes (ITQB/UNL).
CONFLICT OF INTEREST
R.I.M. is founder, shareholder, and paid consultant for Proteostasis Therapeutics Inc. (Cambridge, MA) that is developing small molecule therapeutics for protein misfolding diseases.
References
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 3.Baker MJ, Tatsuta T, Langer T. Quality control of mitochondrial proteostasis. Cold Spring Harbor Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annual review of biochemistry. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 5.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–4. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 6.Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5(3):e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gidalevitz T, Kikis EA, Morimoto RI. A cellular perspective on conformational disease: the role of genetic background and proteostasis networks. Curr Opin Struct Biol. 2010;20 (1):23–32. doi: 10.1016/j.sbi.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968):905–9. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 9.Fan JQ, Ishii S, Asano N, Suzuki Y. Accelerated transport and maturation of lysosomal alpha-galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat Med. 1999;5(1):112–5. doi: 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- 10.Qu BH, Strickland EH, Thomas PJ. Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J Biol Chem. 1997;272(25):15739–44. doi: 10.1074/jbc.272.25.15739. [DOI] [PubMed] [Google Scholar]
- 11.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 12.Desnick RJ, Schuchman EH. Enzyme replacement and enhancement therapies: lessons from lysosomal disorders. Nat Rev Genet. 2002;3(12):954–66. doi: 10.1038/nrg963. [DOI] [PubMed] [Google Scholar]
- 13.Kalinderi K, Fidani L, Katsarou Z, Bostantjopoulou S. Pharmacological treatment and the prospect of pharmacogenetics in Parkinson’s disease. Int J Clin Pract. 2011;65(12):1289–94. doi: 10.1111/j.1742-1241.2011.02793.x. [DOI] [PubMed] [Google Scholar]
- 14.Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52(1):39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Walker FO. Huntington’s disease. Lancet. 2007;369(9557):218–28. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 16.Lindquist SL, Kelly JW. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: progress and prognosis. Cold Spring Harb Perspect Biol. 2011;3(12) doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo W, Sun W, Taldone T, Rodina A, Chiosis G. Heat shock protein 90 in neurodegenerative diseases. Mol Neurodegener. 2010;5:24. doi: 10.1186/1750-1326-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodsky JL, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6(11):1215–25. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 19.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99(16):10417–22. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, Masliah E, Dillin A. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139(6):1157–69. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106(35):14914–9. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci. 2009;64(2):167–70. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen E, Du D, Joyce D, Kapernick EA, Volovik Y, Kelly JW, Dillin A. Temporal requirements of insulin/IGF-1 signaling for proteotoxicity protection. Aging Cell. 2010;9(2):126–34. doi: 10.1111/j.1474-9726.2009.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress development and lifespan. Nat Rev Mol Cell Biol. 2010;11(8):545–55. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280(39):33097–100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 26.Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol. 2003;23(8):2953–68. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem. 2001;276(43):40263–7. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 28.Sandqvist A, Bjork JK, Akerfelt M, Chitikova Z, Grichine A, Vourc’h C, Jolly C, Salminen TA, Nymalm Y, Sistonen L. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol Biol Cell. 2009;20 (5):1340–7. doi: 10.1091/mbc.E08-08-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurivich DA, Sistonen L, Kroes RA, Morimoto RI. Effect of sodium salicylate on the human heat shock response. Science. 1992;255(5049):1243–5. doi: 10.1126/science.1546322. [DOI] [PubMed] [Google Scholar]
- 30.Lee BS, Chen J, Angelidis C, Jurivich DA, Morimoto RI. Pharmacological modulation of heat shock factor 1 by antiinflammatory drugs results in protection against stress-induced cellular damage. Proc Natl Acad Sci U S A. 1995;92(16):7207–11. doi: 10.1073/pnas.92.16.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurivich DA, Sistonen L, Sarge KD, Morimoto RI. Arachidonate is a potent modulator of human heat shock gene transcription. Proc Natl Acad Sci U S A. 1994;91(6):2280–4. doi: 10.1073/pnas.91.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amici C, Sistonen L, Santoro MG, Morimoto RI. Antiproliferative prostaglandins activate heat shock transcription factor. Proc Natl Acad Sci U S A. 1992;89(14):6227–31. doi: 10.1073/pnas.89.14.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov. 2011;10(12):930–44. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dick LR, Fleming PE. Building on bortezomib: second-generation proteasome inhibitors as anti-cancer therapy. Drug Discov Today. 2010;15(5–6):243–9. doi: 10.1016/j.drudis.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Holmberg CI, Illman SA, Kallio M, Mikhailov A, Sistonen L. Formation of nuclear HSF1 granules varies depending on stress stimuli. Cell Stress Chaperones. 2000;5(3):219–28. doi: 10.1379/1466-1268(2000)005<0219:fonhgv>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi A, Elia G, Santoro MG. Activation of the heat shock factor 1 by serine protease inhibitors. An effect associated with nuclear factor-kappaB inhibition. J Biol Chem. 1998;273(26):16446–52. doi: 10.1074/jbc.273.26.16446. [DOI] [PubMed] [Google Scholar]
- 37.Corson TW, Crews CM. Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell. 2007;130(5):769–74. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethi G, Ahn KS, Pandey MK, Aggarwal BB. Celastrol a novel triterpene potentiates TNF-induced apoptosis and suppresses invasion of tumor cells by inhibiting NF-kappaB-regulated gene products and TAK1-mediated NF-kappaB activation. Blood. 2007;109(7):2727–35. doi: 10.1182/blood-2006-10-050807. [DOI] [PubMed] [Google Scholar]
- 39.Cleren C, Calingasan NY, Chen J, Beal MF. Celastrol protects against MPTP- and 3-nitropropionic acid-induced neurotoxicity. J Neurochem. 2005;94(4):995–1004. doi: 10.1111/j.1471-4159.2005.03253.x. [DOI] [PubMed] [Google Scholar]
- 40.Westerheide SD, Bosman JD, Mbadugha BN, Kawahara TL, Matsumoto G, Kim S, Gu W, Devlin JP, Silverman RB, Morimoto RI. Celastrols as inducers of the heat shock response and cytoprotection. J Biol Chem. 2004;279(53):56053–60. doi: 10.1074/jbc.M409267200. [DOI] [PubMed] [Google Scholar]
- 41.Trott A, West JD, Klaic L, Westerheide SD, Silverman RB, Morimoto RI, Morano KA. Activation of heat shock and anti-oxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Mol Biol Cell. 2008;19 (3):1104–12. doi: 10.1091/mbc.E07-10-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang T, Li Y, Yu Y, Zou P, Jiang Y, Sun D. Characterization of celastrol to inhibit hsp90 and cdc37 interaction. J Biol Chem. 2009;284(51):35381–9. doi: 10.1074/jbc.M109.051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang T, Hamza A, Cao X, Wang B, Yu S, Zhan CG, Sun D. A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Mol Cancer Ther. 2008;7(1):162–70. doi: 10.1158/1535-7163.MCT-07-0484. [DOI] [PubMed] [Google Scholar]
- 44.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, Maloney KN, Clardy J, Hahn WC, Chiosis G, Golub TR. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10(4):321–30. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Sreeramulu S, Gande SL, Gobel M, Schwalbe H. Molecular mechanism of inhibition of the human protein complex Hsp90-Cdc37 a kinome chaperone-cochaperone by triterpene celastrol. Angew Chem Int Ed Engl. 2009;48(32):5853–5. doi: 10.1002/anie.200900929. [DOI] [PubMed] [Google Scholar]
- 46.Chadli A, Felts SJ, Wang Q, Sullivan WP, Botuyan MV, Fauq A, Ramirez-Alvarado M, Mer G. Celastrol inhibits Hsp90 chaperoning of steroid receptors by inducing fibrillization of the Co-chaperone p23. J Biol Chem. 2010;285(6):4224–31. doi: 10.1074/jbc.M109.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klaic L, Morimoto RI, Silverman RB. Celastrol analogues as inducers of the heat shock response. Design and synthesis of affinity probes for the identification of protein targets. ACS Chem Biol. 2012;7(5):928–37. doi: 10.1021/cb200539u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Celastrol a triterpene extracted from the Chinese “Thunder of God Vine” is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66(9):4758–65. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 49.Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134(5):769–81. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–49. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94(4):471–80. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 52.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 53.Sittler A, Lurz R, Lueder G, Priller J, Lehrach H, Hayer-Hartl MK, Hartl FU, Wanker EE. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum Mol Genet. 2001;10(12):1307–15. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 54.McLean PJ, Klucken J, Shin Y, Hyman BT. Geldanamycin induces Hsp70 and prevents alpha-synuclein aggregation and toxicity in vitro. Biochem Biophys Res Commun. 2004;321(3):665–9. doi: 10.1016/j.bbrc.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 55.Batulan Z, Taylor DM, Aarons RJ, Minotti S, Doroudchi MM, Nalbantoglu J, Durham HD. Induction of multiple heat shock proteins and neuroprotection in a primary culture model of familial amyotrophic lateral sclerosis. Neurobiol Dis. 2006;24(2):213–25. doi: 10.1016/j.nbd.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 56.Dou F, Netzer WJ, Tanemura K, Li F, Hartl FU, Takashima A, Gouras GK, Greengard P, Xu H. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci U S A. 2003;100(2):721–6. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Auluck PK, Bonini NM. Pharmacological prevention of Parkinson disease in Drosophila. Nat Med. 2002;8(11):1185–6. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 58.Auluck PK, Meulener MC, Bonini NM. Mechanisms of Suppression of {alpha}-Synuclein Neurotoxicity by Geldanamycin in Drosophila. J Biol Chem. 2005;280(4):2873–8. doi: 10.1074/jbc.M412106200. [DOI] [PubMed] [Google Scholar]
- 59.Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13(13):1389–405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 60.Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J Biol Chem. 2008;283(38):26188–97. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waza M, Adachi H, Katsuno M, Minamiyama M, Sang C, Tanaka F, Inukai A, Doyu M, Sobue G. 17-AAG an Hsp90 inhibitor ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11(10):1088–95. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 62.Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J Biol Chem. 2012;287(2):1406–14. doi: 10.1074/jbc.M111.294801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, Chandran J, Lin X, Lai C, Yang WJ, Moore DJ, Dawson TM, Dawson VL, Chiosis G, Cookson MR, Cai H. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci. 2008;28(13):3384–91. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriwaki Y, Kim YJ, Ido Y, Misawa H, Kawashima K, Endo S, Takahashi R. L347P PINK1 mutant that fails to bind to Hsp90/Cdc37 chaperones is rapidly degraded in a proteasome-dependent manner. Neurosci Res. 2008;61(1):43–8. doi: 10.1016/j.neures.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 65.Falsone SF, Kungl AJ, Rek A, Cappai R, Zangger K. The molecular chaperone Hsp90 modulates intermediate steps of amyloid assembly of the Parkinson-related protein alpha-synuclein. J Biol Chem. 2009;284(45):31190–9. doi: 10.1074/jbc.M109.057240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo W, Dou F, Rodina A, Chip S, Kim J, Zhao Q, Moulick K, Aguirre J, Wu N, Greengard P, Chiosis G. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc Natl Acad Sci U S A. 2007;104(22):9511–6. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, Patterson C, Dickson DW, Nahman NS, Jr, Hutton M, Burrows F, Petrucelli L. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest. 2007;117(3):648–58. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, Seredenina T, Woodman B, Moussaoui S, Frentzel S, Luthi-Carter R, Paganetti P, Bates GP. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Invest. 2011;121(8):3306–19. doi: 10.1172/JCI57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaughan CK, Neckers L, Piper PW. Understanding of the Hsp90 molecular chaperone reaches new heights. Nat Struct Mol Biol. 2010;17(12):1400–4. doi: 10.1038/nsmb1210-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ansar S, Burlison JA, Hadden MK, Yu XM, Desino KE, Bean J, Neckers L, Audus KL, Michaelis ML, Blagg BS. A non-toxic Hsp90 inhibitor protects neurons from Abeta-induced toxicity. Bioorg Med Chem Lett. 2007;17(7):1984–90. doi: 10.1016/j.bmcl.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 71.Yu XM, Shen G, Neckers L, Blake H, Holzbeierlein J, Cronk B, Blagg BS. Hsp90 inhibitors identified from a library of novobiocin analogues. J Am Chem Soc. 2005;127(37):12778–9. doi: 10.1021/ja0535864. [DOI] [PubMed] [Google Scholar]
- 72.Salehi AH, Morris SJ, Ho WC, Dickson KM, Doucet G, Milutinovic S, Durkin J, Gillard JW, Barker PA. AEG3482 is an antiapoptotic compound that inhibits Jun kinase activity and cell death through induced expression of heat shock protein 70. Chem Biol. 2006;13(2):213–23. doi: 10.1016/j.chembiol.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 73.Kimura H, Yukitake H, Tajima Y, Suzuki H, Chikatsu T, Morimoto S, Funabashi Y, Omae H, Ito T, Yoneda Y, Takizawa M. ITZ-1 a client-selective Hsp90 inhibitor efficiently induces heat shock factor 1 activation. Chem Biol. 2010;17(1):18–27. doi: 10.1016/j.chembiol.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 74.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8(1):e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunitspecific interactions. Nat Cell Biol. 2006;8(10):1155–62. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16(12):1279–85. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov. 2011;10(12):930–44. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, Saldanha SA, Hodder P, Tait BD, Garza D, Balch WE, Morimoto RI. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2011;8(2):185–96. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 80.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 81.Volchuk A, Ron D. The endoplasmic reticulum stress response in the pancreatic beta-cell. Diabetes Obes Metab. 2010;12(Suppl 2):48–57. doi: 10.1111/j.1463-1326.2010.01271.x. [DOI] [PubMed] [Google Scholar]
- 82.Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332(6025):91–4. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 83.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307(5711):935–9. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 84.Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A. 2012;109(15):E869–78. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Volkmann K, Lucas JL, Vuga D, Wang X, Brumm D, Stiles C, Kriebel D, Der-Sarkissian A, Krishnan K, Schweitzer C, Liu Z, Malyankar UM, Chiovitti D, Canny M, Durocher D, Sicheri F, Patterson JB. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286(14):12743–55. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, Koong AC. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311–4. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24(24):4368–80. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–7. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 89.Hess DA, Humphrey SE, Ishibashi J, Damsz B, Lee AH, Glimcher LH, Konieczny SF. Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology. 2011;141(4):1463–72. doi: 10.1053/j.gastro.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14(2):152–7. [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;11(2):268–81.5. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang F, Song W, Brancati G, Segatori L. Inhibition of endoplasmic reticulum-associated degradation rescues native folding in loss of function protein misfolding diseases. J Biol Chem. 2011;286(50):43454–64. doi: 10.1074/jbc.M111.274332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ciechanover A, Schwartz AL. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci U S A. 1998;95(6):2727–30. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12(9):1191–7. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 96.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5(7):596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 97.Sarkar S, Rubinsztein DC. Small molecule enhancers of autophagy for neurodegenerative diseases. Mol Biosyst. 2008;4(9):895–901. doi: 10.1039/b804606a. [DOI] [PubMed] [Google Scholar]
- 98.Adams J. The proteasome: structure function and role in the cell. Cancer Treat Rev. 2003;29(Suppl 1):3–9. doi: 10.1016/s0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- 99.Weissman AM, Shabek N, Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol. 2011;12(9):605–20. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Noda T, Ohsumi Y. Tor a phosphatidylinositol kinase homologue controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 101.Hockerman GH, Peterson BZ, Johnson BD, Catterall WA. Molecular determinants of drug binding and action on L-type calcium channels. Annual review of pharmacology and toxicology. 1997;37:361–96. doi: 10.1146/annurev.pharmtox.37.1.361. [DOI] [PubMed] [Google Scholar]
- 102.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O’Kane CJ, Schreiber SL, Rubinsztein DC. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3(6):331–8. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Floto RA, Sarkar S, Perlstein EO, Kampmann B, Schreiber SL, Rubinsztein DC. Small molecule enhancers of rapamycin-induced TOR inhibition promote autophagy reduce toxicity in Huntington’s disease models and enhance killing of mycobacteria by macrophages. Autophagy. 2007;3(6):620–2. doi: 10.4161/auto.4898. [DOI] [PubMed] [Google Scholar]
- 104.Tian Y, Bustos V, Flajolet M, Greengard P. A small-molecule enhancer of autophagy decreases levels of Abeta and APP-CTF via Atg5-dependent autophagy pathway. FASEB J. 2011;25(6):1934–42. doi: 10.1096/fj.10-175158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, Yuan J. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104(48):19023–8. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA, Rubinsztein DC. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4(5):295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose a novel mTOR-independent autophagy enhancer accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282(8):5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 108.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 109.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 110.Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A. 2010;107(39):16982–7. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Perlmutter DH. Alpha-1-antitrypsin deficiency: importance of proteasomal and autophagic degradative pathways in disposal of liver disease-associated protein aggregates. Annu Rev Med. 2011;62:333–45. doi: 10.1146/annurev-med-042409-151920. [DOI] [PubMed] [Google Scholar]
- 112.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, Michalopoulos G, Perlmutter DH. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329(5988):229–32. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 113.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Berridge MJ, Bootman MD, Lipp P. Calcium--a life and death signal. Nature. 1998;395(6703):645–8. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- 115.Michalak M, Robert Parker JM, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. 2002;32(5–6):269–78. doi: 10.1016/s0143416002001884. [DOI] [PubMed] [Google Scholar]
- 116.Egan ME, Glockner-Pagel J, Ambrose C, Cahill PA, Pappoe L, Balamuth N, Cho E, Canny S, Wagner CA, Geibel J, Caplan MJ. Calcium-pump inhibitors induce functional surface expression of Delta F508-CFTR protein in cystic fibrosis epithelial cells. Nat Med. 2002;8(5):485–92. doi: 10.1038/nm0502-485. [DOI] [PubMed] [Google Scholar]
- 117.Grubb BR, Gabriel SE, Mengos A, Gentzsch M, Randell SH, Van Heeckeren AM, Knowles MR, Drumm ML, Riordan JR, Boucher RC. SERCA pump inhibitors do not correct bio-synthetic arrest of deltaF508 CFTR in cystic fibrosis. Am J Respir Cell Mol Biol. 2006;34(3):355–63. doi: 10.1165/rcmb.2005-0286OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Loo TW, Bartlett MC, Clarke DM. Thapsigargin or curcumin does not promote maturation of processing mutants of the ABC transporters CFTR and P-glycoprotein. Biochem Biophys Res Commun. 2004;325(2):580–5. doi: 10.1016/j.bbrc.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 119.Egan ME, Pearson M, Weiner SA, Rajendran V, Rubin D, Glockner-Pagel J, Canny S, Du K, Lukacs GL, Caplan MJ. Curcumin a major constituent of turmeric corrects cystic fibrosis defects. Science. 2004;304(5670):600–2. doi: 10.1126/science.1093941. [DOI] [PubMed] [Google Scholar]
- 120.Song Y, Sonawane ND, Salinas D, Qian L, Pedemonte N, Galietta LJ, Verkman AS. Evidence against the rescue of defective DeltaF508-CFTR cellular processing by curcumin in cell culture and mouse models. J Biol Chem. 2004;279(39):40629–33. doi: 10.1074/jbc.M407308200. [DOI] [PubMed] [Google Scholar]
- 121.Dragomir A, Bjorstad J, Hjelte L, Roomans GM. Curcumin does not stimulate cAMP-mediated chloride transport in cystic fibrosis airway epithelial cells. Biochem Biophys Res Commun. 2004;322(2):447–51. doi: 10.1016/j.bbrc.2004.07.146. [DOI] [PubMed] [Google Scholar]
- 122.Mall M, Kunzelmann K. Correction of the CF defect by curcumin: hypes and disappointments. Bioessays. 2005;27(1):9–13. doi: 10.1002/bies.20168. [DOI] [PubMed] [Google Scholar]
- 123.Wang F, Agnello G, Sotolongo N, Segatori L. Ca2+ homeostasis modulation enhances the amenability of L444P glucosylcerebrosidase to proteostasis regulation in patient-derived fibroblasts. ACS Chem Biol. 2011;6(2):158–68. doi: 10.1021/cb100321m. [DOI] [PubMed] [Google Scholar]
- 124.Wang F, Chou A, Segatori L. Lacidipine remodels protein folding and Ca 2+ homeostasis in Gaucher’s disease fibroblasts: a mechanism to rescue mutant glucocerebrosidase. Chem Biol. 2011;18(6):766–76. doi: 10.1016/j.chembiol.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 125.Mu TW, Fowler DM, Kelly JW. Partial restoration of mutant enzyme homeostasis in three distinct lysosomal storage disease cell lines by altering calcium homeostasis. PLoS Biol. 2008;6(2):e26. doi: 10.1371/journal.pbio.0060026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ong DS, Mu TW, Palmer AE, Kelly JW. Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. Nat Chem Biol. 2010;6(6):424–32. doi: 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323(5917):1063–6. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7(10):854–68. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 129.Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 130.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31(12):605–17. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 131.Hockerman GH, Peterson BZ, Sharp E, Tanada TN, Scheuer T, Catterall WA. Construction of a high-affinity receptor site for dihydropyridine agonists and antagonists by single amino acid substitutions in a non-L-type Ca2+ channel. Proc Natl Acad Sci U S A. 1997;94(26):14906–11. doi: 10.1073/pnas.94.26.14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34(7):1721–32. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 133.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447(7141):178–82. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 134.Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35(4):870–80. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lu J, Yang C, Chen M, Ye DY, Lonser RR, Brady RO, Zhuang Z. Histone deacetylase inhibitors prevent the degradation and restore the activity of glucocerebrosidase in Gaucher disease. Proc Natl Acad Sci U S A. 2011;108(52):21200–5. doi: 10.1073/pnas.1119181109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hutt DM, Herman D, Rodrigues AP, Noel S, Pilewski JM, Matteson J, Hoch B, Kellner W, Kelly JW, Schmidt A, Thomas PJ, Matsumura Y, Skach WR, Gentzsch M, Riordan JR, Sorscher EJ, Okiyoneda T, Yates JR, 3rd, Lukacs GL, Frizzell RA, Manning G, Gottesfeld JM, Balch WE. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat Chem Biol. 2010;6(1):25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137–40. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18(5):601–7. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 139.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25(1):84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 140.Ramalingam SS, Parise RA, Ramanathan RK, Lagattuta TF, Musguire LA, Stoller RG, Potter DM, Argiris AE, Zwiebel JA, Egorin MJ, Belani CP. Phase I and pharmacokinetic study of vorinostat a histone deacetylase inhibitor in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13(12):3605–10. doi: 10.1158/1078-0432.CCR-07-0162. [DOI] [PubMed] [Google Scholar]
- 141.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125(3):443–51. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 142.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27(2):328–35. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wickner S, Hoskins J, McKenney K. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature. 1991;350(6314):165–7. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]
- 144.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356(6371):683–9. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 145.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK DnaJ and GrpE. Proc Natl Acad Sci U S A. 1994;91(22):10345–9. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gamer J, Multhaup G, Tomoyasu T, McCarty JS, Rudiger S, Schonfeld HJ, Schirra C, Bujard H, Bukau B. A cycle of binding and release of the DnaK DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J. 1996;15(3):607–17. [PMC free article] [PubMed] [Google Scholar]
- 147.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A. 1999;96(10):5452–7. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Han W, Christen P. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J Biol Chem. 2003;278(21):19038–43. doi: 10.1074/jbc.M300756200. [DOI] [PubMed] [Google Scholar]
- 149.Misselwitz B, Staeck O, Rapoport TA. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2(5):593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 150.Nadler SG, Tepper MA, Schacter B, Mazzucco CE. Interaction of the immunosuppressant deoxyspergualin with a member of the Hsp70 family of heat shock proteins. Science. 1992;258(5081):484–6. doi: 10.1126/science.1411548. [DOI] [PubMed] [Google Scholar]
- 151.Nadeau K, Nadler SG, Saulnier M, Tepper MA, Walsh CT. Quantitation of the interaction of the immunosuppressant deoxyspergualin and analogs with Hsc70 and Hsp90. Biochemistry. 1994;33(9):2561–7. doi: 10.1021/bi00175a027. [DOI] [PubMed] [Google Scholar]
- 152.Jiang C, Fang SL, Xiao YF, O’Connor SP, Nadler SG, Lee DW, Jefferson DM, Kaplan JM, Smith AE, Cheng SH. Partial restoration of cAMP-stimulated CFTR chloride channel activity in DeltaF508 cells by deoxyspergualin. Am J Physiol. 1998;275(1 Pt 1):C171–8. doi: 10.1152/ajpcell.1998.275.1.C171. [DOI] [PubMed] [Google Scholar]
- 153.Fewell SW, Smith CM, Lyon MA, Dumitrescu TP, Wipf P, Day BW, Brodsky JL. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J Biol Chem. 2004;279(49):51131–40. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 154.Mamelak D, Lingwood C. The ATPase domain of hsp70 possesses a unique binding specificity for 3′-sulfogalactolipids. J Biol Chem. 2001;276(1):449–56. doi: 10.1074/jbc.M006732200. [DOI] [PubMed] [Google Scholar]
- 155.Whetstone H, Lingwood C. 3′-Sulfogalactolipid binding specifically inhibits Hsp70 ATPase activity in vitro. Biochemistry. 2003;42(6):1611–7. doi: 10.1021/bi026735t. [DOI] [PubMed] [Google Scholar]
- 156.Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372(2):167–76. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 157.Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, O’Leary J, Morgan D, Lee DC, Shults CL, Rousaki A, Weeber EJ, Zuiderweg ER, Gestwicki JE, Dickey CA. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J Neurosci. 2009;29(39):12079–88. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rafii MS, Aisen PS. Recent developments in Alzheimer’s disease therapeutics. BMC Med. 2009;7:7. doi: 10.1186/1741-7015-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chang L, Miyata Y, Ung PM, Bertelsen EB, McQuade TJ, Carlson HA, Zuiderweg ER, Gestwicki JE. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol. 2011;18(2):210–21. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wisen S, Bertelsen EB, Thompson AD, Patury S, Ung P, Chang L, Evans CG, Walter GM, Wipf P, Carlson HA, Brodsky JL, Zuiderweg ER, Gestwicki JE. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem Biol. 2010;5(6):611–22. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Johnson SM, Connelly S, Fearns C, Powers ET, Kelly JW. The transthyretin amyloidoses: from delineating the molecular mechanism of aggregation linked to pathology to a regulatory-agency-approved drug. J Mol Biol. 2012;421(2–3):185–203. doi: 10.1016/j.jmb.2011.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Asano N, Ishii S, Kizu H, Ikeda K, Yasuda K, Kato A, Martin OR, Fan JQ. In vitro inhibition and intracellular enhancement of lysosomal alpha-galactosidase A activity in Fabry lymphoblasts by 1-deoxygalactonojirimycin and its derivatives. Eur J Biochem. 2000;267(13):4179–86. doi: 10.1046/j.1432-1327.2000.01457.x. [DOI] [PubMed] [Google Scholar]
- 163.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549–62. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]